Abstract
MicroRNAs (miRNAs) play important roles in modulating the neoplastic process of cancers including head and neck squamous cell carcinoma (HNSCC). A genetic polymorphism (rs2292832, C>T) has been recently identified in the precursor of miR-149; nevertheless its clinicopathological implications remain obscure. In this study, we showed that miR-149 is down-regulated in HNSCC compared to normal mucosa and this is associated with a poorer patient survival. In addition, HNSCC patients with the T/T genotype have more advanced tumors and a worse prognosis. Multivariate analysis indicated that patients carried the T/T genotype have a 2.81-fold (95% CI: 1.58–4.97) increased risk of nodal metastasis and 1.66-fold (95% CI: 1.05–2.60) increased risk of mortality compared to other groups. T/T genotype also predicted the worse prognosis of buccal mucosa carcinoma subset of HNSCC. In vitro analysis indicated that exogenous miR-149 expression reduces the migration of HNSCC cells. Moreover, HNSCC cell subclones carrying the pri-mir-149 sequence containing the T variant show a low processing efficacy when converting the pre-mir-149 to mature miR-149. These findings suggest that miR-149 suppresses tumor cell mobility, and that the pre-mir-149 polymorphism may affect the processing of miR-149, resulting in a change in the abundance of the mature form miRNA, which, in turn, modulates tumor progression and patient survival.
Introduction
Mature microRNAs (miRNAs) are small endogenous non-coding RNAs consisting of 20∼22 nucleotide duplexes. After being transcribed, primary miRNAs (pri-mirs) are converted into precursor miRNA (pre-mirs) by Drosha in the nucleus, and then further processed into mature miRNAs by Dicer in the cytosol. The mature miRNAs complex with RISC and regulate target genes by mRNA cleavage through base pairing, by translational repression via binding to the 3′ untranslated region (3′-UTR) of the mRNA, or by deadenylating the mRNA [1]. MiRNAs are very important in terms of biological functioning and disease pathogenesis because one miRNA can target several genes and one gene can be regulated by several miRNAs. Thus a miRNA may play pluripotent roles in the modulation of a range of pathogenic processes [2].
The contribution of miRNAs to human carcinomas, including those of breast cancer, lung cancer, prostate cancer, stomach cancer and other sites, has been published widely [3]–[9]. Head and neck squamous cell carcinoma (HNSCC) includes SCC of oral cavity, oropharynx, hypopharynx and larynx. This disease is the 6th most common cancer worldwide [10]. The high incidence of HNSCC in South Asia countries is highly associated with the synergistic effects of betel quid (BQ) chewing, tobacco smoking, and other carcinogenic substances [11], [12]. MiRNAs also deregulated the genesis and progression of HNSCC. Up-regulation of the oncogenic miRNAs, miR-184, miR-21 and miR-31, contributes to HNSCC carcinogenesis by targeting tumor suppressor genes [5], [13], [14]. However, miR-200c and miR-375 have been shown to suppress the invasion or stemness properties in HNSCC [15]. Furthermore, down-regulation of miR-205 has been found to be associated with the locoregional recurrence of HNSCC [16]. Preliminary screenings also suggested alterations in other miRNAs occur during neoplastic process of HNSCC [5], [14].
Genetic polymorphisms within a miRNA sequence, a target gene sequence or gene sequences associated with miRNA processing are thought to have a functional impact on how miRNAs are active in tumors [17], [18]. A variant allele of the let-7 miRNA complementary site of the K-ras 3′-UTR has been shown to affect the survival of oral cancer patients [17]. The presence of the homozygous T/T genotype in rs11614913 of pre-mir-196a2 has been shown to reduce the survival of pharyngeal SCC patients [19]. An earlier study of ours also showed that the C variant of rs2910164 in pre-mir-146a is associated among patients with a worse cancer progression [20]. The rs2292832 C/T polymorphism in pre-mir-149 has been reported to be associated with an increased risk of gastric carcinoma among males [21]. However, studies also failed to demonstrate the prediction power of this polymorphism when HNSCC and lung carcinoma are examined [22], 23. The combined genotypic polymorphisms found in four pre-mirs, including a pre-mir-149 polymorphism, have been shown to be associated with an increased risk of HNSCC [22]. However, the predictive role of the rs2292832 polymorphism in pre-mir-149 with respect to tumor progression, and the activity of this polymorphism in the regulation of miR-149 expression remain unclear. miR-149 has been found to be down-regulated in malignancies including renal cell carcinoma, prostate carcinoma and astrocytoma [24]–[26]. Our previous and other’s preliminary studies also demonstrated the down-regulation in oral carcinomas [5], [14]. In this context, the present study’s aim is to investigate the deregulation of miR-149 during head and neck carcinogenesis and to explore the implications of the pre-mir-149 polymorphism with respect to HNSCC susceptibility and progression.
Methods and Materials
Ethics Statement
The study samples were collected after obtaining written informed consent and this study was approved by The Institutional Review Board of National Yang-Ming University Hospital (IRB approval no. 2010A013).
Study Subjects
The study subjects consisted of 122 controls and 273 HNSCC patients. They were recruited from the Mackey Memorial Hospital and the National Yang-Ming University Hospital. The clinicopathological data is presented in Table 1. Among these study subjects, data related to BQ chewing and tobacco smoking are recorded. However, the status of viral infection was not determined and the limitation of this study remained. The control individuals included 62 females and 60 males. They were individuals with diagnosis other than cancer and admitted due to surgery for benign oral lesions, trauma management, pre-prosthetic surgery and removal of impacted teeth. Blood samples were draw pre-operatively from these subjects. Tissue specimens were taken from 70 HNSCC tumors, along with paired non-cancerous matched tissue (NCMT) samples. Among the tissues pairs, 24 pairs were from buccal mucosa, 23 pairs from tongue, 10 pairs from gingiva, 3 pairs from retromolar trigone, 2 pairs from floor of mouth, 2 pairs from lip and 6 pairs from oropharyngeal region. There were also 24 neck lymph node metastatic lesions paired with corresponding primary HNSCC available for analysis. Tissues were stored at −80°C until use. Frozen sections were prepared for microdissection to retrieve pure epithelial cells for analysis [14]. A fraction of each tissue sample was also fixed in formalin and processed to give paraffin sections.
