Abstract
Background
In 1990, Blum and colleagues first reported an association between DRD2 and alcoholism. While there have been subsequent replications of this genetic association, there have also been numerous studies that failed to detect an association between DRD2 and alcohol dependence. We propose that one aspect contributing to this inconsistency is the variation in alcohol phenotype used across studies.
Methods
Within the population based Finnish twin sample, FinnTwin16, we previously performed multivariate twin analyses to extract latent genetic factors which account for the variation across seven measures of alcohol consumption (frequency of drinking, frequency × quantity, frequency of heavy drinking, frequency of intoxication, and maximum drinks in a 24 hour period) and problems (the Rutgers Alcohol Problem Index-RAPI and the Mälmö-modified Michigan Alcohol Screen Test - MmMAST) in 3,065 twins. In the present study, we examined the association between thirty-one DRD2/ANKK1 SNPs and the genetic factor scores generated by twin analyses in a subset of FinnTwin16 (n=602). We focus on two of the genetic factors: a general alcohol consumption and problems factor score which represents shared genetic variance across alcohol measures, and an alcohol problems genetic factor score which loads onto the two indices of problematic drinking (MAST and RAPI).
Results
After correction for multiple testing across SNPs and phenotypes, of the thirty-one SNPs genotyped across DRD2/ANKK1, one SNP (rs10891549) showed significant association with the general alcohol consumption and problems factor score (p=0.004), and four SNPs (rs10891549, rs1554929, rs6275, rs6279), representing 2 independent signals after accounting for LD, showed significant association with the alcohol problems genetic factor score (p=0.005, p=0.005, p=0.003, p=0.003).
Conclusions
In this study, we provide additional positive evidence for the association between DRD2/ANKK1 and alcohol outcomes, including frequency of drinking and drinking problems. Additionally, post hoc analyses indicate stronger association signals using genetic factor scores than individual measures, which suggests that accounting for the genetic architecture of the alcohol measures reduces genetic heterogeneity in alcohol dependence outcomes in this sample and enhances the ability to detect association.
Introduction
Alcohol consumption and problems are complex human behaviors that are influenced by both genetic and environmental risk factors (Kendler et al., 1992; Kendler et al., 1994). One strong candidate gene for alcohol-related outcomes is the dopamine receptor D2 gene (DRD2). In 1989, it was hypothesized that the rewarding effects of alcohol are mediated through the mesolimbic dopamine system (Wise and Rompre, 1989). The association between DRD2 and alcoholism was first reported by Blum and colleagues, who found that an increased frequency of the Taq1A1 restriction fragment length polymorphism was observed in postmortem brain tissue from alcoholics (as compared to nonalcoholic controls) (Blum et al., 1990). Since this initial report, there has been an extensive literature examining the relationship between DRD2 and alcohol-related outcomes. While there have been subsequent replications of this genetic association (Blum et al., 1991; Comings et al., 1991; Parsian et al., 1991; Amadeo et al., 1993; Noble et al., 1994; Higuchi et al., 1994; Neiswanger et al., 1995; Hietala et al., 1997; Kono et al., 1997; Ishiguro et al., 1998; Noble, 2003; Foley et al., 2004; Konishi et al., 2004), there have also been numerous studies across a variety of samples, populations, and study designs which fail to find an association between DRD2 and alcohol outcomes (Arinami et al., 1993; Bolos et al., 1990; Chen et al., 1996, 1997, 2001; Cook et al., 1992; Cruz et al., 1995; Edenberg et al., 1998; Gelernter and Kranzler, 1999; Gelernter et al., 1991; Goldman et al., 1992, 1997; Lee et al., 1999; Lobos and Todd, 1998; Lu et al., 1996; Parsian et al., 2000; Sander et al., 1995, 1999; Schwab et al., 1991; Suarez et al., 1994; Turner et al., 1992; Waldman et al., 1999). Critics have proposed that much of this mixed literature resulted from the limitations of early genetic studies including small sample sizes and limited ability to tag all regions of a gene. However, results from more recent genetic association studies remain inconsistent with both positive (Hack et al., 2010, Filbey et al., 2011; Landgren et al., 2011; Van der Zwaluw et al., 2011; Bhaskar et al., 2011) and negative (Kasiakogia-Worlley et al., 2011; Creemers et al., 2011, Heath et al., 2011, Wang et al., 2011, Luo et al., 2011, Schumann et al., 2011) evidence for association between DRD2 and alcohol problems. Interpreting this literature is further complicated by the 2004 discovery that the Taq1A polymorphism that had been most extensively studied was actually located 10 kb downstream from DRD2 in a neighboring gene, ankyrin repeat and kinase domain containing 1 (ANKK1) (Neville et al., 2004). The Taq1A variant is located within an exon of ANKK1, causing a non-synonymous coding change that may affect the substrate binding specificity of the gene product. It has been hypothesized that ANKK1 may be involved in the dopaminergic reward pathway through signal transduction (Neville et al., 2004). There have been many reviews of the DRD2 literature that provide detailed analysis of the variation across these genetic association studies (Goldman, 1998; Noble et al., 2000, Le Foll et al., 2009). However, little attention has been given to variability in the measurement of alcohol problems across these studies.
Many of the aforementioned studies used standard measures of alcohol use and/or problems including the Diagnostic and Statistical Manual of Mental Disorders (DSM) criteria, the Alcohol Dependence Scale (ADS), the Alcohol Expectancy Scale (AES), and the Alcohol Use Disorders Identification Test (AUDIT). Measures of alcohol problems vary by scientific field, setting (clinical vs. research), historical trend (DSM-III vs. DSM-IV), and availability. However, there is evidence to suggest that genetic association results may vary as a function of the alcohol measure used in the analysis. In 2002, Connor and colleagues tested the association between DRD2 and a variety of alcohol phenotypes, finding association with certain alcohol phenotypes (alcohol quantity, alcohol consumed per week, alcohol dependence scale score) and not others (frequency of alcohol use). This is an example of how even when using an identical sample and method in genetic association analyses the measure of the phenotype can affect the results.
