SUMMARY
The diversity of ubiquitin (Ub)-dependent signaling is attributed to the ability of this small protein to form different types of covalently linked polyUb chains and to the existence of Ub binding proteins that interpret this molecular syntax. We used affinity capture/mass spectrometry to identify ALIX, a component of the ESCRT pathway, that has not been previously reported to possess a Ub binding domain. We report that the V domain of ALIX binds directly and selectively to K63-linked polyUb chains, exhibiting a strong preference for chains composed of more than three Ub. Sequence analysis identified two potential Ub binding sites on a single α-helical surface within the coiled-coil region of the V domain. Mutation of these putative Ub binding sites inhibited polyUb binding to the isolated V domain in vitro and impaired budding of lentiviruses. These data reveal an important role for K63 polyUb binding by ALIX in retroviral release.
INTRODUCTION
Ubiquitin (Ub) is a highly conserved 76 amino acid protein that, when conjugated to cellular substrates, functions in a broad range of signaling pathways (Komander and Rape, 2012). This diversity is achieved in part by Ub’s ability to form polyUb chains in which Ub is conjugated to itself through either its amino terminus or any of its seven internal lysines (Ye and Rape, 2009). This Ub code is read by proteins that contain one ore more Ub binding domains (UBDs) that, individually, bind monoubiquitin with low affinity (300–1000 μM) (Husnjak and Dikic, 2012) and when arranged in tandem within a single polypeptide chain, impart linkage selective binding via an avidity based mechanism (Sims and Cohen, 2009).
In this study we used K63 polyUb affinity chromatography and tandem mass spectrometry (LC-MS/MS) to identify ALIX, a component of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery (Henne et al., 2011) as a K63-selective polyUb binding protein. ALIX comprises three distinct structural elements (Figure 1); an N-terminal Bro1 domain that binds CHMP4 subunits of the ESCRT-III complex (McCullough et al., 2008), a central V domain that binds YPXnL motifs found in viral Gag proteins and some MVB cargoes (Dores et al., 2012; Fisher et al., 2007; Lee et al., 2007), and a C-terminal proline-rich region (PRR) that can fold back onto the upstream domains and autoinhibit ligand binding (Zhai et al., 2011; Zhou et al., 2009). ALIX has been shown to function in retrovirus budding (Strack et al., 2003), cytokinesis (Morita et al., 2007), sorting of membrane proteins into MVBs (Dores et al., 2012), and in the generation of MVB-derived exosomal vesicles (Baietti et al., 2012). Importantly, these ESCRT-dependent pathways are regulated by Ub (Mukai et al., 2008; Pohl and Jentsch, 2008; Shields and Piper, 2011), and there is accumulating evidence that K63-linked chains are the relevant Ub species (Erpapazoglou et al., 2012; Ren and Hurley, 2010; Strack et al., 2002; Weiss et al., 2010).
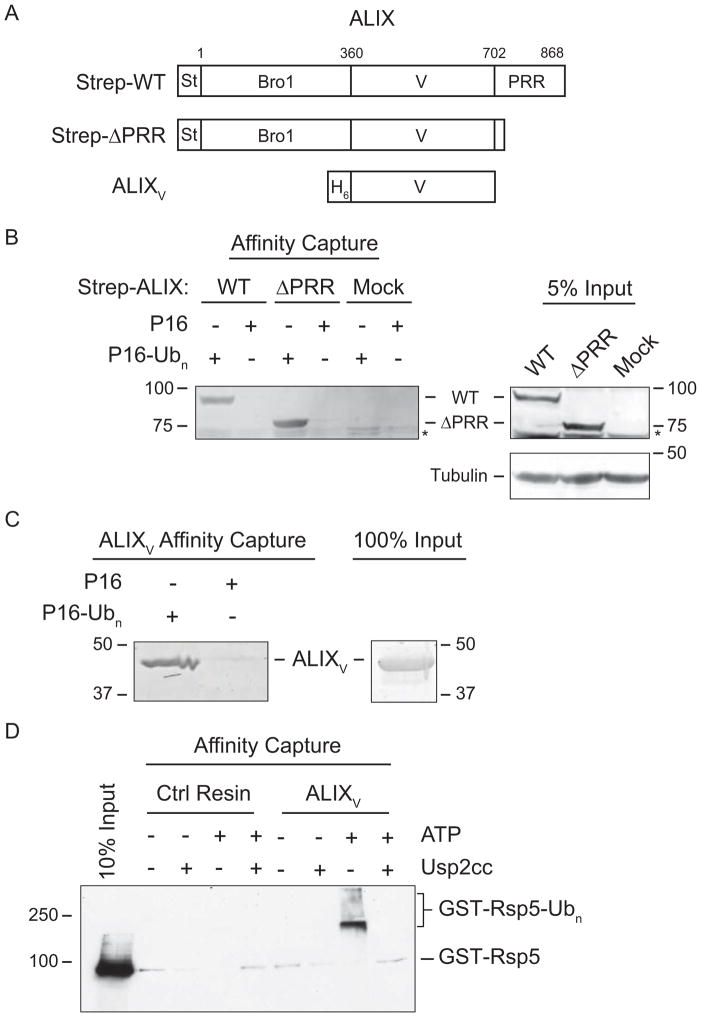
Figure 1. ALIX Binds K63-linked Chains via the V Domain.