Table 1. Clinicopathological variables of the subjects.
| Variables | HNSCC (n = 273) | Control (n = 122) |
| Age (year) | ||
| Mean ± SD | 53.0±11.0 years | 53.5±13.4 years |
| Range | 27–82 years | 21–85 years |
| Gender (Male : Female) | 251∶22 | 60∶62 |
| Site | ||
| Buccal mucosa | 119 | |
| Tongue | 70 | |
| Gingiva | 45 | |
| Retromolar trigone | 7 | |
| Mouth floor | 7 | |
| Oropharynx | 15 | |
| Hypopharynx | 1 | |
| Lip | 7 | |
| Unspecified | 2 | |
| Tumor size | ||
| T1–T3 | 137 | |
| T4 | 136 | |
| Neck lymph node metastasis | ||
| N (−) | 173 | |
| N (+) | 100 | |
| TNM staging | ||
| Stage I–III | 121 | |
| Stage IV | 152 | |
| Perineural invasion | ||
| (−) | 244 | |
| (+) | 29 | |
| Lymphovascular invasion | ||
| (−) | 246 | |
| (+) | 27 | |
| Follow-up period (Mean ± SD) | 50.4±32.7 months | |
The tumor staging was according to American Joint Committee on Caner (AJCC) TNM staging system. The T represents tumor size and the N represents lymph node metastasis. T1: tumor size less than 2 cm; T2: tumor size between 2 to 4 cm; T3: tumor size larger than 4 cm; T4: tumor invading to adjacent tissues or spaces.
DNA Extraction
DNA was prepared from peripheral leukocytes using a genomic DNA extraction kit QIAamp™ DNA Mini Kit (QIAGEN, Valencia, CA). The extraction procedure followed a previously described protocol [20]. The concentration of purified genomic DNA was in the range 4 ng/µl to 12 ng/µl.
Genotyping
The 248-bp sequence encompassing pre-mir-149 was downloaded from a NCBI blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and was amplified using a forward primer (5′-gtcttcactcccgtgcttgt-3′) and a reverse primer (5′-cccgaaacacccgtaagata-3′). Genotyping was performed by direct sequencing of the amplicons (Table 2). The high-throughput genomic analysis was carried out using the thermal cycle sequencing method and a 96-capillary 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). Restriction fragment length polymorphism (RFLP) assay was also used to confirm the results of some cases when there was questionable sequencing result. The PvuII restriction enzyme was utilized to recognize the CAGCTG sequence located within polymorphism. Analysis of the T/T homozygous allele yields 62-bp and 186-bp bands after digestion with PvuII. However, PvuII is unable to digest the C/C homozygous allele and only the undigested 248-bp fragment is found after PvuII digestion.
Table 2. Primers and amplicons.
| Amplification of pre-mir-149 for genotyping | |
| Amplicon | |
| gtcttcactcccgtgcttgtccgaggagggagggagggacgggggctgtgctggggcagc(c)ggaacaacgcaggtcgccgggccggctgggcgagttggccgggcggggctgaggggtcggcgggggaggctgaggcgcgggggccggtgcgcggccgtgagggggtgtgagaggtggctgacggcgccgaggagcccctcagaacctgcaggtggggggagctcccatatcttacgggtgtttcggg | |
| Primers | |
| Forward primer | 5?-gtcttcactcccgtgcttgt-3? |
| Reverse primer | 5?-cccgaaacacccgtaagata-3? |
| Sequencing primer | 5?-acctctcacaccccctcac-3? |
| Amplification of pri-mir-149 and pre-mir-149 for assaying expression | |
| Amplicon | |
| cctcgatccagcctgcccgaggctcccaggccttcgcccgccttgcgtccagcctgccgggggctcccaggccggcgcccgagctctggctccgtgtcttcactcccgtgcttgtccgaggagggagggagggacgggggctgtgctggggcagc(c)ggaacaacgcaggtcgccgggccggctgggcgagttggccgggcggggctgaggggtcggcgggggaggctgaggcgcgggggccggtgcgcggccgtgagggggtgtgagaggtggct | |
| Primers | |
| Pri-mir-149 forward primer | 5?-gccttgcgtccagcct -3? |
| Pri-mir-149 reverse primer | 5?-cccccgtccctccct-3? |
| Pre-mir-149 forward primer | 5?-ctggctccgtgtcttcact-3? |
| Pre-mir-149 reverse primer | 5?-cccccgtccctccct-3? |
(c): Polymorphism site.
qRT-PCR Analysis for miR-149
A TaqMan miRNA assay kit and supplies were used to quantify the expression of miR-149 and miR-149* according to the manufacturer’s instructions (Applied Biosystems). RNU6B and Let-7a were used as internal controls [14]. Ct was the number of cycles at which the fluorescence signal passed the threshold. −ΔCt was the difference in Ct values between the internal controls and miR-149. −ΔΔCt was the difference in ΔCt values between the experimental settings or samples. 2−ΔΔCt represents the fold change in miR-149 expression.
qRT-PCR for Pri-mir-149 and Pre-mir-149
The expression of pri-mir-149 and pre-mir-149 were also determined by TaqMan qRT-PCR analysis. The primer sequences are described in Table 2. The rs2292832 polymorphism is localized outside the pri-mir-149 and pre-mir-149 amplicons. The TaqMan probes were synthesized by Ambion (Carlsbad, CA) and the specificity of the quantitative analysis was tested using synthetic oligonucleotide mimics of pri-mir-149 and pre-mir-149.
Cell Culture, Transient Expression and Knockdown of Expression
HNSCC cell lines, including Fadu, OECM-1 and SAS, as well as 293FT, were cultured as previously described [14], [27]. Chemically modified miR-149 mimic (Applied Biosystems) was used for transient miR-149 expression. miR-149 inhibitor (Applied Biosystems) was used to knockdown miR-149 expression. The controls for the transient expression and knockdown of miR-149 were also purchased from Applied Biosystems. TransFectin™ Lipid Reagent (BioRad Lab, Hercules, CA) was the transfection reagent.
