Abstract
Recently, there have been a lot of intense debates about the acceptance/rejection of paraphyletic groups in biological classification. On the one hand, evolutionary classification states that similarity and common descent are two criteria for biological classification and paraphyletic groups are natural units of biological classification. On the other hand, cladistic classification considers that common descent is the only criterion in biological classification and monophyly should be strictly adhered to. Holcoglossum is used herein as a case to illustrate this problem. Although Holcoglossum is a small orchid genus of less than 20 species, there is little consensus about its generic circumscription since it was established, which leads to confusion in taxonomic treatments in the Aerides-Vanda group. Based on the analyses of molecular and morphological evidence, our results suggest that the clade comprising Holcoglossum s.s., Ascolabium, Penkimia and Ascocentrum himalaicum is strongly supported as a monophyly, and that the three taxa are nested within different subclades of Holcoglossum s.s. Thus, it is reasonable to recognize a monophyletic circumscription of Holcoglossum, which is also well supported by some vegetative and floral characters. The Holcoglossum s.l. would facilitate a better understanding of pollinator-driven floral divergence and vegetative stasis than a paraphyletic and narrowly defined genus.
Introduction
In the era of integrative taxonomy, there is more major congruence in the biological classification between cladistic classification and evolutionary classification except with regard to the acceptance/rejection of paraphyletic groups. Phylogenetic (cladistic) methods and DNA sequences are routinely used in systematics and taxonomy, and both schools of classification have recognized that the principle of common decent plays a major role in biological classification. However, the fundamental question about the acceptance/rejection of paraphyletic groups remains unsolved, and recently there have been many intense debates about this issue [1]–[11]. Evolutionary classification recognizing paraphyletic groups argues the following: (1) there are two criteria for biological classification, i.e., similarity and common descent [12]; (2) many species are paraphyletic [1], [12]–[14]; (3) paraphyletic groups are natural transitional stages in the evolution of taxa and are natural units of biological classification [4]; (4) cladistic classification is incompatible with the Linnean hierarchy system [1], [4], [7], such as diachronous groups and (5) classification based only on common descent often fails to reflect divergence and natural selection. In contrast, cladistic classification states the following: (1) only monophyletic groups in their strictest sense (holophyly) that are evidenced by synapomorphous characters are recognized in biological classification; (2) only species and clades are objective, and supraspecific taxa are terminals in cladistic classification [15], [16]; (3) there are no objective criteria to circumscribe paraphyletic groups, and paraphyletic groups are artificial classes created by taxonomists to emphasize some particular characters or divergence [5], [16]–[18] and (4) cladistic classification can be accommodated within the Linnaean system except for monotypic higher taxa and historic groups [19], with the former being neither paraphyletic nor monophyletic and the latter being dismissed [16], [19]. However, as there are many theorical and lengthy discussions anywhere [1]–[11], a discussion of the merits and fallacies of each school of classification is not the major aim of present paper. Instead, we used the taxonomy of Holcoglossum (Aeridinae, Orchidaceae) as a case to illustrate this problem here.
The orchid subtribe Aeridinae is a large and well-defined horticulturally important group of approximately 1200 species in 120 genera [20], [21]. However, the taxonomy of Aeridinae, particularly the generic delimitations, is difficult and has been considered as “the black pit” [20]–[26]. Seidenfaden [27] (page 8) even stated, “A recurrent dilemma in the study of the Aeridinae is the allotment of a species or a group of species to a genus. − I have several times met with this problem, e.g., in Aerides, Holcoglossum and Ascocentrum.”
Holcoglossum is a small Asian genus, consisting of less than 20 species and mainly distributed in southwestern China and neighboring regions. Nonetheless, Holcoglossum occupies an important systematic position in the informal taxonomic group, the Aerides-Vanda alliance, which includes Aerides, Ascocentrum, Ascolabium, Holcoglossum, Neofinetia, Papilionanthe, Penkimia, Seidenfadenia and Vanda. Holcoglossum has been redefined several times since it was established by Schlechter [27], but it appears that there is little consensus about its generic circumscription, with many species being transferred among genera within the Aerides-Vanda alliance [22]–[24], [28]–[33]. Based on plastid matK and trnL-F and nuclear ITS sequences, two most recent molecular phylogenetic studies of Holcoglossum [32], [33] have helped to clarify this problem; however, their results of each are substantially different, thus further confusing the taxonomy of Vanda-Aeridies alliance [32], [33].
In the present study, phylogenetic relationships were inferred using four DNA markers (plastid matK, trnH-psbA and trnL-F, and nuclear ITS sequences) and combined with morphological analyses with sampling across Aeridinae to (1) illustrate the generic circumscription problem of Holcoglossum, (2) understand the difficulty in accepting/rejecting paraphyletic groups in Holcoglossum, and (3) investigate the evolution of morphological characters within Holcoglossum.
Results
Genetic Distances and Phylogenetic Relationships in Subtribe Aeridinae
The mean genetic distance among Holcoglossum species is 0.010 (ITS), 0.012 (matK) and 0.014 (trnL-F). The genetic distances between Ascocentrum himalaicum, two monotypic genera (Ascolabium and Penkimia) and Holcoglossum s.s. are below 0.020 respectively (Table 1). Penkimia, Ascolabium, and Ascocentrum himalaicum are closer to Holcoglossum s.s. than to any other relatives (Table 1).
Table 1. Average pairwise sequence distances between Holcoglossum s.s. and related genera.
