Abstract
A ∼38kB plasmid (pXF-RIV5) was present in the Riv5 strain of Xylella fastidiosa subsp. multiplex isolated from ornamental plum in southern California. The complete nucleotide sequence of pXF-RIV5 is almost identical to that of pXFAS01 from X. fastidiosa subsp. fastidiosa strain M23; the two plasmids vary at only 6 nucleotide positions. BLAST searches and phylogenetic analyses indicate pXF-RIV5 and pXFAS01 share some similarity to chromosomal and plasmid (pXF51) sequences of X. fastidiosa subsp. pauca strain 9a5c and more distant similarity to plasmids from a wide variety of bacteria. Both pXF-RIV5 and pXFAS01 encode homologues of a complete Type IV secretion system involved in conjugation and DNA transfer among bacteria. Mating pair formation proteins (Trb) from Yersinia pseudotuberculosis IP31758 are the mostly closely related non-X. fastidiosa proteins to most of the Trb proteins encoded by pXF-RIV5 and pXFAS01. Unlike many bacterial conjugative plasmids, pXF-RIV5 and pXFAS01 do not carry homologues of known accessory modules that confer selective advantage on host bacteria. However, both plasmids encode seven hypothetical proteins of unknown function and possess a small transposon-associated region encoding a putative transposase and associated factor. Vegetative replication of pXF-RIV5 and pXFAS01 appears to be under control of RepA protein and both plasmids have an origin of DNA replication (oriV) similar to that of pRP4 and pR751 from Escherichia coli. In contrast, conjugative plasmids commonly encode TrfA and have an oriV similar to those found in IncP-1 incompatibility group plasmids. The presence of nearly identical plasmids in single strains from two distinct subspecies of X. fastidiosa is indicative of recent horizontal transfer, probably subsequent to the introduction of subspecies fastidiosa to the United States in the late 19th century.
Introduction
Horizontal gene transfer plays a critical role in bacterial adaptation and evolution. On average, 81% of the genes in a typical bacterial genome have been involved in a horizontal transfer event at some point in the past [1]. One of the most common mechanisms for DNA exchange is via conjugative plasmids that encode type IV secretion systems (T4SS), a broad class of macromolecular translocation machinery. There are three main types of T4SS: i) conjugation systems that transfer DNA and, in some instances, DNA-binding proteins; ii) effector translocator systems that deliver proteins and other effectors to eukaryotic cells during bacterial infection of eukaryotic hosts; and iii) DNA uptake or release systems that move DNA between the interior of the cell and the extracellular environment [2]. Conjugation systems are found both on self-transmissible or conjugative plasmids, circular DNA molecules that replicate independently of the bacterial chromosome, and on integrative and conjugative elements that integrate into the chromosome, excising and forming a circular intermediate prior to translocation [2]. One well characterized conjugation system is VirB/D4 from Agrobacterium tumefaciens, which is composed of a cell-envelope spanning secretion channel and an extracellular pilus that contacts the recipient cell [3]. For VirB/D4, the translocation system consists of VirB2-11 proteins forming the secretion channel, the VirD4 substrate receptor or type IV coupling protein (T4CP), and proteins for pilus formation and DNA substrate processing [2]. Many conjugative plasmids also contain accessory modules encoding cargo proteins which act as virulence factors, confer resistance to antibiotics/heavy metals, or catabolize toxic organic substances [4].
Xylella fastidiosa is a fastidious, xylem-limited Gram-negative bacterial phytopathogen causing numerous vascular occlusion and water stress diseases including Pierce’s disease of grape, almond leaf scorch, oleander leaf scorch, and other diseases of perennial crops and landscape plants [5]. Four subspecies of X. fastidiosa have been identified based on a multi-locus sequence typing (MLST) phylogeny [6], [7]. Subspecies fastidiosa contains strains of low genetic diversity that cause Pierce’s disease and sometimes almond leaf scorch in the U. S. and diverse strains from Central America; subsp. fastidiosa is thought to have been introduced to the U. S. in the late 19th century [8]. Subspecies multiplex is an endemic North American clade capable of infecting numerous hosts (but generally not grapevine) [9]–[11]. Subspecies pauca contains South American strains causing citrus variegated chlorosis and coffee leaf scorch [12]. Subspecies sandyi consists of closely related strains isolated from oleander in California and Texas and is thought to have been introduced to the United States approximately 30 years ago [6]. Currently, 5 fully sequenced and annotated X. fastidiosa genomes are available (pauca strain 9a5c [13], fastidiosa strain Temecula [14], multiplex strain M12 [15], fastidiosa strain M23 [15], and fastidiosa strain GB514 [16]). Two additional sequences (multiplex strain Dixon, GenBank accession number NZ_AAAL00000000.2, and sandyi strain Ann-1, GenBank accession number NZ_AAAM00000000.3) are incomplete, unassembled shotgun sequences; the Ann-1 sequence may have been derived from a mixed culture and appears to be contaminated with sequences from a multiplex strain [17].
Here, we characterize two closely related 38kB conjugative plasmids of X. fastidiosa. Plasmid pXF-RIV5 was isolated from the Riv5 strain of X. fastidiosa subspecies multiplex [18]; complete sequence of pXF-RIV5 was determined in this current work. Plasmid pXFAS01 is known only as a circular contig discovered during the complete genome sequencing of X. fastidiosa subspecies fastidiosa strain M23 [15]. While minimal annotation of pXFAS01 accompanies the sequence in GenBank, no analysis of the type IV secretion system, origin of transfer or origin of replication from pXFAS01 has been presented previously. This work presents analyses of gene complement and phylogeny of these two closely related plasmids to reveal i) an evolutionary history of recombination among divergent sources to generate the mosaic backbone shared by pXF-RIV5 and pXFAS01, and ii) evidence of recent translocation of plasmid DNA via conjugation among distinct subspecies of X. fastidiosa.