Transwell Migration Assay
Cells were grown in media containing 0.5% fetal bovine serum (FBS, Biological Industries, Kibbutz Beit Haemek, Israel) on a Transwell system (Corning, Acton, MA) with a membrane pore size of 8 µm. Cell migration was induced by the addition of 10% FBS to the culture medium in lower chamber to form a serum gradient. The cell growth was arrested by treatment with 1 µM hydroxyurea (Sigma-Aldrich, St Louise, MO). The migrated cells on the lower surface were counted by fluorescence microscopy after staining with Hoechst 33258 (Sigma-Aldrich).
In situ Hybridization (ISH)
miRCURY LNA™ miR-149 probe labeled with digoxigenin and all associated reagents were purchased from Exiqon (Vedbaek, Denmark). After re-hydration, the paraffin sections were digested by protease K and fixed with 4% paraformaldehyde. Sections were then pre-hybridized for 4 h, and hybridized with the 10 µM miR-149 probe at 45°C overnight. Slides were washed, and then incubated with anti-digoxigenin antibody at room temperature for 3 h. NBT/BCIP (5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium) solution reagent was used to detect the signal.
Plasmid Construction, Viral Infection and Late-RT-qPCR
For exogenous miR-149 expression, pri-mir-149 (sequence marked by arrows in Table 2) with either the T or C genotypic variant was cloned into a lentivirus vector, pLV-EF1a-GFP, which contains a green fluorescent protein (GFP) tag. An empty vector or a vector carrying the inserts were co-transfected with helper plasmids into 293FT cells to produce lentivirus and then stable cell lines were established by lentiviral infection. Late-RT-qPCR to detect reverse transcription of the lentivirus was carried out to identify the genomic integration of viral sequence as a late event of viral infection [28].
Statistical Analysis
The Hardy–Weinberg equilibrium (HWE) was calculated for controls to exclude sampling bias. The last date of follow-up was either the date of death or the last date the patient was contacted. Overall survival was estimated using the Kaplan–Meier analysis, with comparisons between groups made by the log-rank test. The cutoff value for miR-149 was determined using the Youden index obtained from the receiver operating characteristic (ROC) curve [14]. The relationship between clinicopathological variables and genotyping was analyzed by Fisher’s exact test or chi-square analysis. The difference between experiment groups or sample groups was analyzed with t-test. Logistic regression was performed to adjust the relevant covariates of neck lymph node metastases including tumor size, perineural invasion and lymphovascular invasion to achieve an adjusted odds ratio (OR) and 95% confidence interval (95% CI). Cox proportional hazards regression model was performed to determine potential prognostic factors for survival, the hazard ratio (HR) and 95% CI. Differences between values were considered significant when a two-tailed p was <0.05. Statistical analysis was performed with Prism 5 (Graph-Pad, San Diego, CA) or SPSS 17.0 (SPSS Inc., Chicago, IL) software.
Results
Down-regulation of miR-149 Expression in HNSCC Tissues
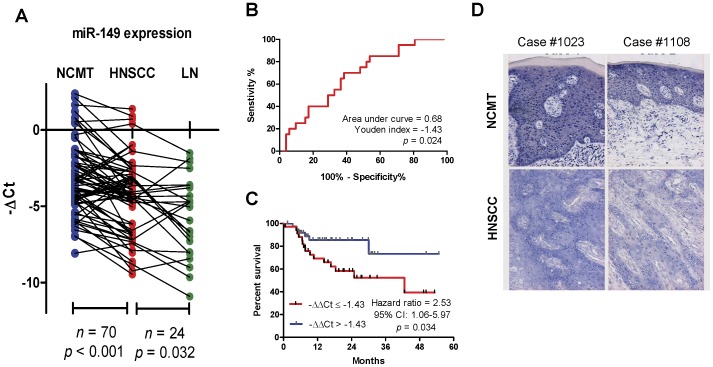
The expression of miR-149 in HNSCC and paired NCMT was determined by qRT-PCR analysis. Paired-t-test indicated a significant down-regulation of miR-149 expression in HNSCC tissue relative to the paired NCMT (p<0.001) (Fig. 1A). Moreover, statistically significant difference in miR-149 expression was noted between primary HNSCC and their paired metastatic lesions (p = 0.032). ROC analysis validated a significant separation of the patient’s survival status when using a –ΔΔCt cutoff value of −1.43 (Fig. 1B). In addition, tumors having a −ΔΔCt ≤−1.43 exhibited a significantly worse prognosis than contrasting group (Fig. 1C). To view the miR-149 transcript in tissue samples, ISH was performed on six NCMT/HNSCC pairs. A representative illustration of two tissue pairs are shown in Fig. 1D and these show strong blue staining in the nucleus and cytosol in epithelial cells of the NCMT. However, the blue staining in the nucleus and cytosol of the tumor cells is less intensive than that in matched NCMT (Fig. 1D). Since the other four tissue pairs exhibited similar staining pattern, down-regulation of miR-149 expression in HNSCC tissue does seem to be confirmed by ISH.
Figure 1. miR-149 expression in HNSCC tissues.
(A) Before-and-after plots of the −ΔCt values from paired NCMT tissue, primary HNSCC and neck lymph node metastasis (LN). Paired t-test. (B) ROC analysis across the –ΔΔCt values from survived patients and dead patients. (C) Kaplan-Meier analysis of overall survival as related to miR-149 expression using −ΔΔCt = −1.43 as a cutoff. (D) ISH. Upper, NCMT, lower HNSCC. Note the miR-149 signals in nucleus and cytosol. NCMT exhibited miR-149 signals that were more intense than HNSCC.
miR-149 Expression is Associated with a Decreased Migration of HNSCC Cells
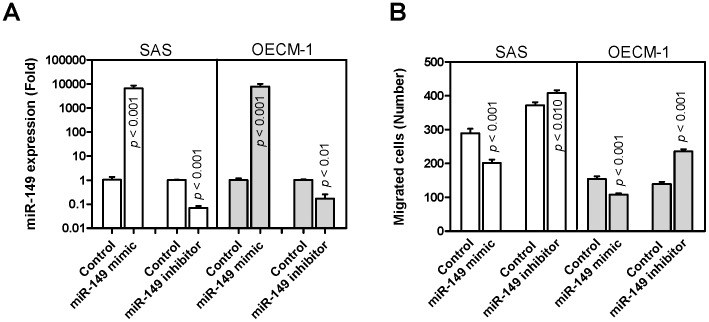
Exogenous expression of miR-149 was carried out by the transfection of miR-149 mimic. Such treatment increased miR-149 expression by >1000-fold in both SAS and OECM-1 cells (Fig. 2A). Knockdown of miR-149 expression by transfection with the miR-149 inhibitor decreased the expression to one-tenth to one-third level of the controls using HNSCC cells. After transient expression or knockdown of expression, the proliferation of HNSCC cells was not affected (not shown). However, transient miR-149 expression decreased and knockdown of miR-149 expression increased the migration of both SAS and OECM-1 cells (Fig. 2B).