(A) ALIX domain organization and constructs. Bro1 domain, V domain, proline-rich region (PRR), StrepTactin tag (St), and His6 tag (H6) are shown.(B) Immobilized P16-Ubn captures Strep-ALIX constructs expressed in mammalian cells. Left, analysis of ALIX proteins bound to immobilized P16-Ubn or P16 from HEK293T cell lysates expressing Strep-WT or Strep-ΔPRR ALIX. Right, 5% input. Proteins were separated by SDS-PAGE and analyzed by immunoblot with anti-StrepTactin or anti-tubulin. Non-specific background band denoted by (*). (C) ALIXV binds directly to immobilized P16-Ubn. Left, analysis of recombinantly expressed and purified ALIXV bound to immobilized P16-Ubn or P16. Right, 100% protein input. Proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. (D) Autoubiquitylated GST-Rsp5 binds to immobilized recombinant ALIXV. GST-Rsp5 was autoubiquitylated (+ATP) and GST-Rsp5-Ubn was deubiquitylated by Usp2cc as indicated. Bound proteins were separated by SDS-PAGE and analyzed by immunoblot with anti-GST. See Figure S1 and Table S1 for details.
There are a number of indications that Ub is involved in retrovirus budding, although its precise role(s) is not yet fully understood (Martin-Serrano, 2007). For example, retroviruses can use Ub E3 ligases of the NEDD4 family to link into the ESCRT pathway (Chung et al., 2008; Martin-Serrano et al., 2005; Rauch and Martin-Serrano, 2011; Usami et al., 2008), and non-budding viral mutants can be rescued by fusing Ub directly to the C-terminal end of the viral Gag protein (Joshi et al., 2008; Patnaik et al., 2000). Freed and coworkers have reported that immobilized monoUb can capture ALIX from crude cell extracts (Joshi et al., 2008). Here, we show that the V domain of ALIX binds directly and preferentially to K63-linked polyUb chains via clusters of three amino acids (termed “triads”), within the ALIX V domain that are similar to triads that contact linear diUb in UBAN (ubiquitin binding in ABIN and NEMO) (Rahighi et al., 2009). Mutation of the ALIX triads impaired retroviral release, indicating that direct Ub binding by ALIX is required for viral budding.
RESULTS
The ALIX V Domain Specifically Interacts with K63 – linked Tetraubiquitin
To identify K63 chain-specific polyUb binding proteins, we used a K63 linked polyubiquitylated protein, P16-Ubn (Figure S1). LC-MS/MS analysis of HEK293 cell lysates identified 87 proteins (Table S1) captured with P16-Ubn, of which 15 were selected as candidate polyUb binding proteins based on their identification in at least two out of three replicates and on their lack of binding to enzymatically deubiquitylated P16-Ubn and non-ubiquitylated P16. This list contains 10 proteins previously shown to interact with Ub, including two polyUb binding receptors associated with the 26S proteasome (RAD23 and RPN10), five proteins associated with ESCRT or endosomal trafficking machinery (hVPS37b, HRS, TSG101, TOM1, and EPS15) as well as a deubiquitinating enzyme (UCHL1) and an autophagy receptor (p62) (Husnjak and Dikic, 2012). The identification of known Ub binding proteins indicated that the P16-Ubn affinity capture procedure worked well, and suggested that the remaining proteins might also bind Ub. The remaining known Ub binding protein, CCDC50, binds selectively to K63 chains via a coiled-coil domain similar to UBAN (Bohgaki et al., 2008). As both ALIX and HD-PTP also contain coiled-coil elements, we considered the possibility that they might also bind polyUb in a similar fashion. This idea was particularly attractive because ALIX was by far the most abundant protein identified in our analysis.
To verify ALIX binding to K63 polyUb, we tested whether immobilized P16-Ubn could capture Strep-tagged ALIX from lysates of HEK293T cells that expressed either the wild type protein (Strep-WT) or a construct that lacked the autoinhibitory C-terminal proline-rich region (Strep-ΔPRR) (Figure 1A). As shown in Figure 1B, Strep-WT and Strep-ΔPRR ALIX both bound P16-Ubn but not P16, confirming our proteomic identification of ALIX (Table S1) as a Ub binding protein and indicating that the C-terminal PRR is dispensable for binding.
As shown in Figure 1C, we found that pure recombinant His6-V domain (ALIXV, residues 360–702; Figure 1C right panel) bound immobilized P16-Ubn but not P16, indicating that the V domain is sufficient for binding polyUb and that the interaction is direct. This conclusion was confirmed in a reciprocal pull-down experiment in which immobilized ALIXV bound autoubiquitylated GST-Rsp5 (GST-Rsp5-Ubn; Figure S1F, G) but not enzymatically deubiquitylated GST-Rsp5-Ubn or non-autoubiquitylated GST-Rsp5 (Figure 1D).