| Average genetic distance | ITS | matK | trnL-F |
| Holcoglossum s.s. | 0.010 | 0.012 | 0.014 |
| Penkimia nagalandensis vs. Holcoglossum s.s. | 0.010 | 0.013 | 0.027 |
| Ascocentrum himalaicum vs. Holcoglossum s.s. | 0.011 | 0.019 | 0.018 |
| Ascolabium pumilum vs. Holcoglossum s.s. | 0.011 | 0.018 | 0.013 |
| Holcoglossum s.s. vs. Aerides | 0.052 | 0.016 | 0.028 |
| Holcoglossum s.s. vs. Ascocentrum | 0.035 | 0.014 | 0.031 |
| Holcoglossum s.s. vs. Papilionanthe | 0.020 | 0.038 | 0.010 |
| Holcoglossum s.s. vs. Vanda | 0.035 | 0.010 | 0.041 |
| Ascocentrum himalaicum vs. Aerides | 0.045 | 0.023 | 0.027 |
| Ascocentrum himalaicum vs. other Ascocentrumspecies | 0.032 | 0.022 | 0.031 |
| Ascocentrum himalaicum vs. Papilionanthe | 0.015 | 0.045 | 0.009 |
| Ascocentrum himalaicum vs. Vanda | 0.032 | 0.018 | 0.040 |
| Penkimia nagalandensis vs. Aerides | 0.045 | 0.017 | 0.032 |
| Penkimia nagalandensis vs. Ascocentrum | 0.029 | 0.016 | 0.022 |
| Penkimia nagalandensis vs. Papilionanthe | 0.015 | 0.038 | 0.017 |
| Penkimia nagalandensis vs. Vanda | 0.029 | 0.018 | 0.031 |
| Ascolabium pumilum vs. Aerides | 0.054 | 0.022 | 0.036 |
| Ascolabium pumilum vs. Ascocentrum | 0.033 | 0.020 | 0.040 |
| Ascolabium pumilum vs. Papilionanthe | 0.020 | 0.049 | 0.017 |
| Ascolabium pumilum vs. Vanda | 0.038 | 0.016 | 0.050 |
Within Aeridinae, the ITS dataset for 138 taxa consisted of 854 characters of which 321 were parsimony informative. The Bayesian trees of each dataset were congruent with the MP trees, except for some weakly supported nodes. The interrelationships among most of the genera in Aeridinae were unresolved. Holcoglossum s.s. was not monophyletic: Ascolabium, Penkimia and Ascocentrum himalaicum were nested within different clades of Holcoglossum (Figure S1). Four out of the five sampled Ascocentrum species (except for A. himalaicum) were nested in the clade that included Vanda, Neofinetia and Christensonia (Figure S1).
Phylogenetic Analyses within a Reduced Holcoglossum Dataset
The ITS dataset displayed 749 characters, 118 of which were parsimony informative. The combined matrix of plastid matK, trnH-psbA and trnL-F sequences indicates 4287 sites of which 258 are parsimony informative (Table S1). The monophyly of the clade including Holcoglossum s.s., Ascolabium, Penkimia and Ascocentrum himalaicum is moderately or weakly supported based on the ITS and the combined plastid datasets, respectively (results not shown).
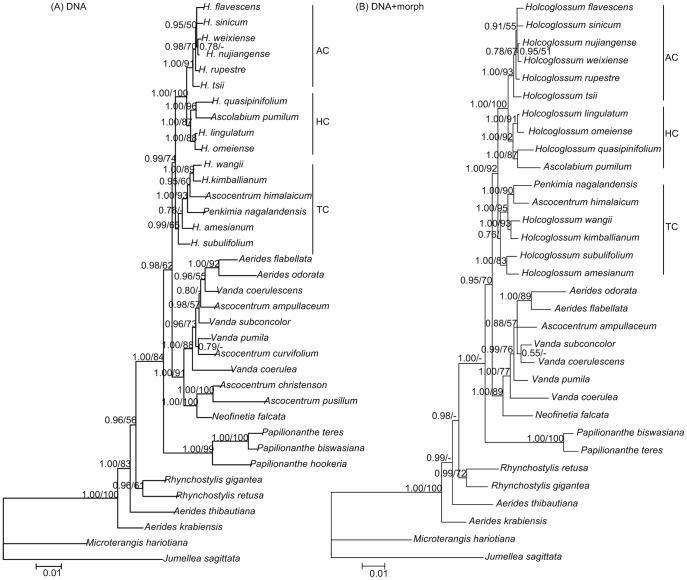
The combined matrix of the nuclear and plastid datasets has 5036 sites, 376 of which are parsimony informative (Table S1). The Bayesian analysis yielded trees having topologies that are consistent with those obtained using MP analysis. The clade including Holcoglossum s.s., Ascolabium, Penkimia and Ascocentrum himalaicum is a monophyly with moderate support (BS = 74, PP = 0.99). Moreover, similar to the results of Fan et al. [32], this clade subdivided into three subclades, i.e., the tropical (TC), the alpine (AC) and the intermediate subclades (HC). Penkimia and Ascocentrum himalaicum are nested in TC (BS = 93, PP = 1.00), and Ascolabium is sister to H. quasipinifolium in HC (BS = 96, PP = 1.00) (Figure 1A).
Figure 1. The 50% consensus Bayesian Inference and strict consensus maximum parsimony tree of Holcoglossum s.l.
A. the combined DNA dataset, B. DNA and morphology dataset. The bootstrap percentages and posterior probability of >50% are shown above each branch.
In total, forty-five gross morphological characters are examined (Table S2). The cladograms from the morphological data based on the MP analysis were poorly resolved (Figure S2). Based on the combination of the morphological and molecular evidence, the clade comprising Holcoglossum s.s., Ascolabium, Penkimia and Ascocentrum himalaicum was strongly supported (BS = 92, PP = 1.00) (Figure 1B).
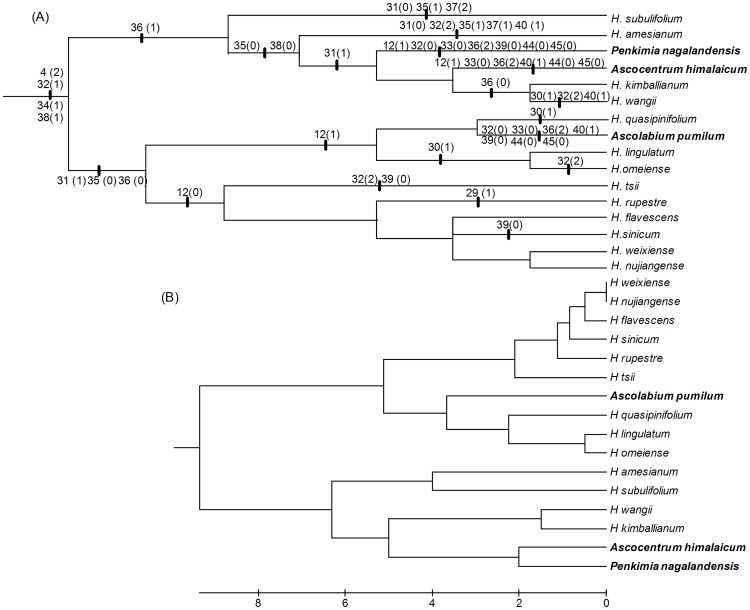
The cladistic and patristic distances are shown in Table S3. The patrocladistic analysis revealed that Ascolabium, Penkimia and Ascocentrum himalaicum remain embedded within different subclades of Holcoglossum s.s. (Figure 2).