Figure 2. miR-149 expression and HNSCC cell migration.
SAS and OECM-1 cells were transfected with miR-149 mimic and miR-149 inhibitor. (A) qRT-PCR analysis. This detected increased miR-149 expression and decreased miR-149 expression following transfecting with miR-149 mimic and miR-149 inhibitor, respectively, relative to the controls. (B) Transwell migration assay. This indicated that transient miR-149 expression decreased cell migration, and knockdown of miR-149 expression increased cell migration. The results are means ± SE from at least triplicate analysis; un-paired t-test.
Genotype and HNSCC Risk
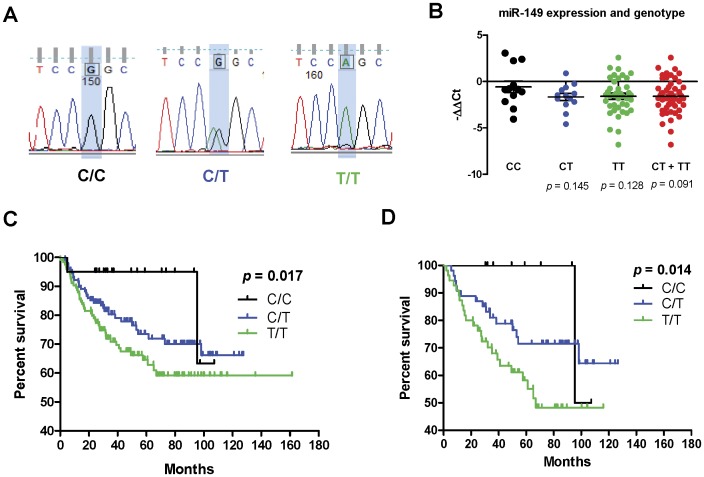
Since miR-149 expression is able to suppress HNSCC cell mobility and this should affect patient survival through regulating invasion or metastasis, the presence of the rs2292832 polymorphism in pre-mir-149 across a non-cancer control cohort and a cancer patient cohort was analyzed. Genotyping by direct sequencing of amplicons was carried out. The analysis is shown in Fig. 3A. There were 15 subjects being additionally validated with RFLP to resolve questionable sequencing data. The genotypic distribution of 122 healthy controls fulfills the HWE (p2+2pq+q2 = 1: chi-square = 0.653, p = 0.722). The genotypic distribution of 273 HNSCC patients also fulfills the HWE (chi-square = 1.510, p = 0.470). These examinations may exclude the sampling bias (Table 3). In univariate analysis, there was significantly higher frequency of the T/T genotype and the T allele in HNSCC patients compared to controls, which suggest a potential association between the pre-mir-149 polymorphism and HNSCC susceptibility (Table 3 and 4). Dominant and recessive models for multivariate analysis were carried out. HNSCC susceptibility was unequivocally shown associated with gender and smoking status in both models. Analysis using dominant model analysis did not reveal an association between T/T genotype and HNSCC susceptibility. Although analysis using recessive model revealed a protective trend of C/C genotype for HNSCC susceptibility, the statistical difference was not significant (Table 4). This polymorphism may not be associated with HNSCC susceptibility.
Figure 3. Analysis of the pre-mir-149 polymorphism.
(A) Sequencing analysis. Representative sequencing results showing the genotypes of pre-mir-149 polymorphisms using blood DNA. (B) Scatter spot charts to show the levels of miR-149 expression in HNSCC patients with various genotypes. The comparison was made against the C/C genotype. Un-paired t-test. (C, D) Kaplan-Meier analysis of overall survival in all HNSCC (C) and HNSCC occurring on buccal mucosa (D).
Table 3. HNSCC susceptibility as related to pre-mir-149 polymorphism (univariate analysis).
| HNSCC susceptibility and pre-mir-149 polymorphism | |||||
| Genotype | C/C | C/T | T/T | p value | |
| Control | 21 | 52 | 49 | 0.012* | |
| HNSCC | 20 | 129 | 124 | ||
| Allelotype | C | T | p value | ||
| Control | 94 (38.5%) | 150 (61.5%) | 0.041# | ||
| HNSCC | 169 (31.0%) | 377 (69.0%) | |||
Chi-square analysis.
Fisher’s exact test.
Table 4. HNSCC susceptibility as related to pre-mir-149 polymorphism (multivariate analysis).
| Logistic regression model for HNSCC susceptibility (dominant model)# | |||
| Variables | OR | 95% CI | p value |
| Genotype: T/T vs. C/C+C/T | 0.91 | 0.51 to 1.61 | 0.748 |
| Age | 1.00 | 0.98 to 1.02 | 0.968 |
| Gender: Male vs. Female | 6.98 | 3.18 to 15.31 | <0.001 |
| BQ chewer vs. Non-BQ chewer | 0.93 | 0.43 to 2.00 | 0.850 |
| Smoker vs. Non-smoker | 2.63 | 1.20 to 5.78 | 0.016 |
| Logistic regression model for HNSCC susceptibility (recessive model) # | |||
| Variables | OR | 95% CI | p value |
| Genotype: T/T+C/T vs. C/C | 2.37 | 0.97 to 5.80 | 0.059 |
| Age | 1.00 | 0.97 to 1.02 | 0.710 |
| Gender: Male vs. Female | 6.50 | 2.96 to 14.26 | <0.001 |
| BQ chewer vs. Non-BQ chewer | 0.84 | 0.39 to 1.83 | 0.669 |
| Smoker vs. Non-smoker | 2.57 | 1.16 to 5.67 | 0.020 |
OR, odds ratio; CI, confidence interval.