Materials and Methods
Culture and MLST of X. fastidiosa subspecies multiplex strain Riv5
Isolation of X. fastidiosa strain Riv5 from ornamental plum (Prunus cerasifera) was described previously [18]. Strain Riv5 cultures were grown in liquid periwinkle wilt (PW) medium for 7–10 days at 28C and used to inoculate plates containing solid PW medium. After 7–10 days of growth at 28C, bacterial colonies were washed from 10 PW plates and extracted for total genomic DNA [11]. Genomic DNA was used as template for PCR amplification of seven housekeeping genes (cysG, gltT, holC, malF, leuA, nuoL, petC) and pilU [7]; consensus sequences were determined for each amplified region based on sequences of three independent clones per PCR product. Consensus sequences for each amplified region were concatenated into a single sequence. MLST was performed as described [7] with concatenated Riv5 sequences aligned with the corresponding concatenated sequences from multiple strains representative of each X. fastidiosa subspecies available in GenBank using CLUSTALX. Phylogenetic placement of strain Riv5 was determined based on a neighbor-joining tree (1000 bootstrap replications) using the multiple alignment of concatenated MLST sequences as input data.
Plasmid DNA isolation and sequencing
Previously, strain Riv5 was shown to harbor a large plasmid (designated here as pXF-RIV5), yielding multiple products when digested with HindIII [18]. Purification of pXF-RIV5 DNA was as described [18] from Riv5 cultures grown under the same regime as that used to extract genomic DNA. Purified plasmid DNA was digested with HindIII; each resulting fragment was gel purified and ligated into HindIII digested pGEM7Zf+ (Promega, Madison, WI). Ligation products were transformed into Escherichia coli JM109; recombinant plasmids bearing HindIII inserts of pXF-RIV5 were sequenced using a combination of universal (M13 forward and reverse) and custom primers. As preliminary sequences of insert termini obtained with universal primers indicated a very close relationship with pXFAS01, custom primers were based on the known sequence of pXFAS01 (GenBank Accession NC_010579.1) associated with the X. fastidiosa subspecies fastidiosa strain M23 genome sequence [15]. The complete nucleotide sequence of pXF-RIV5 (GenBank Accession JX548317) was assembled using the sequence of pXFAS01 as a scaffold.
DNA sequence analysis and annotations
Open reading frames were identified based on similarity to pXFAS01 and using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Annotations are derived from similar sequences identified using BLAST [19] and NCBI conserved domain searches [20]. The origin of conjugative transfer (oriT) was identified by similarity to the oriT regions of pRP4 and pR751 [21]. The tandem repeats in oriV were found using Repfind (http://zlab.bu.edu/repfind/index.shtml). A map of pXF-RIV5 was drawn using GENtle (http://gentle.magnusmanske.de/) open software package.
Phylogenetic analysis of pXF-RIV5
Phylogeny of three proteins encoded by pXF-RIV5, representing three distinct genetic modules resident on pXF-RIV5 (and pXFAS01), were selected for examination. RepA represents a protein involved in DNA replication; TraI is a relaxase homologue representing the tra module (conjugative transfer functions); and TrbG is predicted to be an outer membrane protein and represents the trb module (mating pair formation functions).
Taxa selected for inclusion were based on results of BLAST P searches of the GenBank nonredundant protein database using the corresponding homologues of RepA, TraI and TrbG encoded by pXF-RIV5 as queries. A neighbor-joining tree (1000 bootstrap replicates) for each protein was constructed based on a multiple alignment of amino acid sequences generated using CLUSTALX. Nodes bearing <70% bootstrap support were considered unreliable and collapsed to polytomies.
Results
Riv5 is a strain of subspecies multiplex
Genomic DNA sequences used for MLST of strain Riv5 were deposited as GenBank Accessions JX679700-JX67907. Phylogeny of concatenated sequences of the eight genes examined by MLST indicated that strain Riv5 clustered with X. fastidiosa strains of subspecies multiplex, including the fully-sequenced multiplex strain M12 (data not shown). Phylogeny of each individual gene used for MLST also clustered strain Riv5 with strains of subspecies multiplex (data not shown). These results are consistent with phylogenetic placement of strain Riv5 based on 16S-23S rRNA spacer sequences [18].