Figure 2. Dendrograms of relationships among Holcoglossum.
A. Cladogram from Bayesian inference, B: Corresponding patrocladogram with equal weight.
Gross Morphology and Micromorphology
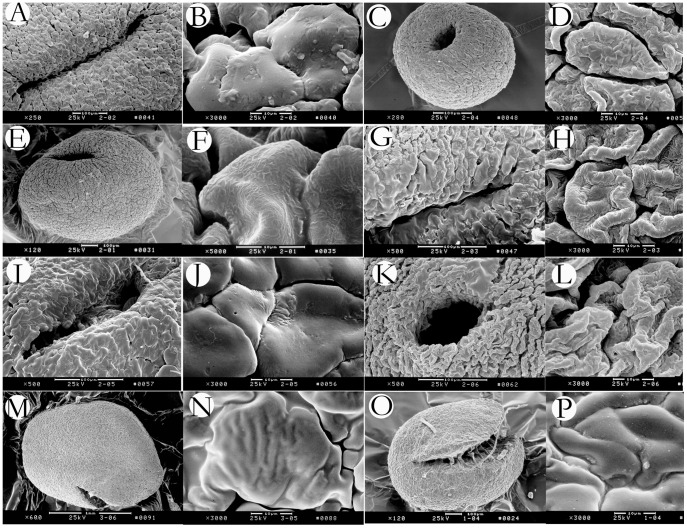
The pollinium micromorphology indicated that the pollinia of all eight examined species of Holcoglossum and Ascocentrum are porate, while the pollinia of Vanda pumila and Aerides rosea are uneven-split (Figure 3). The exine is psilate-scabrate in Ascocentrum ampullaceum, Vanda pumila and most species of Holcoglossum,and striato-reticulate in four alpine species: H. sinicum, H. rupestre, H. nujiangense, H. weixiense (Figure 3).
Figure 3. Pollinia of Holcoglossum and its alliance.
A–B. H. amesianum; C–D. H. himalaicum; E–F. H. kimabllianum; G–H. H. nujiangense; I–J. H. omeiense; K–L. H. rupestre; M–N. Vanda pumila; O–P. Aerides rosea.
Discussion
Monophyly Versus Paraphyly − Definition of Holcoglossum
Based on the morphological and molecular evidence, our results indicated that Holcoglossum Schltr. (Holcoglossum s.s.) is paraphyletic; however, the clade including Holcoglossum s.s., Ascolabium, Penkimia and Ascocentrum himalaicum is strongly supported as a monophyly (Figure 1). The red to yellow tiny flowers misplaced members, namely, Penkimia, the neglected Ascolabium [34], and Ascocentrum himalaicum, have independently evolved at least twice in Holcoglossum s.s. (Figure S3). We are in a dilemma to circumscribe Holcoglossum. It appears that there are three proposals for the circumscription of Holcoglossum. (1) The first is to narrow Holcoglossum, as Liu et al. [33] did: based on this, at least six genera, Holcoglossum, Ascolabium, Penkimia, Paraholcoglossum, Tsiorchis, and another new genus containing Ascocentrum himalaicum, would be recognized, yet Holcoglossum would still be paraphyletic. (2) The second is to circumscribe Holcoglossum according to Jin [30] and Jin & Wood [35]: under this scenario, two monotypic or oligotypic genera, Penkimia (with Ascocentrum himalaicum) and Ascolabium, have to be recognized in addition to the paraphyletic Holcoglossum s.s. (3) The third is to define Holcoglossum sense lato: if Holcoglossum is redefined as monophyletic, then the other four genera, i.e., Penkimia, Ascolabium, Paraholcoglossum, and Tsiorchis, will be included, and Ascocentrum himalaicum will be transferred into it.
Regarding option 1, there are no distinct morphological characters to distinguish Tsiorchis and Paraholcoglossum from Holcoglossum (Figure S3), and the narrowly redefined Holcoglossum is still paraphyletic. Furthermore, the subtribe Aeridinae is abundant in many aberrant species and plagued by the many monotypic genera; therefore, this proposal would set a precedent to separate many aberrant species as new genera. Accordingly, it seems that the recognition of many similar monotypic genera while keeping Holcoglossum as paraphyletic provides no practicability or similarity and does not reflect the maximum evolutionary information.
With regard to option 2, all the members of the paraphyletic Holcoglossum s.s. are consistent in both the vegetative characters [22], [27], [30], [36] and floral characters, which epitomize bee-pollination syndrome [36], making it very good in practicability, high in similarity and information content; however, two small genera have to recognized, as their evolutionary information cannot be related to Holcoglossum.
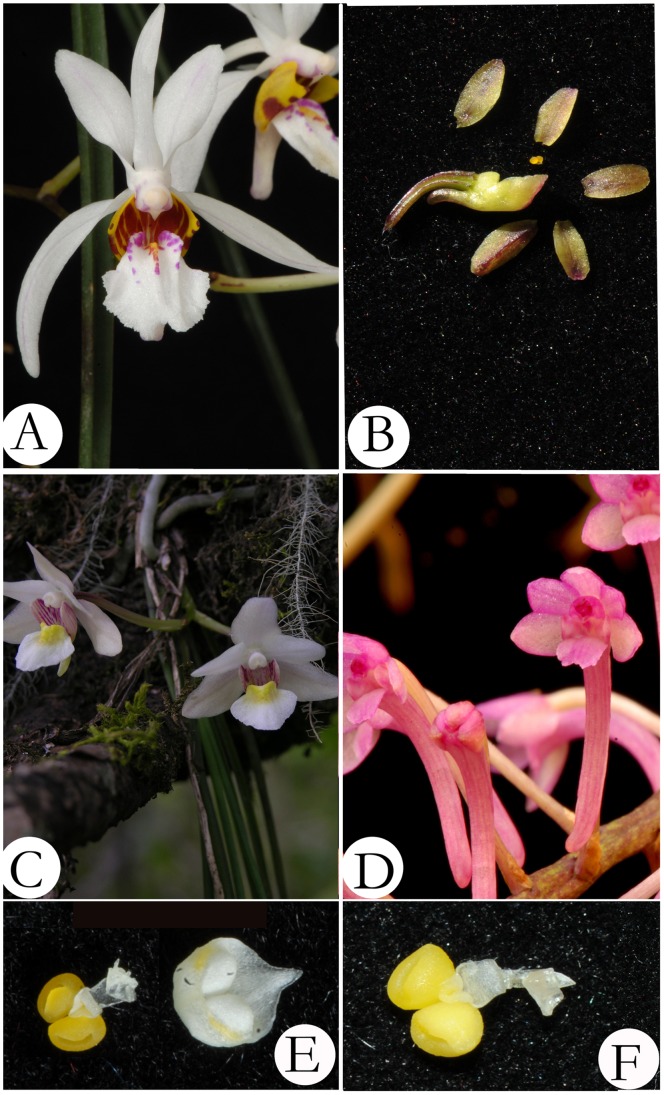
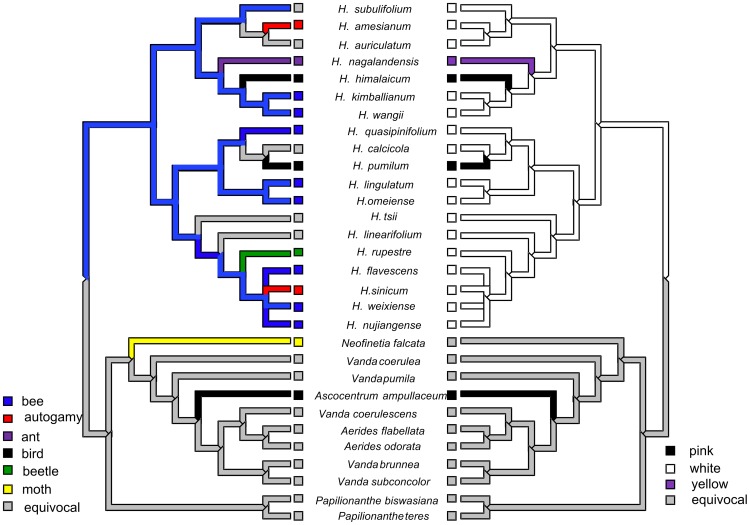