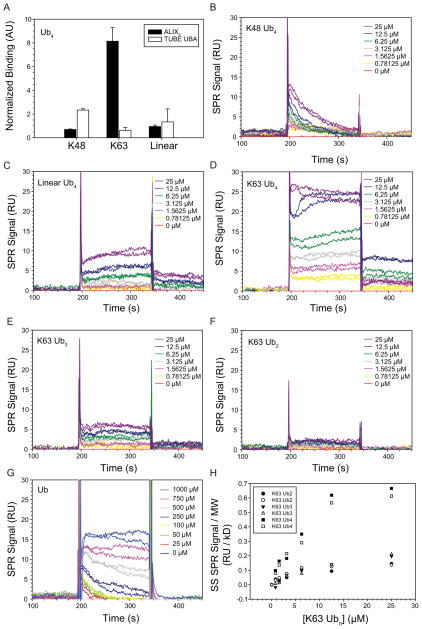
To examine polyUb linkage specificity, we used ALIXV to capture unanchored Ub homopolymers from a mixture of pure K48, K63 and linear Ub4. Recombinant biotin-tagged ALIXV (Figure S2A) was incubated with the Ub4 mixture and bound Ub molecules were analyzed by quantitative MS using synthetic heavy isotope labeled (AQUA) standards. Normalization of the raw binding data to input and subtraction of signal attributed to resin alone (Table S2) revealed that ALIXV bound approximately 8-fold more K63 Ub4 than K48 or linear Ub4 (Figure 2A). By contrast, a control tandem Ub binding domain (TUBE UBA) bound all three types of Ub4 linkages to a roughly equal extent (Figure 2A). Thus, ALIXV exhibits strong linkage specificity for K63 over K48 and linear Ub4 under conditions where all three forms can compete for binding.
Figure 2. ALIX V Domain Preferentially Recognizes K63-linked Tetraubiquitin.
(A) ALIXV selectively binds unanchored K63 Ub4 polyUb chains. Ub AQUA pulldown analysis of biotin-tagged ALIXV (Figure S2A) (black bars) or TUBE UBA (white bars, control) from a mixture of Ub4 (K48, K63, and linear). Normalized binding data are shown (data calculations in Table S2). Measurements represent mean ± s.d. (n=3). (B–G) Interaction of immobilized ALIXV with K48 Ub4 (B), Linear Ub4 (C), K63 Ub4 (D), K63 Ub3 (E), K63 Ub2 (F), and Ub (G). Sensorgrams are shown for ALIXV immobilized at density of 150 RU. Ub chains (0.78–25 μM; twofold serial dilutions) and Ub (25–1000 μM; twofold serial dilutions) were injected in duplicate. (H) ALIXV prefers to bind to K63 polyubiqutin chains containing more than 3 Ub moieties. Equilibrium biosensor binding data from D–F normalized to molecular weight of Ub4, Ub3, and Ub2. See Figure S2 and Table S2 for details.
We used biosensor analyses to measure the interaction of immobilized ALIXV with pure unanchored K48, linear, or K63 Ub4 homopolymers. To minimize avidity effects on the biosensor chip (Zimmerman and Minton, 1993), biotin-tagged ALIXV was immobilized at surface densities of 150 RU (Figure 2B–D) or 220 RU (Figure S2B–D). These experiments again indicate that ALIXV binds preferentially to K63 Ub chains because the equilibrium binding responses were invariably higher for K63 Ub4 (Figure 2D and Figure S2D) than for equivalent concentrations of either K48 Ub4 (Figure 2B and Figure S2B) or linear Ub4 (Figure 2C and Figure S2C).
Biosensor analyses were also used to examine the length dependence of ALIXV binding to K63-linked chains of 1–4 Ub molecules. As shown in Figures 2D–G, ALIXV bound all of the Ub-containing species, including monoUb, but bound much more strongly to tetraubiquitin than to the shorter chains, even when the response signals were corrected for the molecular weight differences (Figure 2H). We conclude that ALIXV has a preference for binding to chains that contain at least four K63-linked Ub molecules.
ALIXV Contains a UBAN-like Coiled-Coil Region Required for Polyubiquitin Binding
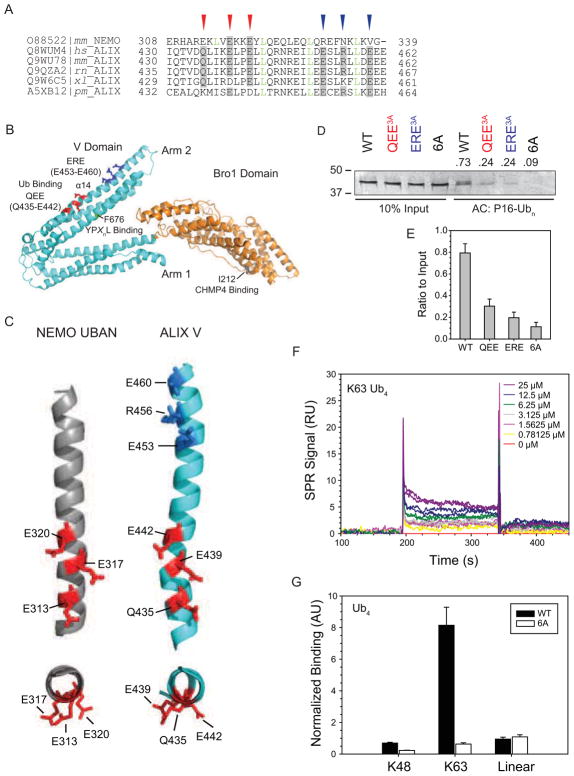
We compared the ALIXV sequence with that of the Ub-binding UBAN domain of NEMO, the only structurally defined coiled-coil UBD (Rahighi et al., 2009) (Figure 3A). UBAN is composed of a parallel dimeric coiled-coil that binds preferentially to linear polyUb chains (Rahighi et al., 2009). Linear chains share a similar open conformation and inter-Ub distance with K63 (Komander et al., 2009). Alignment of the NEMO and ALIX sequences identified several features, including a precise heptad repeat (green letters) and a triad of Glu residues (E313, E317, and E320; red arrowheads) that are conserved between NEMO and metazoan ALIX homologs but not apparently, with yeast Bro1p. In NEMO, the Glu triad makes extensive polar contacts with an atypical surface on the proximal Ub, while the region between E289 and E313 engages the canonical I44 hydrophobic patch of the distal Ub (Rahighi et al., 2009). Among UBAN family members, the presence of a Glu triad in this position is highly conserved, but the motif is partially degenerate in that Gln is occasionally substituted for Glu. In ALIX, the corresponding QEE triad (Q435, E439, E442) is also degenerate, in that Glu can be replaced by Gln or Asp. The side chains of the Glu/Gln triads in NEMO and ALIX are displayed in similar orientations along the α14 strand in Arm 2 of ALIXV (Figures 3B,C).