All predictor variables are adjusted.
The T/T Genotype is Associated with HNSCC Progression
The relationship between clinicopathological variables and genotypes in HNSCC subjects was further analyzed. The results showed that the proportion of patients with the T/T genotype is significantly higher than that of the other genotypes among the more advanced disease groups, including larger tumors, tumors with neck nodal involvement, and stage IV patients (Table 5). Nodal status is the most important clinical event correlated with pre-mir-149 polymorphism. This polymorphism was not associated with other variables. In HNSCC cases receiving both expression analysis and genotyping, subjects carrying the T/T and/or C/T genotypes tended to have lower miR-149 expression relative to subjects carrying the C/C genotype, however, the difference is not statistically significant (Fig. 3B).
Table 5. Clinicopathological variables as related to pre-mir-149 polymorphism.
| Variables | C/C | C/T | T/T | p value |
| T1–3 | 15 | 72 | 50 | 0.003 |
| T4 | 5 | 57 | 74 | |
| N0 | 15 | 93 | 65 | 0.003 |
| N+ | 5 | 36 | 59 | |
| Stage I–III | 14 | 63 | 44 | 0.006 |
| Stage IV | 6 | 66 | 80 | |
| Perineural invasion (−) | 16 | 114 | 114 | 0.241 |
| Perineural invasion (+) | 4 | 15 | 10 | |
| Lymphovascular invasion (−) | 16 | 116 | 114 | 0.503 |
| Lymphovascular invasion (+) | 4 | 13 | 10 |
Chi-square analysis.
The T/T Genotype is Associated with Nodal Metastasis and a Poor Prognosis Among HNSCC Patients
Kaplan-Meyer analysis indicated that patients carrying T/T genotype had worse prognosis than other genotypes (Fig. 3C). Multivariate modules were used to specify the power of the T/T genotype when predicting nodal metastasis and patient survival by adjusting various cofounding factors. Logistic regression analysis indicated that the T/T genotype was an independent factor when predicting nodal metastasis with an odds ratio of 2.81 (95% CI: 1.58 to 4.97) (Table 6). Cox-proportional hazard model identified that the T/T genotype was a risk factor to predict patient survival with a hazard ratio of 1.66 (95% CI: 1.05 to 2.60) (Table 7). In addition, lymphovascular invasion were found to predict both nodal metastasis and patient’s survival. However, the impact of this polymorphism on survival no longer exists when adjusting for tumor size or nodal status (detailed analysis not shown), implicating that the impact of the polymorphism on survival is mediated by its effects on metastasis (Table 6). As buccal mucosa carcinoma subset of HNSCC is highly associated with endemic BQ chewing in Asians [11], [12], it is also the most frequent HNSCC subset in our samples (n = 119; Table 1). Dissection further identified that T/T genotype was associated with the worst prognosis of buccal mucosa carcinoma (Fig. 3D). However, this polymorphism was not associated with HNSCC localized at non-buccal mucosa regions. Logistic regression specified that T/T genotype predicted the nodal metastasis (odds ratio of 2.73; 95% CI: 1.12 to 6.62) (Table 8). Cox-proportional hazard model identified that the T/T genotype also predicted patient survival (hazard ratio of 2.19; 95% CI: 1.13 to 4.24) (Table 9). Similarly, the influence of the polymorphism on survival is also mediated by its effects on nodal metastasis.
Table 6. Multivariate analysis for the prediction of neck nodal metastasis in HNSCC.
| Logistic regression for prediction of neck nodal metastasis# | ||||
| Variables | Subgroups (n) | OR | 95% CI | p value |
| pre-mir-149 polymorphism | T/T (124) vs. C/C+C/T (149) | 2.81 | 1.58 to 4.97 | <0.001 |
| Age | 0.99 | 0.97 to 1.02 | 0.636 | |
| Gender | Male (251) vs. Female (22) | 6.52 | 1.31 to 32.50 | 0.022 |
| Tumor size | T4 (136) vs. T1–3 (137) | 2.14 | 1.20 to 3.82 | 0.010 |
| BQ chewer | Yes (236) vs. No (37) | 0.92 | 0.33 to 2.55 | 0.867 |
| Smoker | Yes (246) vs. No (27) | 0.60 | 0.22 to 1.62 | 0.310 |
| Perineural invasion | Positive (29) vs. Negative (244) | 1.12 | 0.35 to 3.53 | 0.853 |
| Lymphovascular invasion | Positive (27) vs. Negative (246) | 19.47 | 4.91 to 77.28 | <0.001 |
OR, odds ratio; CI, confidence interval.
All predictor variables are adjusted.
Table 7. Multivariate analysis for survival in HNSCC.
| Cox-proportional hazard model for prediction of patient’s survival# | ||||
| Variables | Subgroups (n) | HR | 95% CI | p value |
| pre-mir-149 polymorphism* | T/T (124) vs. C/C+C/T (149) | 1.66 | 1.05 to 2.60 | 0.030 |
| Age | 1.01 | 0.99 to 1.03 | 0.387 | |
| Gender | Male (251) vs. Female (22) | 0.76 | 0.22 to 2.64 | 0.664 |
| BQ chewer | Yes (236) vs. No (37) | 1.21 | 0.47 to 3.13 | 0.699 |
| Smoker | Yes (246) vs. No (27) | 2.08 | 0.72 to 6.01 | 0.180 |
| Perineural invasion | Positive (29) vs. Negative (244) | 1.99 | 0.94 to 4.24 | 0.075 |
| Lymphovascular invasion | Positive (27) vs. Negative (246) | 2.94 | 1.43 to 6.04 | 0.003 |
HR, hazard ratio; CI, confidence interval.
All predictor variables are adjusted.
The impact of the polymorphism on survival is no longer significant when adjusting for tumor size or nodal status.