For option 3, Holcoglossum s.l., including Holcoglossum s.s., two monotypic genera (Ascolabium and Penkimia ), Ascocentrum himalaicum and two newly described genera, Paraholcoglossum and Tsiorchis, is strongly supported as a monophyly by the molecular evidence and patrocladistic analyses (Figure 1 and 2). Indeed, the genetic distances revealed that these species show closer relationships to Holcoglossum s.s. than to other genera within Aeridinae (Table 1). In fact, Ascocentrum himalaicum was included in Holcoglossum by Tsi [28] and Averyanov [37] based on morphological characters. However, Holcoglossum s.l. is greatly heterogeneous in floral traits. The floral autapomorphies of Ascolabium, Penkimia and Ascocentrum himalaicum, such as red or purple, small-sized flowers, short and densely flowered inflorescences, and cylindrical spur, evolved independently at least twice in Holcoglossum and most likely were driven by the pollinator-mediated selection (Figure 4 and 5).
Figure 4. Flowers of Holcoglossum.
A. H. wangii; B. H. nagalandensis; C. H. nujiangense; D. H. himalaicum; E. Pollinarium of H. flavescens; F. Pollinarium of H. wangii.
Figure 5. Relationships between pollination and flower color in Holcoglossum.
The left panel shows diversity of the pollination types, and the right panel indicates the evolution of flower color.
Based on these data, we tentatively propose to recognize a monophyletic circumscription of Holcoglossum that comprises approximately 17 species, including Holcoglossum s.s., Ascolabium, Penkimia, and Ascocentrum himalaicum, whereas Paraholcoglossum and Tsiorchis are placed within Holcoglossum s.s. for the purposes of practicability, similarity and providing the maximum evolutionary information.
Morphological Character Evolution within Holcoglossum s.l
In comparison to the conserved vegetative characters, Holcoglossum s.l. is greatly divergent with regard to its floral characters (Table S2). Recent studies of pollination in Holcoglossum indicated that the pollination systems are more divergent than previously expected. To date, four pollination systems, autogamy [38], beetle pollination [39], bee pollination [40], and ant pollination [41], have been recorded in Holcoglossum, whereas bird pollination in H. himalaicum and H. pumilum remains to be confirmed [42]. Generally, pink flowers are considered bird pollination syndrome, and white flowers with a colored lip are considered bee pollination syndrome [43]. Two pink-flower species, H. himalaicum and H. pumilum, had previously been placed in Ascocentrum due to their floral similarities with A. ampullaceum, but our results indicated that pink flowers have independently evolved at least twice from white flowers in Holcoglossum s.l. (Figure 5, right). It appears that this shift is the result of pollinator-mediated selection (Figure 5, left). Therefore, Holcoglossum s.l. would be better to understand pollinator-driven floral divergence and vegetative stasis than a paraphyletic and narrowly defined genus.
Conclusions
Based on the analyses of molecular and morphological characters, Holcoglossum s.s. is paraphyletic; the clade consisting of elements from five other genera, Ascocentrum, Ascolabium, Paraholcoglossum, Penkimia and Tsiorchis, is strongly supported as a monophyly. Hence, it would be better to retain Holcoglossum as monophyletic by the inclusiveness of three red to yellow species for the sake of information content, practicality and similarity. Therefore, we tentatively propose to recognize the monophyletic Holcoglossum s.l. (∼ 17 species).
Taxonomic Treatment
Holcoglossum Schltr., Repert Spec. Nov. Regni Veg. Beih. 4: 285. 1919;
Type. Holcoglossum quasipinifolium (Hayata) Schltr.
Synonyms
Paraholcoglossum Z.J. Liu, S.C. Chen & L.J. Liu, PLoS One 6(10): e24864. 2011. syn. nov. Type. Paraholcoglossum amesianum (Rchb.f.) Z.J. Liu, S.C. Chen & L.J. Chen.
Tsiorchis Z.J. Liu, S.C. Chen & L.J. Liu, PloS One 6(10): e24864. 2011. syn. nov. Type. Tsiorchis kimballianum (Rchb.f.) Z.J. Liu, S.C. Chen & L.J. Chen.
Ascolabium S.S. Ying, Coloured Ill. Indig. Orchids Taiwan 1: 54. 1977. syn. nov. Type. Ascolabium pumilum (Hayata) S.S. Ying.
Penkimia Phukan & Odyuo, Orchid Rev. 114: 331. 2006. syn. nov. Type. Penkimia nagalandensi s Phukan & Odyuo.
Chenorchis Z. J. Liu, K.W. Liu et L. J. Chen, Acta Ecologica Sinica 28(6):2435. 2008. syn. nov. Type. Chenorchis singchii Z.J. Liu, K.W. Liu et L.J. Chen.