pXF-RIV5 contains a type IV secretion system
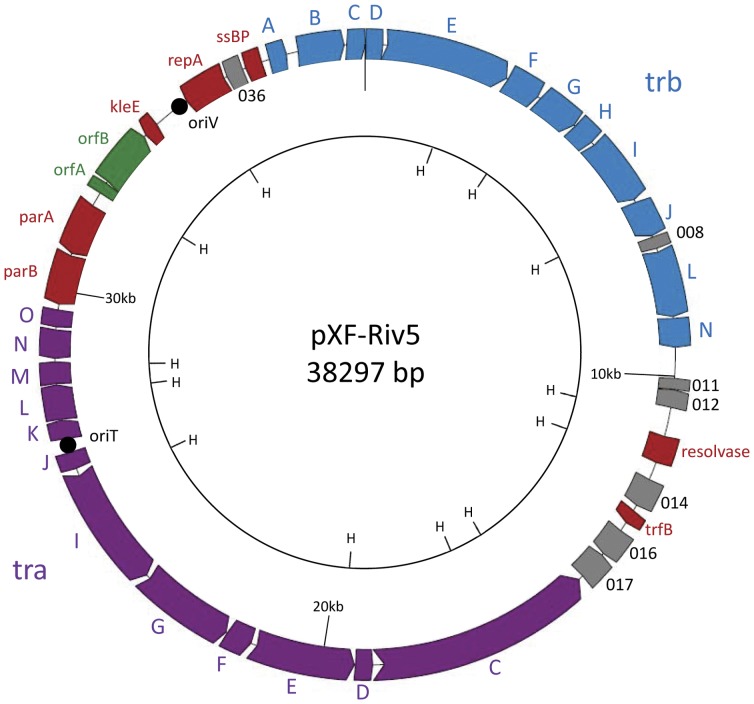
The complete nucleotide sequence of pXF-RIV5 is 38,297 bp in length with a G+C content of 49.2%, which is similar to the 51–52% G+C content of sequenced X. fastidiosa genomes [13], [15]. As detailed in Table 1 and Figure 1, pXF-RIV5 has 33 ORFs encoding proteins similar to characterized proteins of known function from other organisms while seven ORFs encode hypothetical proteins for which functions of homologues identified in GenBank are unknown. The two largest groups of genes are the conjugative transfer (tra) and mating pair formation (trb) modules. Together, these two genetic modules encode homologues of all proteins necessary for a functional T4SS [2]. Genes for plasmid replication (repA, kleE, and ssBP) and partition (parA and parB) also are present. Two genes, orfA and orfB, encode proteins similar to transposon-associated recombinase and transposase, respectively; no other transposon-like elements on pXF-RIV5 were identified. Unlike many conjugative plasmids from animal pathogens or environmental samples, no accessory modules containing homologues of known virulence factors, antibiotic/heavy metal resistance, or catabolism of toxic organic compounds were encoded by pXF-RIV5.
Table 1. Annotation of open reading frames from pXF-RIV5.
| Name | start | stop | # aa | strand | product | most closely related gene product | % aa identity |
| TrbD | 3 | 323 | 107 | + | conjugal transfer protein, ATPase, VirB3 family | conjugation protein TrbD [Azoarcus sp. EbN1] YP_195564.1 | 71 |
| TrbE | 311 | 2875 | 855 | + | conjugal transfer protein, ABC transporter-like, VirB4 family | conjugal transfer protein TrbE [Yersinia pseudotuberculosis IP 31758] YP_001393245.1 | 69 |
| TrbF | 2872 | 3588 | 239 | + | conjugal transfer protein, inner membrane protein, VirB8 family | conjugal transfer protein TrbF [Yersinia pseudotuberculosis IP 31758] YP_001393246.1 | 61 |
| TrbG | 3605 | 4495 | 297 | + | conjugal transfer protein, periplasmic or outer membrane protein, VirB9 family | conjugal transfer protein TrbG [Yersinia pseudotuberculosis IP 31758] YP_001393247.1 | 68 |
| TrbH | 4498 | 4974 | 159 | + | conjugal transfer protein, putative lipoprotein, VirB7 family | conjugal transfer protein TrbH [Yersinia pseudotuberculosis IP 31758] YP_001393248.1 | 41 |
| TrbI | 4980 | 6380 | 467 | + | conjugal transfer protein, inner membrane protein, VirB10 family | conjugal transfer protein TrbI [Yersinia pseudotuberculosis IP 31758] YP_001393249.1 | 53 |
| TrbJ | 6399 | 7172 | 258 | + | conjugal transfer protein, periplasmic or outer membrane protein, VirB5 family | conjugative transfer protein TrbJ [Burkholderia pseudomallei Pakistan 9] ZP_03794994.1 | 66 |
| pRiv5_008 | 7189 | 7425 | 79 | + | hypothetical protein, putative lipoprotein attachment site | lipoprotein [Aggregatibacter actinomycetemcomitans D11S-1] YP_003966129.1 | 44 |
| TrbL | 7447 | 8817 | 457 | + | conjugal transfer protein, inner membrane protein, VirB6 family | conjugal transfer protein TrbL/VirB6 [Yersinia pseudotuberculosis IP 31758] YP_001393251.1 | 54 |
| TrbN | 8823 | 9422 | 200 | + | conjugal transfer protein, lytic transglycosylase domain, possible VirB1 | conjugal transfer protein TrbN [Yersinia pseudotuberculosis IP 31758] YP_001393252.1 | 63 |
| pRiv5_011 | 10035 | 10262 | 76 | - | conserved hypothetical protein, Pfam06156/DUF972 family | hypothetical protein XMIN_4535 [Xanthomonas citri pv. mangiferaeindicae LMG 941] ZP_09883050.1 | 37 |
| pRiv5_012 | 10259 | 10627 | 123 | - | hypothetical protein | hypothetical protein EGYY_28500 [Eggerthella sp. YY7918] YP_004712228.1 | 44 |
| resolvase | 11170 | 11733 | 188 | + | site-specific serine recombinase family protein | putative resolvase [Methylomicrobium alcaliphilum] YP_004901765.1 | 76 |
| pRiv5_014 | 12064 | 12657 | 198 | + | hypothetical protein | hypothetical protein MYA_6037 [Burkholderia sp. KJ006] YP_006337102.1 | 43 |
| TrfB | 12725 | 13039 | 105 | + | probable TrfB transcriptional repressor protein | hypothetical protein BBR47_02790 [Brevibacillus brevis NBRC 100599] YP_002769760.1 | 61 |
| pRiv5_016 | 13216 | 13809 | 198 | + | hypothetical protein | hypothetical protein MYA_6037 [Burkholderia sp. KJ006] YP_006337102.1 | 54 |
| pRiv5_017 | 13850 | 14437 | 196 | - | hypothetical protein | hypothetical protein Acife_3030 [Acidithiobacillus ferrivorans SS3] YP_004785429.1 | 55 |