Twin studies provide a method for examining the genetic relationship between different measures of alcohol use and problems. While some twin studies indicate that the genetic correlation between measures of regular alcohol consumption and problems is strong (Grant et al., 2009; Kendler et al., 2010), there is also evidence that there are genetic risk factors unique to alcohol problems (Dick et al. 2011). Additionally, recent twin studies examining the genetic relationship between the DSM-IV alcohol dependence criteria have indicated that the seven items are not genetically homogeneous (Kendler et al, 2011). Therefore, different measures of alcohol use and problems may be mediated by different genetic factors. This has implications for gene identification studies in that there are valid reasons why true genetic findings may not replicate across studies that have assessed different aspects of alcohol use and dependence.
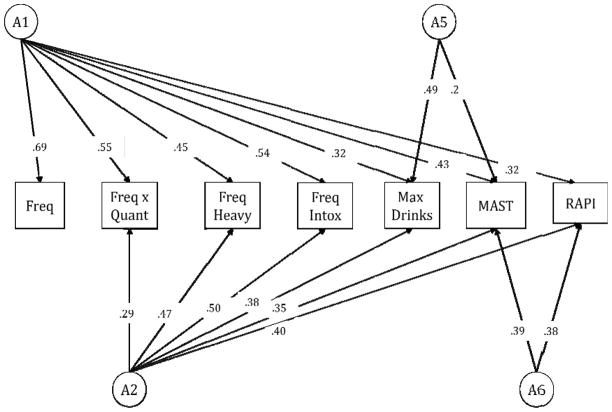
We previously reported analyses conducted within the Finnish population-based twin sample, FinnTwin16, to examine the genetic architecture across seven measures of alcohol consumption (frequency of drinking, frequency × quantity, frequency of heavy drinking, frequency of intoxication, and maximum drinks in a 24 hour period) and problems (the Rutgers Alcohol Problem Index-RAPI and the Mälmö-modified Michigan Alcohol Screen Test - MmMAST) (Dick et al., 2011). Our results yielded a model suggesting four latent factors that account for the genetic variance across the measures of alcohol consumption and measures of problems. The first two latent genetic factors loaded onto all of the drinking measures (consumption and problems), the third latent genetic factor loaded exclusively onto maximum drinks in a 24 hr period and the MmMAST, and the fourth latent genetic factor loaded onto the two indices of problems (the MmMAST and the RAPI). Using comparable measures of alcohol consumption and problems, data from an independent twin sample, the Virginia Adult Twin Study of Psychiatric and Substance Use Disorders, also indicated a parallel genetic architecture (Dick et al., 2011). This previously reported model from the Finntwin16 sample is depicted in Figure 1.
Figure 1.
Best Fitting Model of the Genetic Architecture of Measures of Alcohol Consumption and Problems in the Full Finntwin16 Sample (previously published in Dick et al. 2011)
In the present study, we extended these twin study results to examine the relationship between these measures of alcohol use/problems and DRD2/ANKK1. We hypothesized that examining association with genetic factor scores (previously implicated by the twin analyses within the same sample) would decrease the genetic heterogeneity and consequently increase power to detect genetic association between DRD2/ANKK1 and alcohol outcomes. We were primarily interested in the shared genetic variance across all alcohol measures (Figure 1. latent genetic factor A1) and the shared genetic variance across the two indices of problematic alcohol use (Figure 1. latent genetic factor A6). Additionally, we conducted post hoc analyses of the association between DRD2/ANKK1 and multiple measures of both alcohol consumption and problems in an effort to evaluate whether using genetic factor scores was an improvement upon using individual measures of alcohol consumption and problems.
Methods
Sample
Finntwin16 (FT16) is a population-based study consisting of five consecutive birth cohorts of twins born between 1975 and 1979 (Kaprio et al., 2002). The five birth cohorts contained 3,065 families of twins in which both twins were living and residing in Finland at the age of 16. Details about data collection have been previously published (Kaprio et al., 2002; Kaprio et al., 2006). All twins were identified through Finland’s Population Register Center, permitting exhaustive and unbiased ascertainment. Zygosity was determined using a well-validated questionnaire completed by both co-twins at the baseline (Kaprio et al., 1991). Here we focus on assessments of alcohol consumption and alcohol problems in young adulthood. The average age for the respondent twins at this assessment was 24.4 years (SD=1.50, range 22.8–27.2), with a response rate of 88.1%. Of these individuals, genotypic data was collected on 602 subjects, 36.0% were monozygotic (MZ) twins (n=216), 63.5% were dizygotic (DZ) twins (n=382). The 602 genotyped individuals were selected from twin pairs extremely discordant and concordant (EDAC selection) for alcohol-related problems, using a 22- item version of the Rutgers Alcohol Problem Index (RAPI; White & Labouvie, 1989) administered at age 18.5. EDAC selection has the benefit of enhancing statistical power by focusing on the most informative sibpairs, and it has been suggested as an advantageous sampling method for genetic linkage analyses (Gu et al. 1996). A full description of this selection is described in Latvala et al., 2011 (Latvala et al., 2011).
This was supplemented by parental information and comparisons of school photographs for the 2.9% (n=14) of twins whose zygosity could not be determined definitively from information in the questionnaires (Kaprio et al., 2002; Kaprio et al., 2006b) and later by DNA confirmation.
Measures
Measures of alcohol consumption and problems are described in detail in (Dick et al. 2011). Briefly, consumption measures included: Frequency (how often do you drink alcohol at all?), Frequency × Quantity (the frequency of reported use in the past 28 days multiplied by the quantity of drinks consumed per drinking day during the past 28 days; drinks defined as 1 beer, 1 glass of wine, or 1 mixed drink containing hard liquor equivalent to 10 grams of ethanol), Frequency of Heavy Drinking (at the present, how often do you within one occasion consume more than five bottles of beer, or more than a bottle of wine, or more than half a bottle of hard liquor?), Frequency of Intoxication (how often do you use alcohol to get drunk?), and Max Drinks (the maximum number of drinks twins reported ever consuming in a 24 hour period). Alcohol problem measures included: The Mälmö-modified MAST (Mm-MAST;(17)), a 9-item self-report scale of drinking patterns and problems designed for application in Nordic cultures (18)) and the 22 items from the Rutgers Alcohol Problem Index (RAPI)(19), a reliable scale designed to assess problematic drinking. Parallel to current practice in gene identification efforts for alcohol dependence, only individuals who had evidence of alcohol exposure were included in twin analyses, so that genetic/environmental influences on the decision to initiate alcohol are not confounded with genetic/environmental influences on alcohol consumption or problems. Altogether 2% of the sample had never had a full alcoholic beverage and were excluded from analyses. All measures were coded so that higher scores indicated more frequent drinking or more drinking problems.