Description
Epiphytic, small to moderate-size plants. Roots arising from the base of stem, white and fleshy, tip reddish. Stem short, usually clustered, enclosed by persistent leaf sheaths. Leaves articulate at base, condensed along stem, fleshy, subterete to terete, channeled adaxially. Inflorescences lateral. Flowers white to purple; pedicel and ovary long; dorsal sepal usually erect; lip 3-lobed, spurred or saccate; lateral lobes erect; middle lobe arising from spur; column winged; pollinia two, porate, usually with tapering stipe; anther cap beaked.
Species
Holcoglossum amesianum (Rchb.f.) Christenson, Notes Roy. Bot. Gard. Edinburgh 44(2): 255. 1987. Basionym. Vanda amesiana Rchb.f. in Gard. Chron. 3 ser., 1: 764. 1887. TYPE. Myanmar (Burma). Shan States, comm. imp. Low anno 1887. Herb. Reichenbach 37196 (Holotype, W). Homotypic synonym. Paraholcoglossum amensianum (Rchb.f.) Z.J. Liu, S.C. Chen & L.J. Chen, PLoS One 6(10): e24864. 2011. syn. nov.
Holcoglossum calcicola Schuit. & P. Bonnet, Orchideen J. 16(1): 6. 2009. TYPE. Laos. Bolikhamxai Province, D. Barthélémy, P. Bonnet, A. Schuiteman, V. Lamxay PB 451 (Holotype, Herbarium of the Faculty of Sciences of the National University of Laos).
Holcoglossum flavescens. (Schltr.) Z.H. Tsi, Acta Phytotax. Sin. 20(4): 441. 1982. Basionym. Aerides flavescens Schltr., Fedde Repert. Sp. Nov. 19; 282. 1924. TYPE. China. Yunnan, Yunpe (Current Yongsheng), Simeon Ten 23 (Holotype, BD). Homotypic synonyms. Papilionanthe flavescens (Schltr.) Garay Bot. Mus. Leafl. Harvard Univ. 23(4):270. 1974. Saccolabium yunpeense T. Tang et F. T. Wang, Acta Phytotax. 1: 97. 1951.
Holcoglossum himalaicum. (Deb, Sengupta & Malick) Aver., Bot. Zhurn. (Moscow & Leningrad) 73: 432. 1988. Basionym. Saccolabium himalaicum Deb, Sengupta & Malick, Bull. Bot. Soc. Bengal 22: 213. 1968. TYPE. Myanmar. Sima, Shalik Mokin 13 (Holotype, CAL). Heterotypic synonym. Holcoglossum junceum Z.H. Tsi, Acta Phytotax. Sin. 20: 442.1982. TYPE. China. Yunnan, M.G. Li 1798 (Holotype, PE!); Z.H. Tsi 76 (Paratype, PE!).
Holcoglossum kimballianum. (Rchb.f.) Garay, Bot. Mus. Leafl. 23(4): 182. 1972. Basionym. Vanda kimballiana Rchb.f. in Gard. Chron. 3 ser., 5: 232. 1889. TYPE. Myanmar. Southern Shan States (comm. imp. s.n., Herb. Reichenbach 37216 (Holotype, W). Homotypic synonyms. Tsiorchis kimballianum (Rchb.f.) Z.J. Liu, S.C. Chen & L.J. Chen, PLoS One 6(10): e24864. 2011. syn. nov. Heterotypic synonyms. Vanda saprophytica Gagnep. in Bull. Soc. Bot. Fr. 79: 37. 1932. TYPE. Laos. Between Nong Het and Muang Seng, Tranninh 1400 m, Poliane 16918 (Holotype, P!). H. saprophytica (Gagnep.) Christenson in Not. Bot. Gard. Edinb. 44(2): 255. 1987.
Holcoglossum lingulatum. (Aver.) Aver., Consp. Sosud. Rast. Fl. Vietnama 1: 110. 1990. Basionym. H. kimballianum var. lingulatum Averyanov in Bot. J. (Leningr.) 73(3): 426. fig. 4. 1988. TYPE. Vietnam. Between Chapa and Hoan Lien Song, Takhtajan 0745 (Holotype, LE). Heterotypic synonyms. H. tangii Christenson in Lindleyana 13(2): 121–124. 1999. TYPE. China. Yunnan, without precise locality, Hort. Mountain Orchids s.n. (Holotype, K [spirit]!).
Holcoglossum nagalandensis. (Phukan & Odyuo) X.H. Jin, com. nov. Basionym. Penkimia nagalandensis Phukan & Odyuo, Orchid Rev. 114: 331. 2006. syn. nov. TYPE. India. Nagaland. Odyuo 102808A (Holotype, CAL).
Holcoglossum nujiangense X.H. Jin & S.C. Chen, Nordic J. Bot. 25(1–2): 127. 2008. TYPE. China. Yunnan, Fugong County, X.H. Jin 6930 (Holotype, PE!). Heterotypic synonym. Holcoglossum linearifolium Z.J. Liu, S.C. Chen & L. J. Chen, PLoS One 6(10): e24864. 2011. syn. nov. TYPE. China. Yunnan, Malipo, Z.J. Liu 4865 (Holotype, NOCC).
Holcoglossum omeiense. X.H. Jin & S.C. Chen, Kew Bull. 59(4): 633 (−635). 2005. TYPE. China. Sichuan, Mt. Omei, from Qing-yin Temple to Hongchun Ping, alt. 720–1000 m, 15 September 1963, K.H. Shing et K.Y. Lang 1365A (Holotype, PE!; Isotypes, PE!).
Holcoglossum pumilum. (Hayata) X.H. Jin, com. nov. Basionym. Saccolabium pumilum Hayata, Bot. Mag. (Tokyo) 20: 77. 1906. TYPE. China. Taiwan. Biōritsu, K. Fujii s.n.(Holotype, ?).
Holcoglossum quasipinifolium. (Hayata) Schltr. in Fedde. Repert. Sp. Nov. Beih. 4:285.1919. Basionym. Saccolabium quasipinifolium Hayata, Icon. Pl. Formos. 2: 144. 1912. TYPE. China. Taiwan, Nimandaira, Mt. Arisan, Hayata and Sasaki sine no. (Holotype, TI!; Isotype, TAI F!).
Holcoglossum rupestre (Hand.-Mazz.) Garay in Bot. Mus. Leafl. Harvard Univ. 23(4): 182. 1972. Basionym. Vanda rupestris Hand.-Mazz. in Anz. Akad. Wiss. Wiem. Math.-Nat. 62:241. 1925; Hand.-Mazz., Symb. Sin. 7: 1359. 1936. TYPE. China. Yunnan, Zhongdian (now Shang-ri-la), Hand.-Mazz. 8802 (Holotype, W; Isotypes, WU!, E!, K!).