| TraC | 14505 | 18992 | 1496 | - | conjugal transfer protein, topoisomerase/primase-like | TraC DNA primase [Plasmid QKH54] YP_619864.1 | 40 |
| TraD | 18998 | 19360 | 121 | - | conjugal transfer protein, inner membrane protein | TraD protein [IncP-1 plasmid pKJK5] YP_709180.1 | 49 |
| TraE | 19363 | 21420 | 686 | - | conjugal transfer protein, topoisomerase-primase domain | TraE [Pseudomonas putida] YP_003162628.1 | 73 |
| TraF | 21449 | 21985 | 179 | - | conjugal transfer protein, peptidase/pilin processing protease | TraF protein of DNA transfer system [Methylophaga sp. JAM7] YP_006297569.1 | 68 |
| TraG | 21982 | 23916 | 645 | - | conjugal transfer protein, coupling protein, VirD4 family | conjugal transfer protein TraG [Yersinia pseudotuberculosis IP 31758] YP_001393286.1 | 79 |
| TraI | 23913 | 26405 | 831 | - | conjugal transfer protein, relaxase/mobilization domain | conjugal transfer protein TraI [Yersinia pseudotuberculosis IP 31758] YP_001393287.1 | 45 |
| TraJ | 26440 | 26793 | 118 | - | conjugal transfer protein, relaxosome component | conjugal transfer relaxosome component TraJ [Aeromonas caviae Ae398] ZP_08522309.1 | 54 |
| TraK | 27018 | 27413 | 132 | + | conjugal transfer protein, putative oriT binding protein | TraK protein [IncP-1 plasmid pKJK5] YP_709187.1 | 47 |
| TraL | 27413 | 28138 | 242 | + | conjugal transfer protein, contains P-loop nucleotide binding domain | TraL protein [Pseudomonas sp. ADP] NP_862455.1 | 71 |
| TraM | 28138 | 28590 | 151 | + | conjugal transfer protein, transcriptional activator | traM gene product [Methylomicrobium alcaliphilum] YP_004901800.1 | 54 |
| TraN | 28652 | 29248 | 199 | - | conjugal transfer protein, mating pair stabilization protein | hypothetical protein pKJK5_51 [IncP-1 plasmid pKJK5] YP_709190.1 | 48 |
| TraO | 29275 | 29628 | 118 | - | conjugal transfer protein, putative membrane protein | putative conjugation protein TraO [Azoacrus sp. EbN1] YP_195664.1 | 53 |
| parB-like | 29726 | 30757 | 344 | - | contains parB-like nuclease domain, putative partition site DNA binding protein | ParB equivalent nuclease [uncultured bacterium] YP_112421.1 | 71 |
| parA-like | 30754 | 31833 | 360 | - | putative ATPase involved in plasmid replication and partition | IncC1 protein [uncultured bacterium] NP_598102.1 | 62 |
| orfA | 32062 | 32286 | 75 | + | orfA family, site-specific serine recombinase, transposon-assoc. | transposon IS605 OrfA [Methylacidiphilum infernorum V4] YP_001941027.1 | 85 |
| orfB | 32280 | 33476 | 399 | + | orfB family, helix-turn-helix domain, probable transposase | transposon IS605 OrfB [Methylacidiphilum infernorum V4] YP_001941028.1 | 72 |
| KleE | 33517 | 33837 | 107 | - | probable KleE stable plasmid inheritance protein | KleE protein [Plasmid pB3] YP_133959.1 | 46 |
| RepA | 34560 | 35471 | 304 | - | protein involved in plasmid replication, exact role unknown | RepA [Acidithiobacillus caldus SM-1] YP_004750509.1 | 75 |
| pRiv5-036 | 35493 | 35849 | 119 | - | hypothetical protein | hypothetical protein pSB102_p07 [Plasmid pSB102] NP_361021.1 | 54 |
| ssBP | 35893 | 36252 | 120 | - | single-strand DNA binding protein | single-strand DNA-binding protein [uncultured bacterium] YP_112367.1 | 61 |
| TrbA | 36370 | 36726 | 119 | + | conjugal transfer protein, helix-turn-helix containing tx regulator | conjugal transfer protein TrbA [Yersinia pseudotuberculosis IP 31758] YP_001393241.1 | 73 |
| TrbB | 36930 | 37892 | 321 | + | conjugal transfer protein, ATPase, VirB 11 family | conjugal transfer protein TrbB [Yersinia pseudotuberculosis IP 31758] YP_001393242.1 | 70 |
| TrbC | 37905 | 38297 | 131 | + | conjugal transfer protein, subunit of bacterial pilus, VirB2 family | conjugal transfer protein TrbC [Yersinia pseudotuberculosis IP 31758] YP_001393243.1 | 71 |
The most closely related gene product was identified using BLAST P and excludes other proteins from X. fastidiosa.
Figure 1. Genetic map of pXF-RIV5.
The open reading frames are colored according to presumed function: red, plasmid replication and partition; purple, conjugative transfer (tra); blue, mating pair formation (trb); green, transposon associated; gray, hypothetical proteins of unknown function. The origin of replication (oriV) and the origin of transfer (oriT) are indicated by black circles. An inner circle marks HindIII restriction sites (H) used in subcloning.