Twin Modeling
The twin model we employed has been described in detail elsewhere (Dick et al., 2011). Briefly, a multivariate Cholesky model was fit to the measures of alcohol consumption and problems in order to estimate (1) the magnitude of genetic and environmental influences on each phenotype and (2) the extent to which these influences contributed to the covariation between the phenotypes. Using the statistical software package Mx (Neale and Cardon, 1992), we generated individual scores for each subject weighted by the loadings implicated by the genetic architecture from the best fitting twin model. When the best fitting model (Figure 1) from the full sample (n=2,500) was fit in the genotyped subset (n=602), there was not a significant decrease in model fit (χ2=3.28, p=1.00), however a model with two genetic factors (A1 and A6) fit the genotyped subsample best (AIC = −352.334). Thus, we moved two genetic factors forward in creating individual genetic factor scores for each person within the genotyped sample; (1) A general factor which loads onto measures of alcohol consumption and problems and (2) an alcohol problems factor which loads onto the Mm-MAST and the RAPI. This genetic factor score is similar to a phenotypic factor score in that it encompasses all shared variance across various measures. It differs in that it incorporates genetic information gained from twin data, therefore partitioning this shared variance into shared genetic variance across various measures. Thus, if an individual has an increased score on the specific alcohol measures that are loaded on by the latent genetic factor (e.g., Mm-MAST and RAPI), that individual will also to have an increased score on the genetic factor score (e.g., Alcohol Problems Genetic Factor, which loads onto Mm-MAST and RAPI).
Genotyping
A total of 602 individuals were genotyped using Sequenom’s homogeneous Mass Extend (hME) and iPLEX Gold technology (Sequenom, San Diego, CA, USA). Thirty-one tagging single-nucleotide polymorphisms (SNPs) in DRD2/ANKK1 were selected based on the HapMap Project (http://www.hapmap.org) and NCBI (http://www.ncbi.nlm.nih.gov) databases. The selected variants were bi-allelic and had a minor allele frequency (MAF) >10% in the Caucasian population. The ability to amplify the flanking regions of each SNP was determined by using the applications SNPper (http://www.snpper.chip.org) and RealSNP (http://www.realsnp.com), which define the most reliable regions for designing primers and the quality of the amplicons, respectively. All tagging SNPs failing during the procedure were replaced by newly generated tagging SNPs proposed by Haploview (Barrett, Fry, Maller, & Daly, 2005). The PCR and extension primers were designed using Sequenom’s Mass ARRAY Assay Design software (version 2.0). SNPs were genotyped in 384-well plates according to manufacturer’s instructions. For quality controls, each plate contained at least eight water controls and 22 duplicate samples. PCR reactions were performed in a total reaction volume of 5μl using 20ng of genomic DNA obtained by blood draw (Kettunen et al, Nat Genet 2012). The alleles were automatically called by Sequenom’s Mass ARRAY Typer Analyzer software and verified by two independent persons. Further marker-specific quality controls included a call rate >80% and a Hardy-Weinberg equilibrium (HWE) p-value >0.01 (estimated using unrelated individuals). Mendelian errors were excluded using PedCheck (O’Connell & Weeks, 1998).
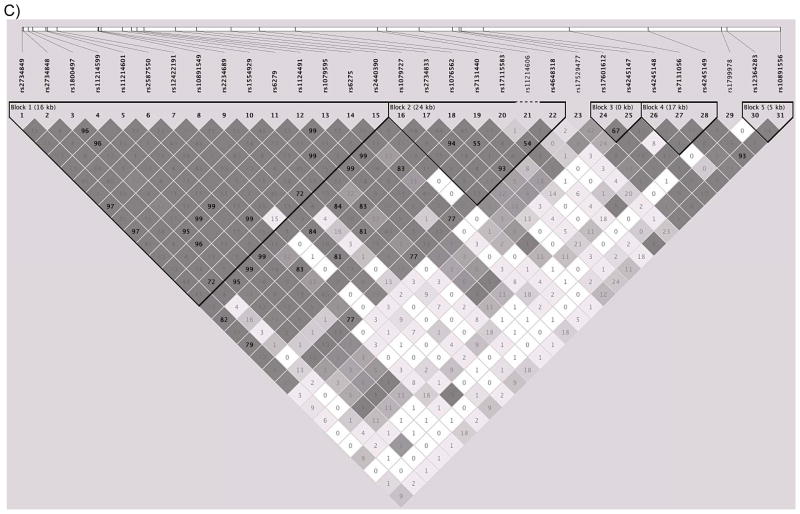
Once data were cleaned for quality control, genotypic data was available on 580 individuals of Finnish descent. An analysis of the population structure of the sample conducted in Eigenstrat (Price et al., 2006), a software program that uses principal components analysis to detect population stratification in genome-wide association studies, indicated a single ethnicity factor; thus all individuals were included in association analyses. Information on the genotyped SNPs, including chromosomal location and minor allele frequency is provided in Table 1. These thirty-one SNPs represent five different haplotype blocks across DRD2/ANKK1 (Figure 2). These SNPs are correlated (r2 range from .21–.93) yet represent five semi-independent signals (defined by r2<0.5) across DRD2/ANKK1 as indicated by a Nyholt correction for related SNPs (Nyholt et al., 2004).
Table 1.