Holcoglossum sinicum Christenson, Notes Roy. Bot. Gard. Edinburgh 44(2): 255. 1987. TYPE. China. Yunnan, Yangbi, SEBC 380 (Holotype, E; Isotypes, KUN!, AMES!).
Holcoglossum subulifolium (Rchb.f.) Christenson, Notes Roy. Bot. Gard. Edinburgh 44(2): 255. 1987. Basionym. Vanda subulifolia Rchb.f., Flora 69: 552. 1886. TYPE. Myanmar. Veitch comm. imp. Herb. Reichenbach 37215 (Holotype, W!). Homotypic synonym. Paraholcoglossum subulifolium (Rchb.f.) Z.J. Liu, S.C. Chen & L. J. Chen, PLoS One 6(10): e24864. 2011. syn. nov. Heterotypic synonyms. Holcoglossum auriculatum Z.J. Liu, S.C. Chen & X.H. Jin, J. Wuhan Bot. Res. 23(2): 154. 2005. TYPE. China. Yunnan, Malipo, Z.J. Liu 2758 (Herbarium, Shenzhen City Wutongshan Nursries). Vanda watsonii Rofle, Gard. Chron. 3.s. 37: 82, 123. fig. 52. 1906. TYPE. Vietnam. Annam, Micholitz s. n. (Holotype, K!).
Holcoglossum tsii T. Yukawa, Ann. Tsukuba Bot. Gard. 19: 1. 2000. TYPE. China. Yunnan, without precise locality, TNS 9512285 (Holotype, Hort. Tsukuba Botanical Garden!; Isotype, PE!).
Holcoglossum wangii Christenson, Lindleyana 13(2): 123. 1998. TYPE. China. Yunnan, Hort. Mountain Orchids s.n. (Holotype, K [spirit]!).
Holcoglossum weixiense X.H. Jin & S.C. Chen, Novon 14(2): 178 (−179; fig. 1). 2004. TYPE. China. Yunnan, Weixi, HK Kadoorie PT 3490 (Holotype, PE!).
Materials and Methods
Ethics Statement
The species collected here are not included in the checklist of Chinese Protected Species. The fieldwork was conducted under the permission of the authority of each natural reserve, specifically Gaoligongshan National Nature Reserve (Yunnan, China), Dali Cangshan-Erhai National Natural Reserve (Yunnan, China), Jianfengling National Nature Reserve (Hainan, China) and Wuzhishan National Nature Reserve (Hainan, China). No specific permits were required for the described field studies.
Taxon Sampling
To determine the systematic positions of Holcoglossum and related genera, we sampled 78 genera and 138 samples, representing all of the major clades in the subtribe Aeridinae, based on previous molecular work (Table S4) [26], [44]. Secondly, we sampled a reduced matrix containing 36 species to analyze the intraspecific relationships within Holcoglossum (Table S5).
To facilitate consistency and convenience, Paraholcoglossum and Tsiorchis were referred to Holcoglossum throughout the Results.
DNA Extraction, PCR and Sequencing
The total DNA was extracted from silica gel-dried materials using the modified CTAB method [45]. The ITS, matK and trnH-psbA primers used for the amplification and sequencing are listed in Xiang et al. [46], and the trnL-F primers are from Taberlet et al. [47]. The selected DNA regions were amplified using a standard polymerase chain reaction (PCR). The sequencing reactions were performed using the ABI Prism Bigdye Terminator Cycle Sequencing Kit (Applied Biosystems, ABI).
Genetic Distance Analyses
To estimate the variation of the ITS, matK and trnL-F sequences across Holcoglossum and related genera, we calculated the pairwise genetic p-distance for each region using MEGA v. 4 [48]. These distances were initially used to evaluate the interspecific divergence with the Kimura 2-Parameter model (K2P). TrnH-psbA was excluded under this analysis because all of the sequences were from Holcoglossum and there was a paucity of related genera.
Phylogenetic Analyses
Clustal X 1.83 [49] was used to obtain an initial alignment of the DNA sequences, followed by manual adjustment using BioEdit [50].
The phylogenetic analyses for each matrix were performed using the maximum parsimony (MP) and Bayesian inference (BI) methods in PAUP v4.0b10 [51] and MrBayes v3.0b4 [52], respectively.
For the MP analyses, heuristic searches were conducted with 1,000 replicates of random addition, with one tree held at each step during the stepwise addition, tree-bisection-reconnection (TBR) branch swapping, MulTrees in effect, and the steepest descent off. All of the characters were unordered and equally weighted, and the gaps were coded as missing data. To access the node support, bootstrap analyses [53] were performed using 1,000 replicates, with 10 random taxon additions and heuristic search options.
Prior to the Bayesian analysis, the Akaike Information Criterion (AIC) implemented in ModelTest version 3.7 [54], [55] was used to select the best-fit model of molecular evolution for each dataset. For the BI analyses, four chains of the Markov Chain Monte Carlo (MCMC) were run, sampling one tree every 1,000 generations for 5,000,000, starting with a random tree. Majority rule (>50%) consensus trees were constructed after removing the “burn-in period” samples (the first 20% of the sampled trees).
Gross Morphology and Micromorphology
To understand the morphology of Holcoglossum and relative genera, we performed herbarium examination of specimens (Table S6) and fieldwork observations for gross morphology, and scanning electric microscope (using KYKY-1000B) for the pollinium micromorphology (Table S7). However, the morphological characters of following not Chinese native five species, Aerides krabiensis, A. thibautiana, Holcoglossum calcicola, Jumellea sagittata, Microterangis hariotiana, are from literature.
Character Mapping
To identify the synapomorphies that are congruent with each of the major clades of Holcoglossum retrieved in the molecular tree and to assess the value of the characters used in the classifications, 45 morphological characters were selected. We used the exemplar method, scoring the morphological characters in the morphological matrix for the same species as used in the molecular analyses. The complete morphological matrix, coding 45 characters for the 35 taxa, is available in Table S2. We also prepared a combined morphological and molecular matrix that included the 32 taxa for character mapping.
Character evolution was reconstructed onto a 50% major consensus tree generated in PAUP using the parsimony ancestral state reconstruction in Mesquite v.2.75 [56]. All of the morphological characters were considered unordered and unweighted.