Table 8. Multivariate analysis for the prediction of neck nodal metastasis in buccal mucosa carcinoma.
| Logistic regression for prediction of neck nodal metastasis# | ||||
| Variables | Subgroups (n) | OR | 95% CI | p value |
| pre-mir-149 polymorphism | T/T(56) vs. C/C+C/T (63) | 2.73 | 1.12 to 6.62 | 0.027 |
| Age | 0.98 | 0.94 to 1.03 | 0.386 | |
| Gender | Male (114) vs. Female (5) | 4.60 | 0.09 to 242.21 | 0.451 |
| Tumor size | T4 (58) vs. T1–3 (61) | 4.01 | 1.58 to 10.17 | 0.003 |
| BQ chewer | Yes (109) vs. No (10) | 0.35 | 0.02 to 6.79 | 0.485 |
| Smoker | Yes (106) vs. No (13) | 0.30 | 0.04 to 2.25 | 0.242 |
| Perineural invasion | Positive (6) vs. Negative (113) | 0.00 | 0.00 to 0.00 | 0.995 |
| Lymphovascular invasion | Positive (7) vs.Negative (112) | 0.00 | 0.00 to 0.00 | 0.994 |
OR, odds ratio; CI, confidence interval.
All predictor variables are adjusted.
Table 9. Multivariate analysis for survival in buccal mucosa carcinoma.
| Cox-proportional hazard model for prediction of patient’s survival# | ||||
| Variables | Subgroups (n) | HR | 95% CI | p value |
| pre-mir-149 polymorphism* | T/T(56) vs. C/C+C/T (63) | 2.19 | 1.13 to 4.24 | 0.021 |
| Age | 1.01 | 0.98 to 1.05 | 0.516 | |
| Gender | Male (114) vs. Female (5) | 1.95 | 0.12 to 30.84 | 0.638 |
| BQ chewer | Yes (109) vs. No (10) | 0.45 | 0.08 to 2.46 | 0.361 |
| Smoker | Yes (106) vs. No (13) | 2.09 | 0.34 to 12.91 | 0.428 |
| Perineural invasion | Positive (6) vs. Negative (113) | 4.59 | 0.60 to 35.34 | 0.146 |
| Lymphovascular invasion | Positive (7) vs.Negative (112) | 0.48 | 0.05 to 4.74 | 0.529 |
HR, hazard ratio; CI, confidence interval.
All predictor variables are adjusted.
The impact of the polymorphism on survival is no longer significant when adjusting for tumor size or nodal status.
The Expression Level of miR-149 is Associated with the Pre-mir-149 Polymorphism
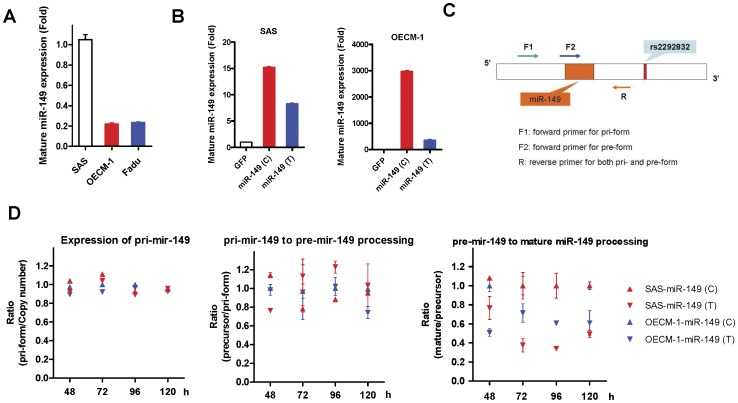
SAS, Fadu and OECM-1 cells carry C/T, T/T and T/T genotype, respectively. miR-149 expression in SAS cells was higher than in Fadu and OECM-1 cells (Fig. 4A). To address if the pre-mir-149 polymorphism may underlie differential miR-149 expression, pre-mir-149 sequences with either the C variant or the T variant were amplified, cloned into a vector to produce lentivirus and then infected into SAS and OECM-1 cells to achieve stable subclones (Table 2), which were designated SAS-GFP, SAS-miR-149(C), SAS-miR-149(T), OECM-1-GFP, OECM-1-miR-149(C), and OECM-1-miR-149(T). Interestingly, SAS-miR-149(C) and OECM-1-miR-149(C) exhibited higher miR-149 expression than SAS-miR-149(T) and OECM-1-miR-149(T) (Fig. 4B). It has been reported that genotypic variation in the precursor region might influence the processing of mature miRNA [18]. Thus an approach was designed to quantify the integrated copy number of the various lentiviruses together with the expression of pri-mir-149, pre-mir-149 and miR-149 in the subclones (Fig. 4C). Quantification was performed on cells at different confluence after seeding for 48 h to 120 h. The ratio of pri-mir-149 to viral integration copy was similar across the subclones at different time point (Fig. 4D, Lt). The ratio of pri-mir-149 to pre-mir-149 seemed to slightly fluctuate at different time points. Whereas, no consistent difference was observed between clones carrying the C variant and T variant (Fig. 4D, middle). However, the ratio of mature miR-149 to pre-mir-149 in SAS-miR-149 (T) and OECM-1-miR-149 (T) subclones were consistently lowered than the (C) subclones (Fig. 4D, Rt). These findings suggest that the T variant in pre-mir-149 may hinder the processing of pre-mir-149 to mature miR-149, which then changes the abundance of miR-149 in HNSCC cells.
Figure 4. Analysis of miR-149 expression in HNSCC subclones.
(A) miR-149 expression in SAS, Fadu and OECM-1 cells carrying the C/T, T/T and T/T phenotypes, respectively. (B) miR-149 expression in the SAS-GFP, SAS-miR-149(C) and SAS-miR-149(T) subclones (Lt); and OECM-1-GFP, OECM-1-miR-149(C) and OECM-1-miR-149(T) subclones (Rt). T variant seemed to be associated with decreased miR-149 expression. Data shown are the means ± SE from triplicate analysis. (C) Schematic diagram to illustrate the analytical strategies used to quantify the expression of the pri-mir-149, pre-mir-149 and miR-149 in subclones. (D) Processing efficiency at different time points from 48 h to 120 h. Lt, processing of pri-mir-149 (normalized against viral integration copy revealed by Late-RT qPCR); Middle, pri-mir-149 to pre-mir-149 processing; Rt, pre-mir-149 to mature miR-149 processing. Both SAS and OECM-1 unequivocally display a decrease in pre-mir-149 to miR-149 processing in the T variant subclones. All ratios (Y axis) were normalized against the OECM-1-miR-149(C) ratio at the same time point. Data shown are the means ± SE from duplicate analysis.