Linear Regression of DRD2/ANKK1 SNPs on Genetic Factor Scores
| DRD2 SNP Information | Genetic Factor Scores | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Chr | Gene | SNP | Base Pair Location | Alleles Major; Minor | MAF | Alcohol Consumption and Problems | Alcohol Problems (MAST and RAPI) | ||
|
| |||||||||
| Beta | p-value | Beta | p-value | ||||||
| 11 | ANKK1 | rs2734849 | 112775370 | A;G | 0.282 | 0.094 | 0.047 | 0.127 | 0.006 |
| 11 | ANKK1 | rs2734848 | 112775584 | T;C | 0.220 | −0.040 | 0.401 | −0.040 | 0.391 |
| 11 | ANKK1 | rs1800497 | 112776038 | G;A | 0.330 | −0.003 | 0.945 | −0.035 | 0.451 |
| 11 | DRD2 | rs11214599 | 112776570 | C;T | 0.330 | −0.007 | 0.886 | −0.043 | 0.353 |
| 11 | DRD2 | rs11214601 | 112777972 | C;T | 0.330 | −0.004 | 0.936 | −0.041 | 0.373 |
| 11 | DRD2 | rs2587550 | 112778135 | A;G | 0.120 | −0.096 | 0.042 | −0.103 | 0.026 |
| 11 | DRD2 | rs12422191 | 112779220 | G;A | 0.900 | 0.001 | 0.981 | 0.034 | 0.460 |
| 11 | DRD2 | rs10891549 | 112783657 | T;C | 0.235 | 0.098 | 0.004 | 0.130 | 0.005 |
| 11 | DRD2 | rs2234689 | 112783693 | C;G | 0.220 | 0.040 | 0.401 | 0.040 | 0.391 |
| 11 | DRD2 | rs1554929 | 112783974 | C;T | 0.235 | 0.098 | 0.039 | 0.130 | 0.005 |
| 11 | DRD2 | rs6279 | 112786283 | C;G | 0.118 | −0.096 | 0.042 | −0.103 | 0.003 |
| 11 | DRD2 | rs1124491 | 112787300 | G;A | 0.330 | 0.005 | 0.914 | −0.042 | 0.367 |
| 11 | DRD2 | rs1079595 | 112787879 | A;C | 0.330 | −0.004 | 0.936 | −0.041 | 0.373 |
| 11 | DRD2 | rs6275 | 112788687 | G;A | 0.117 | −0.099 | 0.038 | −0.102 | 0.003 |
| 11 | DRD2 | rs2440390 | 112792088 | C;T | 0.080 | −0.051 | 0.285 | −0.014 | 0.757 |
| 11 | DRD2 | rs1079727 | 112794392 | T;C | 0.030 | 0.006 | 0.906 | −0.035 | 0.444 |
| 11 | DRD2 | rs2734833 | 112798130 | A;G | 0.241 | −0.098 | 0.038 | −0.108 | 0.019 |
| 11 | DRD2 | rs1076562 | 112801218 | G;A | 0.095 | −0.107 | 0.024 | −0.087 | 0.060 |
| 11 | DRD2 | rs7131440 | 112805120 | T;C | 0.254 | −0.104 | 0.028 | −0.105 | 0.023 |
| 11 | DRD2 | rs17115583 | 112814112 | G;A | 0.043 | −0.091 | 0.056 | −0.081 | 0.081 |
| 11 | DRD2 | rs11214606 | 112815079 | C;T | 0.010 | −0.007 | 0.875 | −0.012 | 0.794 |
| 11 | DRD2 | rs4648318 | 112818599 | T;C | 0.103 | −0.105 | 0.026 | −0.074 | 0.111 |
| 11 | DRD2 | rs17529477 | 112822277 | G;A | 0.033 | 0.052 | 0.267 | 0.042 | 0.359 |
| 11 | DRD2 | rs17601612 | 112822955 | G;C | 0.063 | 0.025 | 0.595 | 0.035 | 0.446 |
| 11 | DRD2 | rs4245147 | 112823217 | T;C | 0.099 | 0.033 | 0.494 | 0.068 | 0.143 |
| 11 | DRD2 | rs4245148 | 112825629 | C;T | 0.060 | 0.033 | 0.491 | 0.089 | 0.053 |
| 11 | DRD2 | rs7131056 | 112834984 | C;A | 0.226 | 0.078 | 0.100 | 0.040 | 0.391 |
| 11 | DRD2 | rs4245149 | 112843567 | G;A | 0.052 | −0.070 | 0.141 | −0.079 | 0.087 |
| 11 | DRD2 | rs1799978 | 112851561 | A;G | 0.050 | −0.044 | 0.255 | 0.019 | 0.684 |
| 11 | DRD2 | rs12364283 | 112852165 | A:G | 0.011 | −0.021 | 0.655 | 0.000 | 0.997 |
| 11 | DRD2 | rs10891556 | 112857971 | G;T | 0.052 | −0.073 | 0.126 | −0.072 | 0.120 |
Note: SNPs that passed Nyholt threshold for significant association (p<0.005) are bolded. The reference build used in this table was HapMap Data Release 28 Phase II+III, August10, on NCBI B36 assesmbly dbSNP b126. The major allele frequencies (MAF) presented in this table were calculated using only one individual per family.
Figure 2.
LD structure of DRD2/ANKK1
Legend: Location of (A) and correlations between (B and C) the single-nucleotide polymorphisms (SNPs) genotyped in the DRD2/ANKK1 gene complex (B) in the CEPH (Centre d’Etude du Polymorphisme Humain) data obtained from the HapMap database (The International HapMap Consortium, 2003) and (C) in the Finntwin16 data, Shading indicates the degree of correlation as measured by D′ (Hedrick & Kumar, 2001); darker shading indicates higher correlations, and white shading indicates that markers are unlinked or uncorrelated. The numbers inside the diamonds are R2 values, another measure of correlation between SNPs. The black triangles grouping subsets of SNPs indicate blocks of SNPs that are highly correlated (as defined by criteria detailed in Gabriel et al., 2002). Not all SNPs genotyped in the Finntwin16 sample were available in the HapMap database; in these cases, proxy SNPs that were the SNPs most highly correlated with the genotyped SNPs are listed. In the Finntwin16 sample, the LD blocks were similar to those in the HapMap CEPH data, and the somewhat stronger LD between markers is in agreement with previous findings from the Finnish population (Service et al., 2006).
Genetic association analyses
Linear regression was used to analyze the association between each of the SNPs and each of the genetic factor scores. The degree of relatedness (~50% for DZ twins and ~100% for MZ twins) was accounted for in the models using the GENMOD command in SAS 8.2 (SAS Institute, 2008). All p-value results from the association analyses were corrected for the number of independent tests conducted; the Nyholt correction indicated a significant threshold of p<0.005. Male and female data were collapsed in the genotypic analyses in order to maximize power to detect genetic association and to mirror the best fitting model from the twin analyses. Additionally, we conducted post hoc analyses of the association between DRD2/ANKK1 and the seven individual measures of alcohol consumption and problems in order to test whether using genetic factor scores would result in different conclusions than had we analyzed multiple individual measures of alcohol use/problems. When evaluating results for the seven alcohol phenotypes, the Nyholt correction indicated a significant threshold of a p<0.001 to take into account the additional tests.