Figure 3. UBAN-like Ub Binding Triads Mediate V Domain K63-linked Polyubiquitin Binding.
(A) ALIXV shares a conserved sequence element with UBAN. Alignment of amino acid sequence of NEMO UBAN from Mus musculus(mm) and of ALIXV from Homo sapiens (hs), Mus musculus (mm), Rattus norvegicus (rn), Xenopus laevis (xl), and Penaeus monodon (pm). UniProtKB accession numbers are indicated. Conserved (red arrow heads) and proposed (blue arrow heads) Ub binding triads are indicated. Leu residues corresponding to the heptad repeat are indicated in green letters. (B) Structural organization of human ALIXBro1-V (PDB code 2OEV). Bro1 domain (orange), V domain (cyan), and sites of CHMP4 binding (I212, dark grey), YPXnL motif binding (F676, green), and Ub binding triads QEE (red) and ERE (blue), corresponding to the identification and color code in panel A are indicated. (C) The disposition of the first ALIXV triad is similar to that of UBAN. Structural view of the conserved regions of NEMO UBAN (grey, PDB code 3F89) from Mus musculus and ALIXV (cyan, PDB code 2OEX) from Homo sapiens. Side (top panels) and end-on (bottom panels) views of α-helices containing conserved (red) and putative (blue) triad residues. Color code corresponds to panel A. (D) ALIXV triad mutants are impaired in binding to immobilized P16-Ubn. Bound proteins were eluted in SDS sample buffer, separated by SDS-PAGE and visualized by Coomassie blue staining. (E) Quantification of the levels of ALIXV captured by P16-Ubn in D as a ratio to the levels of each respective input. Measurements represent mean ± s.d. (n=3) (F) Impaired binding of immobilized ALIXV 6A mutant to K63 Ub4. Biosensor responses are shown for ALIXV 6A immobilized at density of 150 RU. Ub chains (0.78–25 μM; twofold serial dilutions) were injected in duplicate. (G) Impaired binding of ALIXV 6A to unanchored Ub4 polyUb chains of the indicated linkages. Ub AQUA pulldown analysis was performed as in Figure 2A (data calculations in Table S2) for ALIXV WT (black bars) or 6A (white bars). Measurements represent mean ± s.d. (n=3). Experimental data used for ALIXV WT (black bars) was from Figure 2A for comparison. See Figure S3 for details.
Linkage-specific polyUb binding is commonly achieved by precise spatial arrangement of multiple UBDs, often in tandem (Sims and Cohen, 2009), and we noted the presence of an additional Glu-rich triad (designated ERE; E453, R456, E460; Figures 3A–C, blue arrows/residues). To test whether these Glu-rich clusters contribute to Ub recognition, we tested P16-Ubn binding to pure ALIXV harboring Ala substitution mutations of QEE (QEE3A), ERE (ERE3A), or both (6A). Mutations in either triad substantially reduced binding of purified ALIXV to immobilized P16-Ubn (Figure 3D,E) and mutation of both triads reduced binding to near-background levels (Figure 3D). Similarly, binding of K63 Ub4 to immobilized ALIXV was also substantially reduced by the 6A mutation as assessed in a biosensor binding analysis (compare Figure 3F with Figure 2D) or by Ub4 affinity capture/AQUA under conditions of competitive equilibrium binding (Figure 3G). The impaired Ub4 binding resulting from these Ala substitutions was not due to gross disruption of the helical structure or impaired thermal stability of the V domain because the circular dichroism spectra of the mutant proteins were similar to wildtype ALIXV (Figure S3A). Furthermore, Ala substitution of residues between the two triads (designated RE; R446, E449) also substantially reduced binding of purified ALIXV to immobilized P16-Ubn (Figure S3B) while not disrupting the helical structure or thermal stability of the V domain (Figure S3A). These data lead us to propose that a repeating Glu-rich motif, arranged along the α14 helix between amino acids Q435 and E460 of ALIX, contributes to linkage-specific polyUb binding.
Ub Binding Mutations Inhibit ALIX Functions in Retrovirus Budding
HIV-1 budding is promoted primarily through an interaction between the p6Gag PTAP late assembly domain and the cellular TSG101/ESCRT-I complex (Martin-Serrano and Neil, 2011; Usami et al., 2009). Viral infectivity and virion release from 293T cells are therefore severely impaired by a p6Gag PTAP motif mutation in the R9 proviral expression construct (termed HIV-1ΔPTAP). However, ALIX binds a YPXnL late assembly domain within p6Gag, and can substantially rescue the budding defects in the HIV-1ΔPTAP construct when overexpressed (Fisher et al., 2007; Usami et al., 2007). ALIX overexpression therefore increases HIV-1ΔPTAP titers (Figure 4A, panel 1) and virus release, as reflected by an increase in the release of virion-associated viral MA and CA proteins into the culture supernatant (panel 2). This system therefore provides a convenient assay for ALIX function in HIV-1 budding, and was used to test whether the ALIX 6A mutation that blocked Ub binding in vitro altered the ability of ALIX to stimulate HIV-1 budding.