Patrocladistic Analysis
We displayed the patrocladistic analysis according to Stussey and König [57]. This cladogram with high support based on Bayesian inference served as a structure for the patristic distance. The cladistic distances were calculated from the selected cladogram, and these values were placed in a new cladistic matrix. The patristic distance was defined as the number of apomorphic step changes separating two taxa on the cladogram. The patristic distance was then added to the cladistic distance to form the combined patrocladistic data matrix (Table S3). These combined distances are used as input into UPGMA (packages in MEGA) to construct the patrocladogram.
Supporting Information
Bayesian inference tree of subtribe Aeridinae based on ITS. The bootstrap percentages and posterior probability of >50% are shown above each branch. “−” = no value. “*”represents data from Fan et al. [32], and “#”represents data from Liu et al. [33].
(JPG)
The strict consensus maximum parsimony tree of Holcoglossum s.l. based on the morphological data. The bootstrap percentages of >50% are shown above each branch.
(JPG)
Character mapping of Holcoglossum s.l. and related genera. See Table S2 for the character numbers and states.
(JPG)
Statistics from the phylogenetic analyses of the various datasets.
(DOC)
Morphological data for the phylogenetic analyses.
(DOC)
Matrix of cladistic (lower left) and patristic (upper right) distances among taxa.
(DOC)
Taxa and GenBank accession numbers for the ITS sequences in phylogenetic analysis of subtribe Aeridinae. A dash indicates missing data; *represent the sequences obtained in this study, and the remaining sequences are from GenBank.
(DOC)
Taxa and GenBank accession numbers for the ITS, matK , trnL-F and trnH - psbA sequences in phylogenetic analysis of Holcoglossum alliance. A dash indicates missing data; *represent the sequences obtained in this study, and the remaining sequences are from GenBank.
(DOC)
Samples used in the gross morphology investigation.
(DOC)
Samples used in the micromorphology.
(DOC)
Acknowledgments
We thank Dr. Chung Shi-Wen (Division of Forest Biology, Taiwan Forestry Research Institute, Taipei) for the material of Ascolabium pumilum and his assistance during the fieldwork in Taiwan, and Dr. Wang Hong (Kunming Institute of Botany, Chinese Academy of Sciences) for micromorphology experiments.
Funding Statement
This research was supported by a grant from the National Natural Science Foundation of China (31170176 for Jin). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hörandl E (2006) Paraphyletic versus monophyletic taxa - evolutionary versus cladistic classifications. Taxon 55: 564–570. [Google Scholar]
- 2. Hörandl E (2007) Neglecting evolution is bad taxonomy. Taxon 56: 1–5. [Google Scholar]
- 3. Hörandl E (2010) Beyond cladistics: Extending evolutionary classifications into deeper time levels. Taxon 59: 345–350. [Google Scholar]
- 4. Hörandl E, Stuessy TF (2010) Paraphyletic groups as natural units of biological classification. Taxon 59: 1641–1653. [Google Scholar]
- 5. Ebach MC, Williams DM, Morrone JJ (2006) Paraphyly is bad taxonomy. Taxon 55: 831–832. [Google Scholar]
- 6. Brummitt RK (2006) Am I a bony fish? Taxon 55: 268–269. [Google Scholar]
- 7. Brummitt RK (2002) How to chop up a tree. Taxon 51: 31–41. [Google Scholar]
- 8. Brummitt RK (2008) Evolution in taxonomic perspective. Taxon 57: 1019–1050. [Google Scholar]
- 9. Nordal I, Stedje B (2005) Paraphyletic taxa should be accepted. Taxon 54: 5–6. [Google Scholar]
- 10. Stuessy TF (2010) Paraphyly and the origin and classfication of angiosperum. Taxon 59: 689–693. [Google Scholar]
- 11. Podani J (2010) Monophyly and paraphyly: A discourse without end. Taxon 59: 1011–1015. [Google Scholar]
- 12. Mayr E, Bock WJ (2002) Classifications and other ordering systems. Journal of Zoological Systematics and Evolutionary Research 40: 169–194. [Google Scholar]
- 13. Crisp MD, Chandler GT (1996) Paraphyletic species. Telopea 6: 813–844. [Google Scholar]
- 14. Rieseberg LH, Bouillet L (1994) Are many plant species paraphyletic? Taxon 43: 21–32. [Google Scholar]
- 15. Bininda-Emonds ORP (1998) Superspecific taxa as terminals in cladistic analysis, implicit assumptions of monophyly and a comparison of methods. Biological Journal of the Linnean Society 64: 101–133. [Google Scholar]
- 16. Schmidt-Lebuhn AN (2012) Fallacies and false premises-a critical assessment of the argument for the recognition of paraphyly taxa in botany. Cladistics 28: 174–187. [DOI] [PubMed] [Google Scholar]
- 17. Donoghue MJ, Cantino DC (1988) Paraphyly, ancestors and the goals of taxonomy, a botanical defense of cladism. Botanical Review 54: 107–128. [Google Scholar]
- 18. Humphries CJ, Chappil JA (1988) Systematics as science, a response to Cronquist. Botanical Review 54: 129–144. [Google Scholar]
- 19. Freudenstein JV (1998) Paraphyly ancestors and classification. Taxon 47: 95–104. [Google Scholar]
- 20.Dressler RL (1981) The orchids: natural history and classification: Harvard University Press, Cambridge.
- 21.Dressler RL (1993) Phylogeny and classification of the orchid family: Dioscorides, Portland.