Results
Twin Analyses
The phenotypic correlations across the measures of alcohol consumption and problems ranged from .45–.99 and were virtually identical to those previously reported in the full sample (Dick et al. 2011). Polychoric correlations were computed on only one twin from each pair, chosen randomly. MZ and DZ twin correlations for each of the measures were described previously (Dick et al. 2011). For the first genetic factor score (General Alcohol Consumption and Problems), scores ranged from −2.50 to 4.25 (mean=0, SD= 0.86). For the second genetic factor score (Alcohol Problems), scores ranged from −0.28 to 1.54 (mean=0, SD=0.52).
Genetic Association Analyses
Recall that the Nyholt threshold for a significant p-value for the two genetic factor scores is p<0.005. Of the thirty-one SNPs genotyped across DRD2/ANKK1, one SNP (rs10891549) showed significant association with the general alcohol consumption and problems factor score (p=0.004). Four SNPs (rs10891549, rs1554929, rs6275, rs6279) showed significant association with the alcohol problems genetic factor score (p=0.005, p=0.005, p=0.003, p=0.003, respectively). These results are detailed in Table 1. In addition, we conducted post hoc analyses in which we examined the association between DRD2/ANKK1 SNPs and the individual seven phenotypic measures of alcohol consumption and problems. These results are detailed in Table 2. Recall that the Nyholt corrected p-value for the seven alcohol outcomes is p<0.001. Using this criterion, none of the DRD2/ANKK1 SNPs were significantly associated with any of the individual alcohol measures.
Table 2.
Linear Regression of DRD2/ANKK1 SNPs on Individual Measures of Alcohol Consumption and Problems
| Alcohol Measures (p-values)
| |||||||
|---|---|---|---|---|---|---|---|
| SNP | Frequency Of Drinking | Frequency × Quantity | Frequency of Heavy Drinking | Frequency of Intoxication | Max Drinks 24 hr. Period | MAST | RAPI |
| rs2734849* | .016 | .443 | .032 | .032 | .278 | .012 | .007 |
| rs2734848* | .119 | .637 | .379 | .329 | .455 | .378 | .522 |
| rs1800497* | .662 | .668 | .839 | .925 | .802 | .650 | .337 |
| rs11214599 | .816 | .593 | .706 | .729 | .718 | .512 | .278 |
| rs11214601 | .777 | .654 | .749 | .776 | .667 | .538 | .293 |
| rs2587550 | .005 | .695 | .045 | .036 | .335 | .027 | .046 |
| rs12422191 | .404 | .541 | 1.00 | .732 | .739 | .225 | .981 |
| rs10891549 | .012 | .441 | .028 | .024 | .230 | .009 | .006 |
| rs2234689 | .119 | .637 | .379 | .329 | .455 | .378 | .522 |
| rs1554929 | .012 | .441 | .028 | .024 | .230 | .009 | .006 |
| rs6279 | .005 | .695 | .045 | .036 | .335 | .027 | .046 |
| rs1124491 | .793 | .640 | .723 | .759 | .683 | .549 | .275 |
| rs1079595 | .777 | .654 | .749 | .776 | .667 | .538 | .293 |
| rs6275 | .004 | .616 | .042 | .032 | .407 | .026 | .053 |
| rs2440390 | .221 | .806 | .262 | .108 | .407 | .750 | .718 |
| rs1079727 | .566 | .783 | .885 | .916 | .756 | .632 | .430 |
| rs2734833 | .046 | .345 | .027 | .013 | .226 | .034 | .015 |
| rs1076562 | .010 | .473 | .010 | .011 | .261 | .058 | .073 |
| rs7131440 | .045 | .294 | .018 | .008 | .210 | .034 | .021 |
| rs17115583 | .039 | .332 | .063 | .043 | .579 | .094 | .084 |
| rs11214606 | .937 | .927 | .893 | .752 | .642 | .816 | .755 |
| rs4648318 | .014 | .575 | .028 | .013 | .311 | .103 | .126 |
| rs17529477 | .184 | .388 | .411 | .090 | .229 | .424 | .239 |
| rs17601612 | .632 | .835 | .853 | .534 | .482 | .515 | .327 |
| rs4245147 | .444 | .800 | .912 | .586 | .348 | .209 | .101 |
| rs4245148 | .309 | .298 | .927 | .782 | .343 | .073 | .080 |
| rs7131056 | .037 | .160 | .087 | .075 | .702 | .550 | .368 |
| rs4245149 | .023 | .429 | .258 | .152 | .729 | .115 | .102 |
| rs1799978 | .530 | .528 | .263 | .154 | .325 | .357 | .768 |
| rs12364283 | .568 | .448 | .671 | .656 | .811 | .879 | .935 |
| rs10891556 | .017 | .434 | .228 | .129 | .743 | .140 | .147 |
Located in ANKK1
Conclusions
Two-decades of genetic studies have left the relationship between DRD2/ANKK1 and alcoholism indeterminate. Many reasons have been put forth to explain the mixed association results. Among them, poor DNA extraction techniques, population stratification, and failure to properly screen controls for drug and alcohol disorders. Previous reviews of this literature have detailed the variability and limitations of these studies (Goldman, 1998). A 2000 review by Noble (Noble, 2000) focused on sample size, types of alcoholics analyzed, and the nature of comparative controls employed in a variety of previously published studies. He reviewed several samples each of which used varying measures of alcoholism (The Michigan Alcoholism Screening Test, the presence or absence of medical complications of alcoholism, alcohol consumption, Severity of Alcohol Dependence Questionnaire (SADQ), and the DSM-III-R criteria). In this paper, we focus on the variability in the measure of the phenotype used across this literature in an effort to understand how this variability may effect the conclusions one would draw about the evidence for association with DRD2/ANKK1.