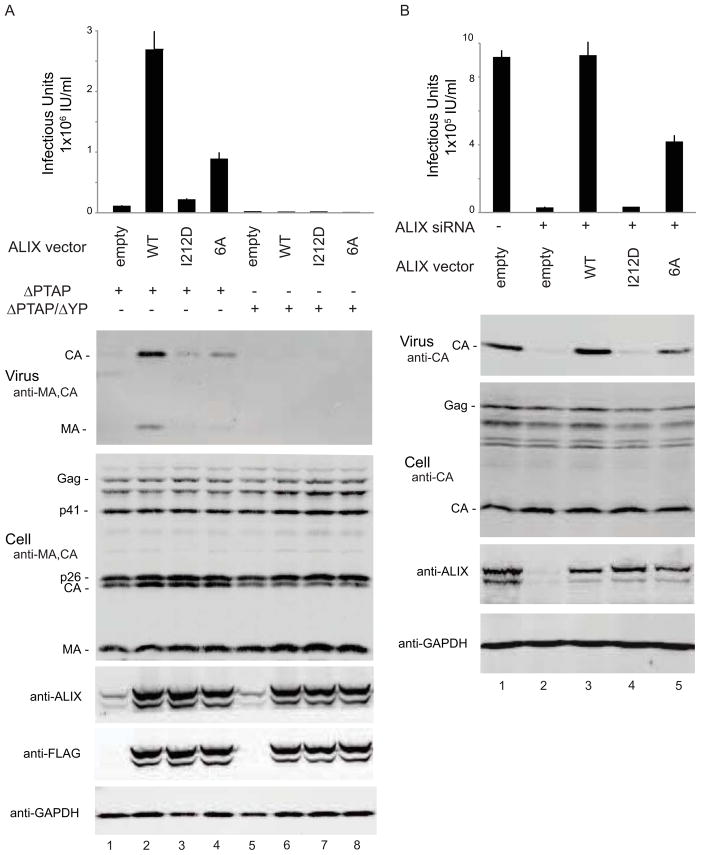
Figure 4. ALIX Ub Binding Mutant is Defective in HIV-1ΔPTAP and EIAV Infectivity and Release.
(A) ALIX6A Ub binding mutant is defective in rescue of HIV-1ΔPTAP virus release and infectivity. Viral infectivity (panel 1) and immunoblots showing virion release (panel 2, “Virus”, anti-CA and anti-MA), cellular Gag protein levels (panel 3, “Cell”, anti-CA and anti-MA), exogenous and endogenous ALIX expression levels (panels 4 and 5, anti-ALIX and anti-FLAG) and cellular GAPDH levels (panel 6, anti-GAPDH). Measurements represent mean ± s.d. (n=6). Lanes 1 to 4 show the proviral HIVΔPTAP expression construct co-transfected with an empty vector (control, lane 1), or with vectors expressing wild type FLAG-ALIX (WT, control, lane 2), FLAG-ALIXI212D (I121D, CHMP4 binding control, lane 3), or FLAG-ALIX6A (6A, Ub binding mutant, lane 4). Lanes 5 to 8 show the same set of vectors co-transfected with the proviral HIVΔPTAP/ΔYP expression construct, which carries an inactivating mutation in the ALIX binding site of p6Gag. (B) ALIX6A Ub binding mutant is defective in EIAV virus release and infectivity. EIAV vector titers (panel 1) and immunoblots showing virion release (panel 2, “Virus”, anti-CA), cellular Gag protein levels (panel 3, “Cell”, anti-CA), endogenous and exogenous ALIX expression levels (panels 4, anti-ALIX) and cellular GAPDH levels (panel 5, anti-GAPDH). Measurements represent mean ± s.d. (n=6). Lane 1 shows EIAV release from control 293T cells expressing endogenous ALIX. Lanes 2 to 5 show EIAV produced in cells depleted of endogenous ALIX and co-transfected with an empty vector (empty, control, lane 2), or with vectors expressing siRNA-resistant wild type ALIX (WT, control, lane 3), ALIXI212D (I212D, CHMP4 binding control, lane 4), or ALIX6A (6A, Ub binding mutant, lane 5). See Figure S4 for details.
Control experiments demonstrated that ALIX stimulation of virion release and infectivity were lost upon mutation of the ALIX binding site for the downstream CHMP4/ESCRT-III proteins (ALIXI212D, Figure 4A) or upon mutation of the ALIX binding site within p6Gag (HIV-1ΔPTAP, ΔYP). In the actual experiment, the ALIX 6A mutant stimulated virus release and infectivity to some extent (Figure 4A), but was less effective than wild type ALIX in stimulating either HIV-1ΔPTAP infectivity (panel 1) or virion release (panel 2). Wild type ALIX and ALIX6A were expressed at equivalent levels (panels 4 and 5), and neither protein altered cellular expression or proteolytic processing of the viral Gag-derived proteins, implying that the differences in virion release and infectivity reflected differences in the efficiency of particle release. Moreover, the 6A mutation did not affect the affinity of ALIXV binding to recombinant GST-p6Gag protein, as measured in a biosensor analysis (Figure S4A). Thus, the inhibitory effects of this mutation on ALIX function in HIV-1 release likely resulted from loss of ALIX-Ub interaction.