Relationship of pXF-RIV5 to other X. fastidiosa plasmids
The sequence of pXF-RIV5 is almost identical to pXFAS01 from X. fastidiosa subsp. fastidiosa strain M23 [15]. For consistency, nucleotide coordinates of pXF-RIV5 were assigned to correspond to nucleotide coordinates designated for pXFAS01. Alignment of pXF-RIV5 and pXFAS01 revealed polymorphism, all of which are transitions, at only six nucleotide positions over the entire ∼38 kB length. Two transitions are located in intergenic regions at nt 9,556 (between trbN and ORF11) and at nt 36,281 (between ssBP and trbA). The other four transitions are located in the traI gene encoding a conjugative relaxase homologue. Two transitions (nts 24,789 and 24,921) in the traI gene were synonymous substitutions that did not alter predicted protein sequence. The remaining two transitions in the traI gene were nonsynonymous substitutions that altered the codon for amino acid 462 (nt 25,019) from proline (pXFAS01) to serine (pXF-RIV5) or altered the codon for amino acid 386 (nt 25,250) from serine (pXFAS01) to proline (pXF-RIV5). Although both nonsynonymous substitutions are not in highly conserved portions of TraI, potential alteration of function cannot be excluded. Clustering of four out of six polymorphic sites in less than 500 bp of a 38 kB plasmid raises the possibility that all substitutions in traI were introduced by a single recombination event between pXF-RIV5 or pXFAS01 and a closely related plasmid.
The plasmid (pXF51) of X. fastidiosa subsp. pauca strain 9a5c encodes a partial trb module [22] which is 96% identical at the nucleotide level over almost 9 kB of pXF-RIV5 and pXFAS01, spanning trbE through trbN (nts 481 – nts 9401). Strain 9a5c also has an extensive cluster of trb genes on the chromosome [22] sharing 97% nucleotide sequence identity with ∼10 kB of pXF-RIV5 and pXFAS01 (nts 37612 – nts 9417). A small region of pXF-RIV5 and pXFAS01 (140 bp; nts 9276 – nts 9414) shares 89% nucleotide sequence identity to 25 kB IncP-1 plasmids (pXF-RIV11, pXF-RIV16, pXF-RIV19, and pXF-RIV25) from mulberry-infecting strains of X. fastidiosa [18]. In pXF-RIV5 and pXFAS01, the homologous region is within the trbN gene; however, in the IncP-1 plasmids, the complete trbN gene is not present.
DNA replication elements and T4SS components of pXF-RIV5 have distinct evolutionary histories
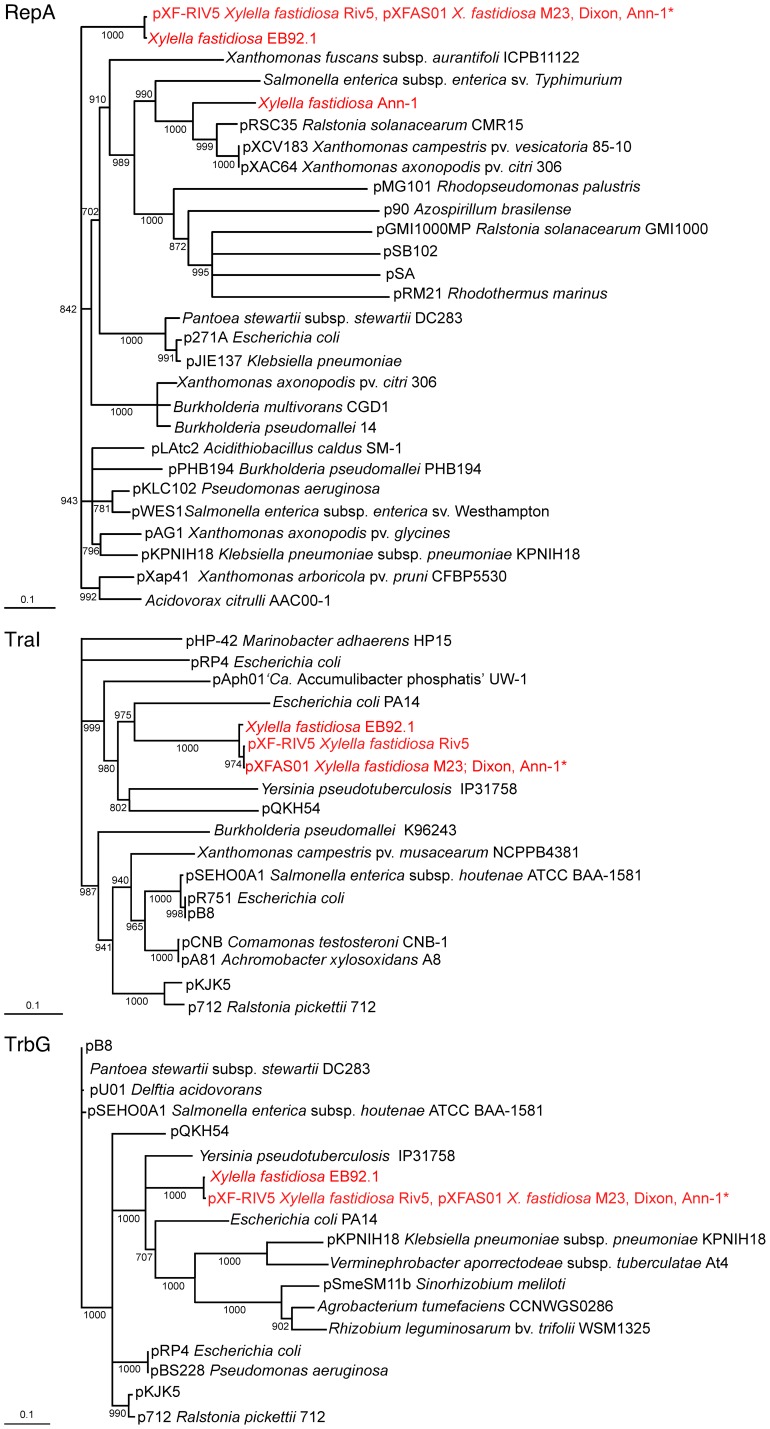
Most proteins encoded by pXF-RIV5 and pXFAS01 are homologues of proteins encoded by numerous bacterial taxa. As shown in Figure 2, neighbor-joining phylogenetic trees were constructed for representative proteins (TraI and TrbG) from the two T4SS modules and for the replication protein RepA. The trees for TraI and TrbG have similar but not identical topology. The most closely related homologues (excluding those from X. fastidiosa) for both TraI and TrbG are from Yersinia pseudotuberculosis IP31758 and E. coli PA14. Because Tra and Trb proteins must work together to form a functional T4SS, it is not surprising that both T4SS gene clusters have similar phylogeny.