The 36 studies published between 1990 and 2011(Table 3), have yielded both positive and negative evidence of association across a variety of alcohol phenotypes. If more weight is placed on the recently published studies (Dick et al., 2004; Hack et al., 2011; Creemers et al., 2011; Schumann et al., 2011), which are presumably better powered to detect genetic association in that they use larger sample sizes and test a greater number of markers across DRD2/ANKK1 gene, and considering the publication bias that leaves many null results unreported, there is little evidence of association between DRD2/ANKK1 and alcohol phenotypes. It does appear however, that most of the studies that used quantitative/continuous measures of alcohol use and problems provide positive evidence of genetic association between DRD2/ANKK1 and alcohol related traits. This may reflect the fact that using quantitative measures can increase power to detect genetic association (Waldman et al., 1999, Kuo et al., 2010). However, it is of note that the largest of the aforementioned studies (Schumann et al., 2011), a meta-analyses of alcohol consumption GWAS on over 21,000 individuals, did not produce a genome wide significant variant in either DRD2 or ANKK1. The association with DRD2/ANKK1 appears to be contingent upon the specific measure of the phenotype, specific SNPs, and specific population used in a study. This is consistent with the implications of our twin studies that indicate that different genetic factors may contribute to risk for different measures of the “same” outcome (Dick et al., 2011). Moreover, while two measures of alcohol problems can both be valid and widely used, they are not necessarily genetically homogenous.
Table 3.
Previously Published Studies on the Genetic Association between DRD2/ANKK1 and Alcohol Phenotypes
| Study | Measure of the Phenotype | Study Design | Sample Size | SNPS | Evidence of Association |
|---|---|---|---|---|---|
| Blum et al., 1990 | DSM-III-R Alcohol Dependence and/or Abuse | Case/Control | Brain tissue from 35 cases; 35 controls | Tag1 A1 | Positive (post mortem samples) |
| Blum et al., 1991 | Severe alcoholics | Case/Control | 96 cases (52 severe) | Taq1 A1 | Positive (post mortem samples) |
| Comings et al., 1991 | Michigan Alcohol Screen Test** × stress exposure | Cross-sectional | 309 Honduran males | Taq1 A1 | Positive (with stress exposure) |
| Gelernter et al., 1991 | DSM-III-R Alcohol Dependence | Case/Control | 44 white cases; 68 controls | Taq1 A1 | Negative |
| Turner et al., 1992 | DSM-III-R Alcohol Dependence; AD+medical complications | Cross-sectional | 47 white males | Taq1 A1 | Negative |
| Amadeo et al., 1993 | DSM-III-R Alcohol Dependence | Case/Control | 69 French Polynesian cases; 57 controls | Taq1 A1 | Positive (combination of ADH2 and DRD2) |
| Arinami et al., 1993 | DSM-III-R Alcohol Dependence; Greater severity | Case/Control | 70 Japanese cases; 100 Japanese controls (unscreened) | Taq1 A1 | Positive |
| Bolos et al., 1990 | DSM-III-R Alcohol Dependence | Case/Control | 40 white cases; 127 controls | Taq1 A1 | Negative |
| Higuchi et al., 1994 | DSM-III-R Alcohol Dependence; Greater severity (Feigner Criteria) | Case/Control | 280 Japanese cases; 289 controls | Taq1 A1 (+) | Positive |
| Noble, 1994 | SADQ (Severity) | Case/Control | 73 cases; 80 controls | Taq1 A1 | Positive |
| Suarez et al., 1994 | Medical complications from Alcoholism | Case/Control | 88 white cases; 89 controls | Taq1 A1 (+) | Negative |
| Geijer et al., 1994 | DSM-III-R Alcohol Dependence | Case/Control | 74 cases; 81 controls | Taq1 A1/B1 | Negative |
| Cruz et al., 1995 | Alcohol Withdrawal Symptoms | Case/Control | 38 Mexican cases; 38 controls | Taq1 A1 | Negative |
| Lu et al., 2001 | DSM-III-R Alcohol Dependence; Conduct Disorder (CD) | Case/Control | 34 cases with CD, 63 cases without CD; 85 controls | Taq1 A1/B1 | Positive |
| Hietala et al., 1997 | SADQ (Severity); MAST | Case/Control | 70 Finnish male cases; 50 controls | Taq1 A1 | Positive |
| Kono et al., 1997 | DSM-III-R Alcohol Dependence; Early onset | Case/Control | 100 Japanese cases; 93 controls | Taq1 A1 | Positive |
| Ishiguro et al., 1998 | DSM-III-R Alcohol Dependence | Case/Control | 209 Japanese cases; 152 controls | Taq1 A1 | Positive |
| Lobos and Todd, 1998 | DSM-III-R Alcohol Dependence; Severity (Feigner Criteria) | Case/Control | 55 cases; 80 controls | 5 SNPs (6 haplotypes) | Negative |
| Edenberg et al., 1998 | DSMIII-R AD and Feigner Criteria | Linkage | 433 cases; 401 controls | Taq1 A1 | Negative |
| Sander et al., 1999 | DSMIII-R AD; Family history of Alcoholism | Case/Control | 310 German cases; 196 controls | TaqI A (+) | Negative |
| Waldman et al., 1999 | Quantitative Alcohol Measures** | TDT | 433 cases; 401 controls (COGA) | Taq1 A1 | Positive |
| Gelernter & Kranzler, 1999 | DSM-III-R Alcohol Dependence | Case/Control | 160 EA cases; 136 controls | Taq1 A1/B1 | Negative |
| Lee et al., 1999 | DSM-III-R Alcohol Dependence | Case/Control | 128 cases; 85 controls | Taq1 A1 | Negative |
| Parsian et al., 2000 | Medical complications from alcoholism; Feigner Criteria; Cloninger Criteria | Case/Control | 173 cases; 88 controls | TaqI A (+) | Negative |
| Chen et al., 2001 | DSM-IV Alcohol Dependence | Case/Control | 203 cases; 213 controls | −141C Ins/Del | Positive |
| Foley et al., 2004 | Alcohol Consumption from medical records** | Taq1 A1/B1 | Positive | ||