Discussion
Previous studies have shown that miR-149 is down-regulated in oral carcinoma and astrocytoma [5], [14], [24]. This is the first study identify its down-regulation in HNSCC and link this down-regulation to a worse patient survival. qRT-PCR analysis performed on microdissected tissues confirmed the down-regulation of the mature form of miR-149 in tumor cells compared to adjacent non-cancerous epithelial cells; furthermore, the ISH assays also confirmed these findings. Although ISH is a powerful tool that allows the detection of the location and amount of a transcript, it was not used as a standard assay in this study as it also is able to detect nuclear signals of pri-mir-149 [29]. The down-regulation of miR-149 expression from primary HNSCC to metastatic lesion further indicates the roles of miR-149 for HNSCC progression. In this study, we delivered miR-149 mimic duplex that had been modified to ensure the right use of the guide strand of this duplex in RISC. miR-149 expression was found to suppress tumor cell migration. The various phenotypic influences were also confirmed by knockdown experiments. Recent reports have indicated that the passenger strand of the miR-149 duplex, miR-149*, acts as a distinct miRNA that exerted a wide variety of apoptotic regulation effects on in cancer cells [30], [31]; therefore, our approach strategy has been to ascertain the impact of miR-149 on cell migration, and in the process bypass any effect from miR-149*. Since the processing of pre-mir-149 through lentiviral delivery will yield a duplex of both miR-149 and miR-149*, phenotypic assays were not performed on the stable subclones. Up to the present, no gene has been reported to be the target of miR-149 and therefore our preliminary assays were unable to validate any gene as a true miR-149 target in HNSCC cells. Further investigations are required to discover any genes that are negatively regulated by miR-149, especially those that are able to promote cell mobility.
Genetic variants in miRNA sequences that might drive differential functional impacts have attracted attention recently [32]. Polymorphism in pre-mir-196a2 has been reported to increase the risk of colorectal carcinoma, hepatocellular carcinoma and HNSCC [19], [33]. Polymorphism in pre-mir-146a has been shown to be a risk factor for breast carcinoma [34], gastric carcinoma [35] and oral carcinoma [36]. rs2292832 polymorphism in pre-mir-149 has been reported to be correlated with susceptibility to gastrointestinal carcinoma, with C/T and C/C individuals showing a protective effect [21]. However, this polymorphism was not correlated with the breast carcinoma and non-small cell lung carcinoma (NSCLC) susceptibility [23], [37]. The current study shows that the pre-mir-149 polymorphism is not associated with susceptibility of HNSCC in our study cohort. Similar result was also reported in non-Hispanic HNSCC population [22]. Polymorphisms in pre-mir-196a2 or pre-mir-146a have also been associated with poor prognosis of NSCLC and oral carcinoma [18], [20]. This is also the first study to show that T/T HNSCC patients seem to have more advanced tumor progression and a poorer prognosis. Multivariate analysis stratified the T/T genotype as an independent predictor and therefore it is a possible biomarker for HNSCC progression. For HNSCC occurring on oropharynx, the presence of HPV in tumor samples is strongly associated with patient’s survival [38]. Our assay identified the association between at T/T genotype in pre-mir-149 and the poor survival of HNSCC occurring on buccal mucosa. Several molecular markers were found to be prognostic predictors of buccal mucosa carcinoma [12], combined use of miR-149 polymorphism with these markers in future could validate more powerful prognostic prediction for buccal mucosa carcinoma [11]. Just as the transient miR-149 expression ablates the mobility of tumor cells, the induction of miR-149 may have a therapeutic efficacy against HNSCC progression. Since the C allelic frequency was only 0.31 in HNSCC patients, and therefore C/C tumor samples were few, we were unable to confirm that T/T tumor samples had lower level of miR-149 expression in this study. Nevertheless, validation of pre-mir-149 polymorphism as biomarker using blood samples may be useful in a preventive capacity and should also help with tumor prognosis assessment; in the latter case the approach is simpler than complicated tumor tissue analysis.
Polymorphisms occurring in the core region of a miRNA would be expected to influence the translation repression of miRNA due to its altered mRNA binding affinity following sequence change [17]. Polymorphism in the promoter region of a miRNA would be expected to affect the synthesis of miRNA and the changes in expression of the miRNA would then indirectly impairs target gene expression subsequently [39]. However, the functional implications of polymorphisms occurring in either the primary or precursor form of a miRNA remain largely unclear. We postulate that this type of genetic variant may influence the processing of the mature miRNA [18]. Experiments have identified that, relative to the C variant, the T variant in precursor form may slow down the processing from pre-mir-149 to mature miR-149. The result would be a decrease in miR-149 that, in turn, may attenuate the suppression effect of miR-149 on cell motility. This type of change may underlie the effect of miR-149 on the susceptibility and progression of HNSCC. Although the actual mechanism still needs further elucidation, potentially the RNA binding proteins involved in miRNA processing may contribute to this difference. The RNA binding protein Lin28 inhibits the processing of let-7 family by uridylation of the 3′ nucleotides of the precursor, which acts as a obstacle to Dicer processing; the uridylated precursor then undergoes degradation [40], [41]. Moreover, the enzyme, TUT4, which is also responsible for precursor uridylation, is able to recognize the sequence GGAG and in concert with Lin28 interferes with the processing of multiple miRNAs [42]. Three GGAG sequences are present in the precursor region of miR-149. However, since the polymorphism site is not within these sequences, the role of Lin28 in miR-149 processing needs further investigation.
This study demonstrated the down-regulation of miR-149 expression is associated with increased tumor cell mobility in HNSCC. The T/T genotype of the pre-mir-149 polymorphism may contribute to the down-regulation of expression, and this, in turn, seems to determine the risk of HNSCC and prognosis of HNSCC patients.