Figure 2. Phylogeny of RepA, TraI, and TrbG proteins encoded by pXF-RIV5.
Neighbor-joining trees (1000 bootstrap replicates) presented are based on a multiple alignment of amino acid sequences. Bootstrap support values are shown for nodes with values >70%; nodes bearing ≤70% bootstrap support were collapsed to polytomies. Scale bar at lower left of each tree indicates a genetic distance of 0.1. Taxa names beginning with “p” are known to be encoded by plasmids, with plasmid designation preceding bacterial host nomenclature; replicon (plasmid versus chromosome) of other taxa is not specified. Taxa in red are encoded by strains of Xylella fastidiosa; multiple taxa assigned to a single branch are 100% identical. X. fastidiosa Ann-1 sequences identical to subspecies multiplex homologues are designated with an asterisk (Ann-1*) and may be derived from sequences contaminating the available Ann-1 strain genome sequence. Protein ID numbers are shown in Table S1.
The phylogenetic history inferred for RepA is quite different from that of the T4SS (Figure 2). Most RepA homologues from X. fastidiosa constituted a clade distinct from RepA homologues encoded by all other taxa identified in BLAST P searches. RepA from X. fastidiosa strains Dixon, EB92.1 and Ann-1* were either identical, or nearly identical to that encoded by pXF-RIV5 and pXFAS01. Dixon and Riv5 are subspecies multiplex strains whereas M23 (host of pXFAS01) and EB92.1 are strains of subspecies fastidiosa. It is noted that two distinct RepA sequences are associated with the Ann-1 genome. One RepA sequence from Ann-1 (designated Ann-1*) is identical to that of pXF-RIV5, pXFAS01, and strain Dixon. The second RepA sequence from Ann-1 (designated Ann-1) shared only 53.2% amino acid sequence identity with RepA of pXF-RIV5 and pXFAS01 and clustered in a different clade with RepA from pRSC35 of Ralstonia solanacearum CMR15 as the most closely related homologue identified. As the Ann-1 strain genome sequence is known to be contaminated with a subspecies multiplex genome sequence [17], the simplest interpretation of these results is that RepA Ann-1* represents the multiplex contaminant and that RepA Ann-1 is the divergent homologue resident in the “true” Ann-1 genome of subspecies sandyi. Confirmation of this interpretation will require sequencing of genomes of additional strains of subspecies sandyi.
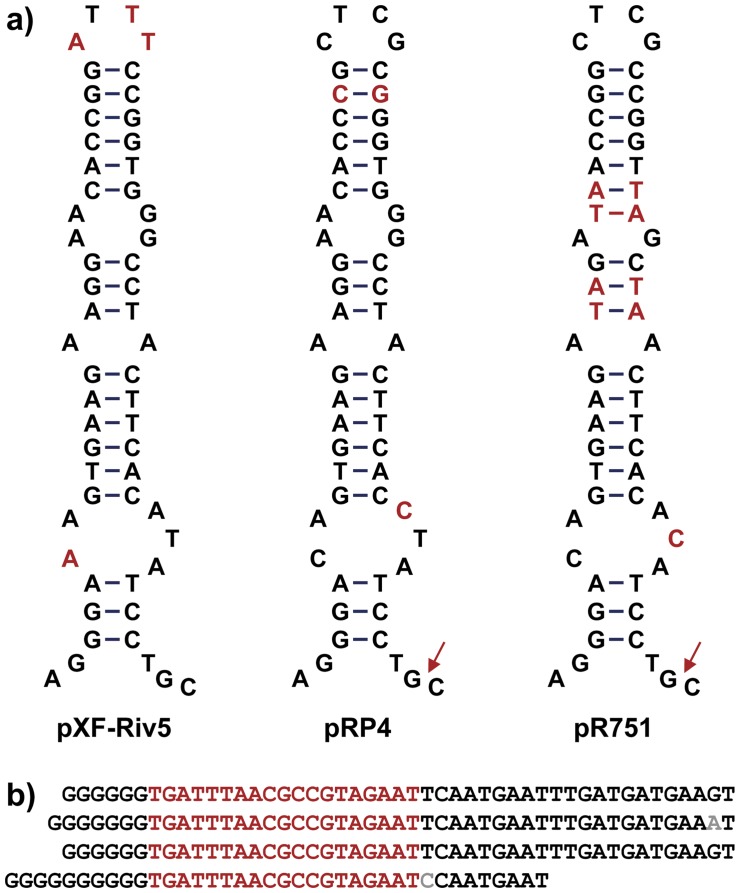
The presumptive origin of transfer (oriT) of pXF-RIV5 and pXFAS01 (nts 26869–26921) are similar to the experimentally verified oriT [21] from two E. coli plasmids (pRP4 and pR751). The stem-loop inverted repeat structure that forms the TraK binding site of pRP4 oriT shares 89% nucleotide sequence identity (56 of 63 nts identical) with the oriT homologue of pXF-RIV5 and pXFAS01 (Figure 3A). Two nucleotide substitutions in the oriT sequences between the X. fastidiosa plasmids and pRP4 are compensatory changes in the stem, preserving secondary structure (Figure 3A); the other five substitutions are in loop regions. The oriT region from pR751 shares less nucleotide sequence identity (79.3%; 50 of 63 bp) with pXF-RIV5 and pXF-AS01 and has one additional stem base pair relative to pXF-RIV5, pXFAS01 and pRP4.