| Konishi et al., 2004 | DSM-IV Alcohol Dependence | Case/Control | 200 Mexican American cases; 351 controls | TaqI A1/B1 | Positive |
| Dick et al., 2007 | DSM-III-R Alcohol Dependence; Feigner Criteria | Family based association | 219 Caucasian families (n = 1,923) (COGA) | 26 single nucleotide polymorphisms (SNPs) across DRD2/ANKK1 | Positive |
| Yang et al., 2008 | DSM-III-R or DSM-IV Alcohol Dependence and/or Drug Dependence | Case/Control | 136 AD+DD cases;166 AD cases; 414 controls | 43 SNPs across DRD2/ANKK1, TTC12, NCAM1 | Positive |
| Hack et al., 2010 | DSM-IV Alcohol Dependence; | Case/Control | 545 Irish cases; 509 controls | 15 DRD2 SNPs (excluding Taq1A1) | Negative |
| Filbey et al., 2011 | Impulsive behavior on the Go/NoGo task Heavy Alcohol Drinking** | Cross-sectional | 53 cases | rs1799732 | Positive |
| Van der Zwaluw et al., 2011 | Adolescent Binge Drinking | Cross-sectional | 282 Dutch adolescent cases | Taq1A | Positive |
| Bhaskar et al., 2011 | Michigan Alcohol Screen Test** | Case/Control | 81 cases; 151 controls | 6 DRD2 SNPs | Positive |
| Creemers et al., 2011 | Adolescent Regular alcohol use | Cross-sectional | 1192 Dutch adolescents | Taq1A1 | Negative |
| Schumann et al., 2011 | Alcohol Consumption | Cross-sectional | 21,607 drinkers | Affymetrix 500K coverage of DRD2 | Negative |
Measure used in the present study
In the present study, we modeled the genetic architecture of the alcohol outcomes available in the Finntwin16 sample in an attempt to examine more genetically homogenous alcohol phenotypes. We found modest evidence of association between DRD2/ANKK1 SNPs and both genetically informed measures of alcohol consumption and problems. As rs10891549 and rs1554929 are highly correlated (r2=.98) and rs6275 and rs6279 are highly correlated (r2=0.87), there were two true independent signals detected in this sample. The first of these signals (rs10891549/rs1554929) is highly correlated with the SNPs within the ANKK1 gene, and may be indirectly associated with ANKK1, the original locus detected in association with alcohol problems. The association between the rs10891549/rs1554929 locus was found with both general alcohol consumption and problems in this sample. The second signal (rs6275/rs6279) may be potentially functional as rs6275 and rs6279 are non-synonymous polymorphisms that are located on the 3′UTR and may have a regulatory effect. This locus was only significantly associated with alcohol problems in the Finntwin16. Perhaps multiple independent signals within the DRD2/ANKK1 gene complex are differentially associated with alcohol outcomes; this may provide some explanation of the inconsistent genetic association findings.
In an effort to assess the utility of the genetic factor score, we also examined the association between DRD2/ANKK1 SNPs and the individual phenotypic measures of alcohol consumption and problems. As the inclusion of seven outcomes required a more stringent statistical test correction, no SNP passed the significance threshold put forth to correct for the multiple tests conducted. These results may suggest that we are indeed reducing genetic heterogeneity in the alcohol measures using the genetic factor scores. Additionally, we increase power to detect association in reducing the number of phenotypes examined (we correct for the analysis of two factor scores versus seven measures of alcohol consumption and problems). Thus, one can increase power to detect genetic association by (1) reducing the number of tests conducted, and (2) modeling the genetic architecture of the trait/disorder within your sample. Further, the need to refine these phenotypes to obtain adequate power to demonstrate association argues against claims of robust association between DRD2/ANKK1 and alcohol dependence. These results should be considered in light of several limitations. First, the generalizability of these results may be limited as analyses were conducted using twin data from a relatively homogenous Finnish population. In addition, the relative youth of the sample is relevant. With an average age of 24.4 years, it is likely that many participants may be just entering, or have not yet aged into the period of highest risk for heavy/problem use.
In summary, we provide modest evidence for the association between DRD2/ANKK1 and alcohol use/problems. In capturing the genetic heterogeneity across alcohol measures in genetic factor scores, we found association between DRD2/ANKK1 SNPs with both regular and problematic drinking. It should be noted that the β values associated with each significant DRD2/ANKK1 SNP range from 0.001–1.30, indicating that a very small portion of the variation in alcohol behavior is accounted for by DRD2/ANKK1 SNPs. In this study, we also demonstrated how to maximize the information obtained by twin analyses and molecular analyses within the same sample. By reducing the genetic heterogeneity inherent in the alcohol phenotype and the number of phenotypes analyzed, we detect a genetic association between DRD2/ANKK1 and alcohol use and problems, which would have been deemed nonsignificant had we not incorporated the genetic architecture across the traits.
Acknowledgments
The Finnish Twin studies have been supported by the National Institute of Alcohol Abuse and Alcoholism (grants AA-12502, AA-00145, and AA-09203 to RJR and AA15416 and K02AA018755 to DMD), the Academy of Finland (grants 100499, 205585, 141054 and 118555 to JK), and the Academy of Finland Centre of Excellence Programme (to LP & JK).