Unlike HIV-1, EIAV cannot recruit TSG101/ESCRT-I and relies on ALIX to bud from cells (Martin-Serrano and Neil, 2011; Usami et al., 2009). Depletion of endogenous ALIX from 293T cells therefore potently inhibits both EIAV infectivity (Figure 4B, panel 1) and virus budding, as measured by release of virion-associated CA protein (panel 2). EIAV release and infectivity were fully rescued upon re-expression of wild type ALIX from a siRNA-resistant construct, demonstrating that the depletion was specific and that the system could be used to test ALIX function. Expression of a siRNA-resistant ALIX6A construct rescued EIAV release, but the mutant protein was less efficient than wild type ALIX. Control experiments demonstrated that ALIXI212D was inactive in EIAV release, that cellular levels of EIAV Gag and CA proteins were comparable, and that the exogenous ALIX proteins were all expressed at similar levels. Thus ALIX function in HIV-1 and EIAV budding is impaired by a mutation in the ALIX V domain that inhibits Ub binding. We conclude that ALIX normally binds Ub when facilitating retrovirus budding and that this interaction contributes to the efficiency of ALIX function.
DISCUSSION
The Coiled-Coil Domain of ALIX Confers Polyubiquitin Binding
ALIX shares structural similarity within a ~50 residue region that is homologous to the helical coiled-coil UBAN domain in NEMO that binds to linear polyUb chains with micromolar affinity. NEMO contacts each of the two Ub moieties within a bound linear Ub2 molecule in very different ways. The distal Ub is bound through the canonical hydrophobic patch centered around Ile44, while the proximal Ub is bound primarily through polar interactions between a surface of Ub adjacent to but distinct from the Ile44 patch and a polar face of NEMO (Rahighi et al., 2009). The latter binding site is composed of two Arg (R309, R312) and a triad of Glu residues (E313, E317, E320) that make extensive contacts with this non-canonical surface of Ub. Despite the aforementioned sequence similarities, there are significant differences between ALIX and NEMO. Notably, UBAN comprises a coiled-coil dimer of two identical parallel strands where each Ub moiety of a linear diubiquitin molecule contacts its binding surface on the opposite strand of the dimeric unit. By contrast, the α14 helix in ALIX that contains the NEMO homology is one strand of a coiled-coil helical bundle composed of 3 dissimilar helical strands and the other two strands do not contain any obvious Ub binding motifs. Our finding that a second triad (ERE) in ALIX, which is absent from NEMO, also contributes to polyUb binding supports the hypothesis that this helical surface of ALIX may contact more than one K63-linked Ub in a polyUb chain and suggests an avidity-based binding modality that magnifies the relatively weak binding affinities intrinsic to the individual UBDs (Ren and Hurley, 2010; Sims and Cohen, 2009). The spacing between the two triads that we have identified in ALIX is compatible with the dimensions of both K63 and linear diubiquitin (Komander et al., 2009).
Our data do not rule-out the possibility, however, that additional Ub binding sites may exist elsewhere in the V domain — a possibility consistent with our biosensor experiments (Figure 3F) which show that K63 tetraubiquitin binding to the 6A mutant, while greatly reduced, is still higher than background. Preliminary principal component analysis of 40,000 ab initio structural simulations of monoubiquitin binding to ALIXV identified five regions with plausible contacts on the V domain at distances feasible to bridge a tetraubiquitin chain (data not shown). Clearly, structural analysis of ALIXV –tetraubiquitin complexes, together with more sophisticated modeling and mutagenesis will be needed to precisely define the mechanism by which ALIXV binds selectively to K63 tetraubiquitin. Finally, although we have not observed a role for ALIXV dimers or higher oligomers in linkage-specific polyUb binding in vitro, we cannot exclude that such a mechanism could contribute to higher avidity polyubiqutin binding in vivo.
ALIX Ub Binding Functions in Retrovirus Budding
The ESCRT pathway facilitates budding of retroviruses and many other enveloped viruses whose structural proteins use late assembly domains to recruit early-acting ESCRT factors, which in turn recruit the ESCRT-III and VPS4 complexes that mediate the membrane fission step (Martin-Serrano and Neil, 2011; Usami et al., 2009). The two best understood late assembly domains are the PTAP motif, which binds directly to the UEV domain of the TSG101 subunit of the ESCRT-I complex, and the YPXnL motif, which binds directly to the V domain of ALIX. TSG101 UEV also binds Ub (Sundquist et al., 2004; Teo et al., 2004), and we show here that the ALIX V domain does too, extending the parallels between these two viral late domain pathways. Furthermore, we find that mutations that inhibit the ALIX-Ub interaction impair the ability of ALIX to function in HIV-1 and EIAV budding, implying that Ub recognition plays an important role(s) during the virus budding process (Figure 4).