Figure 3. Structure of oriT and oriV.
A) oriT inverted repeats from pXF-RIV5, pRP4, and pR751 are shown as stem-loop structures to emphasize potential base parings. Unique bases are shown in red; bases conserved in at least two structures are shown in black. Red arrows indicate experimentally determined cleavage sites in pRP4 and pR751 [21]. B) Tandem repeats in pXF-RIV5 oriV are aligned and the 19-bp core is shown in red. Residues that vary from the consensus are shown in gray. Nucleotides shown (34333 – 34503) are contiguous.
The presumptive vegetative origin of replication (oriV) of pXF-RIV5 and pXFAS01 (nts 34039–34504) consists of an approximately 300 bp A+T rich region followed by four tandem repeats (Figure 3B). While the combination of an A+T rich region followed by tandem repeats is present in oriV of many plasmids, more common arrangements have a larger number of repeats (16 to 20) with the 19 nt core of the repeat forming a perfect 10 base pair palindrome [23]. The 19 nt core repeat sequence in pXF-RIV5 is a partial palindrome with only 6 base pairs possible.
Discussion
As with many bacteria, X. fastidiosa harbors a variety of plasmids. Several have been characterized, including an IncP-1 plasmid [18], a small rolling-circle replicon [24], and pXF51 [22]. Here, we have described distinct attributes of pXF-RIV5 and pXFAS01, resident in strains representing multiple subspecies of X. fastidiosa. Given the probable conjugative abilities of these plasmids, it is possible that similar plasmids may be found in other as yet uncharacterized strains of X. fastidiosa or even in other bacterial species that share ecological niches with X. fastidiosa.
While many broad host range conjugative plasmids belong to the IncP-I incompatibility group and contain trfA homologues for vegetative replication [2], [25], replication of pXF-RIV5 and pXFAS01 uses a RepA-dependent process. Therefore, pXF-RIV5 and pXFAS01 are not readily assigned to a classic incompatibility group. Nonetheless, tra and trb genetic modules of pXF-RIV5 and pXFAS01 do cluster (Figure 2) with a subgroup typified by E. coli pRP4 and other IncPα group plasmids [2], [4]. It is likely that an ancestral recombination event occurred between an IncPα group plasmid similar to pRP4 and a plasmid with RepA-dependent vegetative replication to create the plasmid backbone (e.g., modules controlling replication and conjugative transfer) found in pXF-RIV5 and pXFAS01.
In addition to backbone genetic modules, many conjugative plasmids contain accessory modules encoding host-beneficial functions [26]. These accessory modules are often located at the ends of the tra and trb modules and/or near transposon or resolvase genes. No accessory modules with identifiable function were encoded by pXF-RIV5 and pXFAS01, nor by any other characterized plasmid of X. fastidiosa. Interestingly, ORFs for five of seven hypothetical proteins encoded by pXF-RIV5 and pXFAS01 are located downstream of both tra and trb modules, and in close proximity to a resolvase homologue (Figure 1). Whether these ORFs of unknown function constitute an accessory module conferring selective advantage to X. fastidiosa remains to be determined.
pXF-RIV5 and pXFAS01 are the only characterized plasmids of X. fastidiosa encoding all known factors (e.g., a complete T4SS) required for transfer of DNA from recipient to donor cells via conjugation. DNA transfer among strains/subspecies of X. fastidiosa has occurred, as evidenced by massive introgression events leading to the origin of mulberry-infecting [27] and citrus/coffee-infecting [28] strains of X. fastidiosa. In these cases, the mechanism of DNA transfer leading to homologous recombination appears to be transformation, as lengths of recombinant regions were generally small, albeit numerous. Indeed, recent evidence suggests that X. fastidiosa is naturally competent for acquisition of foreign DNA with intrinsic transformation efficiency higher than that of many other bacterial species [29]. Thus, the identification of pXF-RIV5 and pXFAS01, bearing all known hallmarks of a conjugative plasmid, suggests that X. fastidiosa, a plant pathogen of significant economic concern, also may transfer large segments of DNA via conjugation. Indeed, the presence of almost identical plasmids in two separate subspecies of X. fastidiosa (pXF-RIV5 in multiplex and pXFAS01 in fastidiosa) implies a recent inter-subspecies translocation event. Subspecies multiplex is relatively diverse and, therefore, likely has been present in the U. S. for a considerable time; subspecies fastidiosa in the U. S. exhibits limited genetic diversity. It has been hypothesized that all strains of subspecies fastidiosa in the U. S. are derived from a single introduction from Central America that occurred circa 1880 [8]. If so, the inter-subspecies plasmid translocation event responsible for host associations of pXF-RIV5 and pXFAS01 occurred more recently than 1880. Collectively, these observations suggest that the introduction of exotic subspecies of X. fastidiosa further complicates disease management, as newly introduced X. fastidiosa subspecies not only may cause disease(s) previously not known to occur in a region, they also provide a wealth of genetic diversity to be shared with endemic subspecies.
Supporting Information
GenBank protein ID numbers for all proteins appearing in phylogenetic trees ( Figure 2 ).