- 22. Garay LA (1972) On the systematics of the monopodial orchids I. Bot Mus Leafl. 23: 149–212. [Google Scholar]
- 23. Garay LA (1974) On the systematics of the monopodial orchids II. Botanical Mus Leafl Harv Univ 23: 369–376. [Google Scholar]
- 24. Seidenfaden G (1988) The orchid genera in Thailand XIV. Fifty-nine vandoid genera. Opera Bot 95: 304–308. [Google Scholar]
- 25. Seidenfaden G (1992) The orchids of IndoChinaChina. Opera Bot 114: 441–443. [Google Scholar]
- 26. Topik H, Yukawa T, Ito M (2005) Molecular phylogenetics of subtribe Aeridinae (Orchidaceae): insights from plastid matK and nuclear ribosomal ITS sequences. Journal of Plant Research 118: 271–284. [DOI] [PubMed] [Google Scholar]
- 27. Schlechter R (1919) Orchideologiae Sino-Japonicae Prodromus. Feddes Repert Spec Nov Regni Veg Beih 4: 285. [Google Scholar]
- 28. Tsi ZH (1982) A study of the genus Holcoglossum of Orchidaceae. Acta Phytotaxonomica Sinica 20: 439–444. [Google Scholar]
- 29. Christenson EA (1987) An infrageneric classification of Holcoglossum Schltr. (Orchidaceae: Sarcanthinae) with a key to the genera of the Aerides-Vanda alliance. Notes Roy Bot Gard Edinburgh 44: 249–256. [Google Scholar]
- 30. Jin XH (2005) Generic delimitation and a new infrageneric system in the genus Holcoglossum (Orchidaceae: Aeridinae). Botanical Journal of the Linnean Society 149: 465–468. [Google Scholar]
- 31. Jin XH, Zhang T, Gu ZJ, Li DZ (2007) Cytological studies on the genus Holcoglossum (Orchidaceae: Aeridinae). Botanical Journal of the Linnean Society 154: 283–288. [Google Scholar]
- 32. Fan J, Qin HN, Li DZ, Jin XH (2009) Molecular phylogeny and biogeography of Holcoglossum (Orchidaceae: Aeridinae) based on nuclear ITS, and chloroplast trnL-F and matK. Taxon 3: 849–861. [Google Scholar]
- 33.Liu ZJ, Chen LJ, Chen SC, Cai J, Tsai WC, et al.. (2011) Paraholcoglossum and Tsiorchis, two new orchid genera established by molecular and morphological analyses of the Holcoglossum alliance. PLoS ONE 6 e24864. doi:24810.21371/journal.pone.0024864. [DOI] [PMC free article] [PubMed]
- 34. Ying SS (1977) Ascolabium S.S.Ying, Coloured Ill. Indig.. Orchids Taiwan 1: 54. [Google Scholar]
- 35.Jin XH, Wood JJ (2009) Holcoglossum. In: Wu ZY, Raven PH, editors. Flora of ChinaChina: Beijing: Science Press, St. Louis: Missouri Botanical Garden. 499–502.
- 36. Christenson EA (1998) Two new species of Holcoglossum Schltr. (Orichidaceae: Aeridinae) from ChinaChina. Lindleyana 13: 121–124. [Google Scholar]
- 37. Averyanov LV (1988) New species and nomenclatural changes in the Orchidaceae family of Vietnamses flora. Botanical Journal (Leningrad) 73: 101–107. [Google Scholar]
- 38. Liu KW, Liu ZJ, Huang L, Li LQ, Chen LJ, et al. (2006) Pollination: self-fertilization strategy in an orchid. Nature 441: 945–946. [DOI] [PubMed] [Google Scholar]
- 39. Jin XH, Chen SC, Qin HN (2005) Pollination system of Holcoglossum rupestre (Orchidaceae): a special and unstable system. Plant Systematic and Evolution 254: 31–38. [Google Scholar]
- 40.Jin XH, Chen SC, Li DZ (2007) Holcoglossum nujiangense (Orchidaceae: Aeridinae) - a new species and its pollination system. Nordic Journal of Botany 25.
- 41. Liu ZJ, Chen LJ, Liu K, Li LQ, Ma XY, et al. (2008) Chenorchis, a new orchid genus, and its eco-strategy of ant pollination. Acta Ecologica Sinica 28: 2433–2444. [Google Scholar]
- 42. Stpiczynska M, Davies KL, Kaminska M (2011) Comparative anatomy of the nectary spur in selected species of Aeridinae (Orchidaceae). Ann Bot 107: 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Cingel NA (2001) An atlas of orchid pollination: America, Africa, and Australia: A. A. Balkema, Rotterdam, Netherlands.
- 44. Kocyan A, Vogel EF, de Conti E, Gravendeel B (2008) Molecular phylogeny of Aerides (Orchidaceae) based on nuclear and two plastid markers: A step forward in understanding the evolution of the Aerdinae. Molec Phylogenetics and Evolution 48: 422–443. [DOI] [PubMed] [Google Scholar]
- 45. Doyle JJ, Doyle JL (1987) A rapid isolation procedure from smalll quantities of fresh leaf tissue. Phytochemitry Bulletion 19: 11–15. [Google Scholar]
- 46. Xiang XG, Hu H, Wang W, Jin XH (2011) DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNAbarcode candidates. Molecular Ecology and Research 11: 1012–1021. [DOI] [PubMed] [Google Scholar]
- 47. Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- 48. Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95 / 98 / NT. Nucl Acid Symp Series 41: 95–98. [Google Scholar]
- 51.Swofford DL (2002) PAUP*: Phylogenetic Analysis using Parsimony (* and Other Methods), version 4.0b10. : Sinauer, Sunderland.
- 52. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 53. Felsenstein (1988) Phylogenies from molecular sequences: inference and reliability. Journal of Annal Review Genetics 22: 521–565. [DOI] [PubMed] [Google Scholar]
- 54. Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 55. Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793–808. [DOI] [PubMed] [Google Scholar]
- 56.Maddison WP, Maddison DR (2007) Mesquite: a modular system for evolutionary analysis. Version 2.75. Available: http://mesquiteproject.org. Accessed: 2011 Sep 30.
- 57. Stuessy TF, Konig C (2008) Patrocladistics classifictation. Taxon 57: 594–601. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian inference tree of subtribe Aeridinae based on ITS. The bootstrap percentages and posterior probability of >50% are shown above each branch. “−” = no value. “*”represents data from Fan et al. [32], and “#”represents data from Liu et al. [33].
(JPG)
The strict consensus maximum parsimony tree of Holcoglossum s.l. based on the morphological data. The bootstrap percentages of >50% are shown above each branch.
(JPG)
Character mapping of Holcoglossum s.l. and related genera. See Table S2 for the character numbers and states.
(JPG)
Statistics from the phylogenetic analyses of the various datasets.
(DOC)
Morphological data for the phylogenetic analyses.
(DOC)
Matrix of cladistic (lower left) and patristic (upper right) distances among taxa.
(DOC)
Taxa and GenBank accession numbers for the ITS sequences in phylogenetic analysis of subtribe Aeridinae. A dash indicates missing data; *represent the sequences obtained in this study, and the remaining sequences are from GenBank.
(DOC)
Taxa and GenBank accession numbers for the ITS, matK , trnL-F and trnH - psbA sequences in phylogenetic analysis of Holcoglossum alliance. A dash indicates missing data; *represent the sequences obtained in this study, and the remaining sequences are from GenBank.
(DOC)
Samples used in the gross morphology investigation.
(DOC)
Samples used in the micromorphology.
(DOC)