Acknowledgments
We acknowledged the High-throughput Genome Analysis Core Facility of National Core Facility Program for Biotechnology, Taiwan (NSC-100-2319-B-010-001), for their sequencing services.
Funding Statement
This study was supported by grants from National Yang Ming University Hospital (RD2010-003) and National Science Council (NSC98-2314-B-010-017-MY2; NSC-99-2628-B-195-002-MY3, NSC-99-2628-B-010-012-MY3) in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 2. Kasinski AL, Slack FJ (2011) Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 11: 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, et al. (2008) Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate 68: 1152–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, et al. (2008) MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci U S A 105: 19300–19305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, et al. (2008) Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14: 2588–2592. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, et al.. (2009) microRNA miR-196a-2 and Breast Cancer: A Genetic and Epigenetic Association Study and Functional Analysis. Cancer Res. [DOI] [PMC free article] [PubMed]
- 7.Liu X, Chen Z, Yu J, Xia J, Zhou X (2009) MicroRNA Profiling and Head and Neck Cancer. Comp Funct Genomics: 837514. [DOI] [PMC free article] [PubMed]
- 8. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, et al. (2008) E2F1-Regulated MicroRNAs Impair TGFbeta-Dependent Cell-Cycle Arrest and Apoptosis in Gastric Cancer. Cancer Cell 13: 272–286. [DOI] [PubMed] [Google Scholar]
- 9. Jeong CH, Kim DY, Shin SY, Hong J, Kye SB, et al. (2009) The effect of implant drilling speed on the composition of particle collected during site preparation. J Korean Acad Periodontol 39: 253–259. [Google Scholar]
- 10. Parkin DM, Bray F, Ferlay J, Pisani P (2001) Estimating the world cancer burden: Globocan 2000. Int J Cancer 94: 153–156. [DOI] [PubMed] [Google Scholar]
- 11. Sharan RN, Mehrotra R, Choudhury Y, Asotra K (2012) Association of betel nut with carcinogenesis: revisit with a clinical perspective. PLoS One 7: e42759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu CJ, Chang KW, Chao SY, Kwan PC, Chang SM, et al. (2004) The molecular markers for prognostic evaluation of areca-associated buccal squamous cell carcinoma. J Oral Pathol Med 33: 327–334. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Huang H, Sun L, Yang M, Pan C, et al. (2009) MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res 15: 3998–4008. [DOI] [PubMed] [Google Scholar]
- 14. Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, et al. (2010) miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res 70: 1635–1644. [DOI] [PubMed] [Google Scholar]
- 15. Lo WL, Yu CC, Chiou GY, Chen YW, Huang PI, et al. (2011) MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol 223: 482–495. [DOI] [PubMed] [Google Scholar]
- 16. Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, et al. (2009) Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 174: 736–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, et al. (2009) A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 30: 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu Z, Chen J, Tian T, Zhou X, Gu H, et al. (2008) Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 118: 2600–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, et al. (2010) Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res 16: 3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hung PS, Chang KW, Kao SY, Chu TH, Liu CJ, et al. (2012) Association between the rs2910164 polymorphism in pre-mir-146a and oral carcinoma progression. Oral Oncol 48: 404–408. [DOI] [PubMed] [Google Scholar]
- 21.Mw Z, Mj J, Yx Y, Sc Z, B L, et al.. (2011) Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol Carcinog. [DOI] [PubMed]
- 22. Liu Z, Li G, Wei S, Niu J, El-Naggar AK, et al. (2010) Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116: 4753–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F, et al.. (2011) Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med. [DOI] [PubMed]
- 24. Li D, Chen P, Li XY, Zhang LY, Xiong W, et al. (2011) Grade-specific expression profiles of miRNAs/mRNAs and docking study in human grade I-III astrocytomas. OMICS 15: 673–682. [DOI] [PubMed] [Google Scholar]
- 25. Liu H, Brannon AR, Reddy AR, Alexe G, Seiler MW, et al. (2010) Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell Renal Cell Carcinoma. BMC Syst Biol 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, et al. (2010) Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer 126: 1166–1176. [DOI] [PubMed] [Google Scholar]
- 27. Shieh TM, Lin SC, Liu CJ, Chang SS, Ku TH, et al. (2007) Association of expression aberrances and genetic polymorphisms of lysyl oxidase with areca-associated oral tumorigenesis. Clin Cancer Res 13: 4378–4385. [DOI] [PubMed] [Google Scholar]
- 28. Hung PS, Kao SY, Liu CJ, Tu HF, Wu CH, et al. (2008) Insulin-like growth factor binding protein-5 enhances the migration and differentiation of gingival epithelial cells. J Periodontal Res 43: 673–680. [DOI] [PubMed] [Google Scholar]
- 29. Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, et al. (2010) MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 120: 1298–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin L, Hu WL, Jiang CC, Wang JX, Han CC, et al. (2011) MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci U S A 108: 15840–15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin RJ, Lin YC, Yu AL (2010) miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog 49: 719–727. [DOI] [PubMed] [Google Scholar]
- 32. Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 10: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo J, Jin M, Zhang M, Chen K (2012) A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One 7: e30585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lian H, Wang L, Zhang J (2012) Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs2910164 polymorphism in Europeans. PLoS One 7: e31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kogo R, Mimori K, Tanaka F, Komune S, Mori M (2011) Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res 17: 4277–4284. [DOI] [PubMed] [Google Scholar]
- 36. Xu W, Xu J, Liu S, Chen B, Wang X, et al. (2011) Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis. PLoS One 6: e20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang M, Jin M, Yu Y, Zhang S, Wu Y, et al. (2012) Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 21: 274–280. [DOI] [PubMed] [Google Scholar]
- 38. Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, et al. (2008) Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100: 261–269. [DOI] [PubMed] [Google Scholar]
- 39. Xu Y, Liu L, Liu J, Zhang Y, Zhu J, et al. (2011) A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer 128: 412–417. [DOI] [PubMed] [Google Scholar]
- 40. Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, et al. (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283: 21310–21314. [DOI] [PubMed] [Google Scholar]
- 41. Heo I, Joo C, Cho J, Ha M, Han J, et al. (2008) Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- 42. Heo I, Joo C, Kim YK, Ha M, Yoon MJ, et al. (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]