We do not yet know which ubiquitylated protein(s) are bound by ALIX during virus budding nor understand the mechanistic consequences of Ub binding. Ub binding frequently leads to covalent ubiquitylation (Hurley JH, 2006) and this possibility should now be tested for ALIX, particularly as the protein is monoubiquitylated in vivo (Erpapazoglou et al., 2012; Votteler et al., 2009). Ub has long been known to be involved in retrovirus budding (Martin-Serrano, 2007; Vogt, 2000), and experiments that implicate the importance of K63 Ub chains in this process (Strack et al., 2002; Weiss et al., 2010) fit well with our observation that ALIX binds preferentially to K63-linked chains in vitro. The relevant targets of ubiquitylation during virus budding have remained elusive, however, because both retroviral Gag proteins, such as HIV p6Gag (Gottwein and Kräusslich, 2005; Ott et al., 2000)and ESCRT factors including ALIX itself (Votteler et al., 2009), can be ubiquitylated in vivo. Indeed, Ub appears to play multiple roles in ESCRT-mediated MVB sorting in yeast, where ubiquitylation of both MVB cargoes and ESCRT factors is functionally important (Erpapazoglou et al., 2012). Similarly, we envision that Ub could play multiple roles during viral budding, with Gag ubiquitylation serving to increase the binding affinity and recruitment of late domain binding partners like ESCRT-I and ALIX, and ESCRT factor ubiquitylation serving to enhance ESCRT machinery assembly or other functions. ALIX plays a critical role in nucleating the assembly of downstream CHMP4/ESCRT-III complexes, and this activity is required for ALIX stimulation of HIV budding (Fisher et al., 2007; Usami et al., 2007). To perform this role, ALIX apparently must undergo conformational changes that relieve autoinhibition, expose ligand binding sites, enhance membrane binding, and promote protein oligomerization (Pires et al., 2009; Zhai et al., 2011; Zhou et al., 2009), and we speculate that a subset of these changes are promoted by Ub binding.
EXPERIMENTAL PROCEDURES
Affinity Capture and Immunoblotting
293T Cells were transfected (CaPO4) with 8 μg vector or vector expressing Strep-ALIX wild type or ΔPRRALIX (gifts of M. Maki, Nagoya). After 72h cells were harvested, lysed in capture buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 2mM EDTA, 40 mM NEM, 1X protease inhibitor cocktail (Roche) and 1% (v/v) Triton X-100). DTT (42 mM) was added shortly after lysis. Lysates were cleared by centrifugation at 20,000g and Strep-tagged proteins were captured with immobilized P16-Ubn, P16, or deubiquitylated P16-Ubn as indicated, washed 2x in capture buffer, eluted in SDS loading buffer containing 100 mM DTT and immunoblotted as indicated.
In Vitro Pull-Downs
ALIXV and IsoT (10 μg in 300 μL) were incubated O/N at 4 °C with 20 μL immobilized P16-Ubn, or P16 in binding buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 2mM EDTA, 1 DTT). Resins were washed 2x and eluted in SDS loading buffer containing 100 mM DTT. Reciprocal pull-downs of autoubiquitylated GST-ΔC2Rsp5 were performed in the same fashion using purified ALIXV, IsoT, and hPlic2UBA immobilized on Affigel10 resin at 1 mg ml−1.
Competitive Ub Binding Reactions
Biotin-tagged TUBE UBA or ALIXV (50 μg) were incubated with a mixture of ~8 μg each of K48, K63, and linear Ub4 chains in 300 μL binding buffer overnight at 4 °C. 20 μL of streptavidin-agarose was then added and incubated for 1 hr at 4°C. The resin was washed 2X in binding buffer, 2X in 50 mM NH4HCO3 (pH 8.0) containing 50 μL 0.1% Rapigest (Waters) in 50 mM NH4HCO3 with 10 pmol of synthetic AQUA peptide standards and 1 μg trypsin and incubated overnight at 37°C. After hydrolysis of Rapigest with HCl samples were separated on a C-18 reversed-phase column and directly analyzed by inline ESI-TOF MS Ub AQUA (see Supplemental Experimental Procedures).
Rescue of HIVΔPTAP Release and Infectivity
293T cells were co-transfected (lipofectamine 2000) with 1 μg of a proviral expression construct (HIVΔPTAP, WISP01-108 or HIVΔPTAP/ΔYP, WISP05-124) (Fisher et al., 2007; Garrus et al., 2001) and 1 μg of either empty control pCI-neo or a pCI-neo vector that expressed either wild type or mutant ALIX proteins (wt ALIX, WISP12-84; ALIXI212D, WISP05-117, ALIX6A, WISP12-85) (Fisher et al., 2007). Cells and virion-containing culture supernatants were harvested at 48 hr and viral titers were measured in triplicate using single-cycle MAGIC assays in HeLaP4 cells. Virions were collected by pelleting through 20% sucrose at 15,000g. Cells were lysed with 50mM Tris pH 7.4, 150mM NaCl, 1% Triton X-100, protease inhibitor cocktail (Roche) and cleared by centrifugation at 15,000g. Immunoblots were probed with rabbit anti-HIV CA (UT 416) and MA (UT 556) antisera, rabbit anti-ALIX (UT 324), mouse anti-FLAG-M2, mouse anti-GAPDH. Secondary antibodies were anti-mouse IgG (goat) conjugated to Alexa680 or anti-rabbit IgG (donkey) conjugated to IRdye800.
Rescue of EIAV release and infectivity
293T cells were transfected following the time course: t=0, seed 2×105 cells/well in 6 well plates; t=24h, transfect cells with siRNA using RNAi max (sense sequence GAACAAAUGCAGUGAUAUAtt, 10 nM siRNA, 7.5 μl Lipofectamine RNAiMax, Invitrogen); t=48h, change media and co-transfect cells with siRNA (10 nM siRNA), pCMVΔ3 vector or pCMVΔ3 expressing siRNA-resistant ALIX (wild type pCMVΔ3-ALIX, WISP12-86; pCMVΔ3-ALIXI212D, WISP12-87; or pCMVΔ3-ALIX6A, WISP12-88) and an EIAV vector comprising 0.2 μg pEV53, 0.2 μg pSIN6.1CeGFPW and 0.075 μg phCMV- VSV-G (Olsen, 1998; Yee et al., 1994) using 10 μl lipofectamine 2000; t=72h, media change (2ml); t=96h, harvest cells and culture supernatants for analysis as described above (rabbit anti-EIAV, UT418).