(DOCX)
Acknowledgments
We thank Kunbo Zhang for technical assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer. This article is in the public domain and not copyrightable. It may be freely reprinted with customary crediting of the source.
Funding Statement
This study was supported by United States Department of Agriculture Appropriated Project 5302-22000-008-00D. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dagan T, Artzy-Randrup Y, Martin W (2008) Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proceedings of the National Academy of Sciences of the United States of America 105: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73: 775–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christie PJ (2004) Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta 1694: 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcillan-Barcia MP, Francia MV, de la Cruz F (2009) The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiology Reviews 33: 657–687. [DOI] [PubMed] [Google Scholar]
- 5. Hopkins DL, Purcell AH (2003) Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Disease 86: 1056–1066. [DOI] [PubMed] [Google Scholar]
- 6. Yuan X, Morano L, Bromley R, Spring-pearson S, Stouthamer R, et al. (2010) Multilocus sequence typing of Xylella fastidiosa causing Pierce's disease and oleander leaf scorch in the United States. Phytopathology 100: 601–611. [DOI] [PubMed] [Google Scholar]
- 7. Scally M, Schuenzel EL, Stouthamer R, Nunney L (2005) Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Applied and Environmental Microbiology 71: 8491–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, et al. (2010) Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce's disease of grapevine in the U.S. PLoS ONE. 5: e15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melanson RA, Sanderlin RS, McTaggart AR, Ham JH (2012) A systematic study reveals that Xylella fastidiosa strains from pecan are part of X. fastidiosa subsp.multiplex . Plant Disease 96: 1123–1134. [DOI] [PubMed] [Google Scholar]
- 10. Davis MJ, French WJ, Schaad NW (1981) Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Current Microbiology 6: 309–314. [Google Scholar]
- 11. Chen J, Groves R, Civerolo EL, Viveros M, Freeman M, et al. (2005) Two Xylella fastidiosa genotypes associated with almond leaf scorch disease on the same location in California. Phytopathology 95: 708–714. [DOI] [PubMed] [Google Scholar]
- 12. Almeida RPP, Nascimento FE, Chau J, Prado SS, Tsai CW, et al. (2008) Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Applied and Environmental Microbiology 74: 3690–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simpson AJG, Relnach F, Arruda P, Abreu FA, Acencio M, et al. (2000) The genome sequence of the plant pathogen Xylella fastidiosa: The Xylella fastidiosa consortium of the organization for nucleotide sequencing and analysis, Sao Paulo, Brazil. Nature 406: 151–157. [DOI] [PubMed] [Google Scholar]
- 14. Van Sluys MA, De Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, et al. (2003) Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa . Journal of Bacteriology 185: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Xie G, Han S, Chertkov O, Sims D, et al. (2010) Whole genome sequences of two Xylella fastidiosa strains (M12 and M23) causing almond leaf scorch disease in California. Journal of Bacteriology 192: 4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang S, Flores-Cruz Z, Kumar D, Chakrabarty P, Hopkins DL, et al. (2011) The Xylella fastidiosa biocontrol strain EB92-1 is very similar and syntenic to Pierce's disease strains. Journal of Bacteriology 193: 5576–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nunney L, Elfekih S, Stouthamer R (2012) The importance of multilocus sequence typing: cautionary tales from the bacterium Xylella fastidiosa . Phytopathology 102: 456–460. [DOI] [PubMed] [Google Scholar]
- 18. Stenger DC, Lee MW, Rogers EE, Chen J (2010) Plasmids of Xylella fastidiosa mulberry-infecting strains share extensive sequence identity and gene complement with pVEIS01 from the earthworm symbiont Verminephrobacter eiseniae . Physiological and Molecular Plant Pathology 74: 238–245. [Google Scholar]
- 19. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Research 39: D225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pansegrau W, Ziegelin G, Lanka E (1988) The origin of conjugative IncP plasmid transfer: interaction with plasmid-encoded products and the nucleotide sequence at the relaxation site. Biochimica et Biophysica Acta 951: 365–374. [DOI] [PubMed] [Google Scholar]
- 22. Marques MV, da Silva AM, Gomes SL (2001) Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa. Plasmid 45: 184–199. [DOI] [PubMed] [Google Scholar]
- 23. Klockgether J, Reva O, Larbig K, Tummler B (2003) Sequence Analysis of the Mobile Genome Island pKLC102 of Pseudomonas aeruginosa C. Journal of Bacteriology. 186: 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guilhabert MR, Stewart VJ, Kirkpatrick BC (2006) Characterization of putative rolling-circle plasmids from the Gram-negative bacterium Xylella fastidiosa and their use as shuttle vectors. Plasmid 55: 70–80. [DOI] [PubMed] [Google Scholar]
- 25. Stenger DC, Lee MW (2011) Phylogeny of replication initiator protein TrfA reveals a highly divergent clade of incompatibility group P1 plasmids. Appl Environ Microbiol 77: 2522–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van der Auwera GA, Krol JE, Suzuki H, Foster B, Van Houdt R, et al. (2009) Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek 96: 193–204. [DOI] [PubMed] [Google Scholar]
- 27. Nunney L (2011) Homologous recomination and the invasion of a new plant host by the pathogenic bacterium, Xylella fastidiosa . Phytopathology 101: S130. [Google Scholar]
- 28. Nunney L, Yuan X, Bromley RE, Stouthamer R (2012) Detecting genetic introgression: high levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Applied and Environmental Microbiology 78: 4702–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kung SH, Almeida RP (2011) Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 77: 5278–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank protein ID numbers for all proteins appearing in phylogenetic trees ( Figure 2 ).
(DOCX)