Supplementary Material
Highlights.
The ESCRT protein ALIX binds selectively to K63-linked polyubiquitin chains.
K63 Ub chains bind to conserved amino acid triads in the coiled coil V domain
PolyUb binding by ALIX contributes to budding of human and equine lentiviruses
Acknowledgments
We thank J.A. Olzmann, C.M. Richter, and B. Schrul for critical reading of the manuscript and the members of the Kopito lab and R.E. Cohen for helpful discussion. This work was supported by NIH grants (R01 GM074874 to RRK and NIH RO1 AI051174 to WIS). DPD was supported by a predoctoral training grant from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFRENCES
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012 doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bohgaki M, Tsukiyama T, Nakajima A, Maruyama S, Watanabe M, Koike T, Hatakeyama S. Involvement of Ymer in suppression of NF-κB activation by regulated interaction with lysine-63-linked polyubiquitin chain. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2008;1783:826–837. doi: 10.1016/j.bbamcr.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJK, Paing MM, Montagne WA, Meerloo T, Trejo J. ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol. 2012;197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Léon S, Raposo G, Reggiori F, Haguenauer-Tsapis R. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Molecular Biology of the Cell. 2012;23:2170–2183. doi: 10.1091/mbc.E11-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, Chung H, Zhai Q, Robinson H, Sundquist W, Hill C. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841– 852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Garrus J, von Schwedler U, Pornillos O, Morham S, Zavitz K, Wang H, Wettstein D, Stray K, Cote M, Rich R. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55– 65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Kräusslich H-G. Analysis of Human Immunodeficiency Virus Type 1 Gag Ubiquitination. Journal of Virology. 2005;79:9134–9144. doi: 10.1128/JVI.79.14.9134-9144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT Pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hurley JH, LS, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annual Review of Biochemistry. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. Functional Replacement of a Retroviral Late Domain by Ubiquitin Fusion. Traffic. 2008;9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The Ubiquitin Code. Annual Review of Biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Neil SJD. Host factors involved in retroviral budding and release. Nat Rev Micro. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proceedings of the National Academy of Sciences. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. The EMBO journal. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A, Mizuno E, Kobayashi K, Matsumoto M, Nakayama KI, Kitamura N, Komada M. Dynamic regulation of ubiquitylation and deubiquitylation at the central spindle during cytokinesis. J Cell Sci. 2008;121:1325–1333. doi: 10.1242/jcs.027417. [DOI] [PubMed] [Google Scholar]
- Olsen J. Gene transfer vectors derived from EIAV. Gene Therapy. 1998;5:1481–1487. doi: 10.1038/sj.gt.3300768. [DOI] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Chertova EN, Gagliardi TD, Schubert U. Ubiquitination of HIV-1 and MuLV Gag. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- Patnaik A, Chau V, Wills JW. Ubiquitin is part of the retrovirus budding machinery. Proc Natl Acad Sci U S A. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A Crescent-Shaped ALIX Dimer Targets ESCRT-III CHMP4 Filaments. Structure (London, England : 1993) 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific Recognition of Linear Ubiquitin Chains by NEMO Is Important for NF-κB Activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Rauch S, Martin-Serrano J. Multiple Interactions between the ESCRT Machinery and Arrestin-Related Proteins: Implications for PPXY-Dependent Budding. Journal of Virology. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. The EMBO journal. 2010;29:1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-Specific Avidity Defines the Lysine 63-Linked Polyubiquitin-Binding Preference of Rap80. Molecular Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX Is a Binding Partner for HIV-1 p6 and EIAV p9 Functioning in Virus Budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Gottlinger HG. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J Virol. 2002;76:5472–5479. doi: 10.1128/JVI.76.11.5472-5479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin Recognition by the Human TSG101 Protein. Molecular Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Teo H, Veprintsev DB, Williams RL. Structural Insights into Endosomal Sorting Complex Required for Transport (ESCRT-I) Recognition of Ubiquitinated Proteins. Journal of Biological Chemistry. 2004;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger H. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614– 6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Gottlinger HG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- Vogt VM. Ubiquitin in retrovirus assembly: Actor or bystander? Proceedings of the National Academy of Sciences. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Iavnilovitch E, Fingrut O, Shemesh V, Taglicht D, Erez O, Sorgel S, Walther T, Bannert N, Schubert U, et al. Exploring the functional interaction between POSH and ALIX and the relevance to HIV-1 release. BMC Biochemistry. 2009;10:12. doi: 10.1186/1471-2091-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Göttlinger H. Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag. PLoS Pathog. 2010;6:e1001107. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J, Friedmann T, Burns J. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43:99–1112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. Activation of the Retroviral Budding Factor ALIX. Journal of Virology. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lee YC, Lin SH, Kuang J. The CHMP4b- and Src-docking sites in the Bro1 domain are autoinhibited in the native state of Alix. Biochem J. 2009;418:277–284. doi: 10.1042/BJ20081388. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB, Minton AP. Macromolecular Crowding: Biochemical, Biophysical, and Physiological Consequences. Annual Review of Biophysics and Biomolecular Structure. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.