References
- Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, Madden PA, Todorov A, Trull T, Bucholz KK, Todd RD, Sher K, Heath AC. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs. 2009;70(2):157–168. doi: 10.15288/jsad.2009.70.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadeo S, Abbar M, Fourcade ML, Waksman G, Leroux MG, Madex A, Selin M, Champiat J-C, Brethome A, Leclaire Y, Castelnau D, Venisse J-L, Mallet J. D2 dopamine receptor gene and alcoholism. J Psychiatr Res. 1993;27:173–179. doi: 10.1016/0022-3956(93)90005-m. [DOI] [PubMed] [Google Scholar]
- Arinami T, Itokawa M, Komiyama T, Mitsushio H, Mori H, Mifune H, Hamaguchi H, Toru M. Association between severity of alcoholism and the A1 allele of the dopamine D2 receptor gene TaqI A RFLP in Japanese. Biol Psychiatry. 1993;33:108–114. doi: 10.1016/0006-3223(93)90309-2. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, Edenberg HJ, Rice J. The collaborative study on the genetics of alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Blomqvist O, Gelernter J, Kranzler HR. Family-based study of DRD2 alleles in alcohol and drug dependence. Am J Med Genet B Neuropsychiatr Genet. 2000;96:659–664. doi: 10.1002/1096-8628(20001009)96:5<659::aid-ajmg12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Blum K, Cull JG, Braverman ER, Comings DE. Reward deficiency syndrome. Am Sci. 1996;84:132–145. [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Finley O, Montgomery A, Ritchie T, Ozkaragoz T, Fitch RJ, Sadlack F, Sheffield D. Association of the A1 allele of the D2 dopamine receptor gene with severe alcoholism. Arch Gen Psychiatry. 1991;48:409–416. doi: 10.1016/0741-8329(91)90693-q. [DOI] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, Briggs AH, Cohn JB. Allelic association of human dopamine D2 receptor gene in alcoholism. J Am Med Assoc. 1990;263:2055–2060. [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from pedigree data. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- Bolos AM, Dean M, Lucase-Derse S, Ramsburg M, Brown GL, Goldman D. Population and pedigree studies reveal a lack of association between the D2 receptor gene and alcoholism. JAMA. 1990;264:3156–3160. [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Chen C-H, Huang J, Hsu Y-PP, Seow S-V, Chen C-C, Cheng ATA. Genetic polymorphisms of the promoter region of dopamine D2 receptor and dopamine transporter genes and alcoholism among four aboriginal groups and Han Chinese in Taiwan. Psychiatr Genet. 2001;11:187–195. doi: 10.1097/00041444-200112000-00002. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Chien SH, Hwu HG. Lack of association between TawI A1 allele of dopamine D2 receptor gene and alcohol-use disorders in Atayal natives of Taiwan. Am J Med Genet B Neuropsychiatr Genet. 1996;67:488–490. doi: 10.1002/(SICI)1096-8628(19960920)67:5<488::AID-AJMG10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Lu M-L, Hsu Y-PP, Chen C-C, Yu J-M, Cheng ATA. Dopamine D2 receptor gene and alcoholism among four aboriginal groups and Han in Taiwan. Am J Med Genet B Neuropsychiatr Genet. 1997;74:129–136. [PubMed] [Google Scholar]
- Clayton D. A generalization of the transmission/disequilibrium test for uncertain haplotype transmission. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings DE, Blum K. Reward deficiency syndrome: genetic aspects of behavioral disorders. Prog Brain Res. 2000;126:325–341. doi: 10.1016/S0079-6123(00)26022-6. [DOI] [PubMed] [Google Scholar]
- Comings DE, Comings BG, Muhleman D, Dietz G, Shahbahrami B, Tast D, Knell E, Kocsis P, Baumgarten R, Kovacs BW. The dopamine D2 receptor locus as a modifying gene in neuropsychiatric disorders. JAMA. 1991;266:1793–1800. [PubMed] [Google Scholar]
- Connor JP, Young RM, Lawford BR, Ritchie TL, Noble EP. D(2) dopamine receptor(DRD2) polymorphism is associated with severity of alcohol dependence. Eur Psychiatry. 2002;17(1):17–23. doi: 10.1016/s0924-9338(02)00625-9. [DOI] [PubMed] [Google Scholar]
- Cook BL, Wang ZW, Crowe RR, Hauser R, Freimer M. Alcoholism and the D2 receptor gene. Alcohol Clin Exp Res. 1992;16:806–809. doi: 10.1111/j.1530-0277.1992.tb00683.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ. Genetics of alcohol and other abused drugs. Drug Alcohol Depend. 1998;51:61–71. doi: 10.1016/s0376-8716(98)00066-0. [DOI] [PubMed] [Google Scholar]
- Cruz C, Camarena B, Mejia JM, Paez F, Eroza V, de la Fuente JR, Kershenobich D, Nicolini H. The dopamine D2 receptor gene TaqI A1 polymorphism and alcoholism in a Mexican population. Arch Med Res. 1995;26:421–426. [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Rose D, Kaprio J, Kendler KS. Measures of Current Alcohol Consumption and Problems: Two Independent Twin Studies Suggest A Complex Genetic Architecture. Alcohol Clin Exp Res. 2011;35(12):2152–61. doi: 10.1111/j.1530-0277.2011.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, Koller DL, Goate A, Rice J, Van Eerdewegh P, Reich T, Cloninger CR, Nurnberger JI, Kowalczuk M, Wu B, Li TK, Conneally PM, Tischfield JA, Wu W, Shears S, Crowe R, Hesselbrock V, Schuckit M, Porjesz B, Begleiter H. A family-based analysis of the association of the dopamine D2 receptor (DRD2) with alcoholism. Alcohol Clin Exp Res. 1998;22:505–512. [PubMed] [Google Scholar]
- Finckh U, Rommelspacher H, Kuhn S, Dufeu P, Otto G, Heinz A, Delttling M, Giraldo-Valasquez M, Pelz J, Graf K-J, Harms H, Sander T, Schmidt LG, Rolfs A. Influence of the dopamine D2 receptor (DRD2) genotype on neuroadaptive effects of alcohol and the clinical outcome of alcoholism. Pharmacogenetics. 1997;7:271–281. doi: 10.1097/00008571-199708000-00002. [DOI] [PubMed] [Google Scholar]
- Foley PF, Loh E-W, Innes DJ, Williams SM, Tannenberg AEG, Harper CG, Dodd PR. Association studies of neurotransmitter gene polymorphisms in alcoholic Caucasians. Ann N Y Acad Sci. 2004;1025:39–46. doi: 10.1196/annals.1316.005. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Goldman D, Risch N. The A1 allele at the D2 dopamine receptor gene and alcoholism. JAMA. 1993;269:1673–1677. [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. D2 dopamine receptor gene (DRD2) allele and haplotype frequencies in alcohol dependent and control subjects: no association with phenotype or severity of phenotype. Neuropsychopharmacology. 1999;20:640–649. doi: 10.1016/S0893-133X(98)00110-9. [DOI] [PubMed] [Google Scholar]
- Gelernter J, O’Malley S, Risch N, Kranzler HR, Krystal J, Merikangas K, Kennedy JL, Kidd KK. No association between an allele at the D2 dopamine receptor gene (DRD2) and alcoholism. JAMA. 1991;266:1801–1807. [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Goldman D, Brown GL, Albaugh B, Robin R, Goodson S, Trunzo M, Akhtar L, Lucas-Derse S, Long JC, Linnoila M, Dean M. DRD2 dopamine receptor genotype, linkage disquilibrium, and alcoholism in American Indians and other populations. Alcohol Clin Exp Res. 1993;17:199–204. doi: 10.1111/j.1530-0277.1993.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Goldman D, Dean M, Brown GL, Bolos AM, Tokola R, Virkkunen M, Linnoila M. D2 dopamine receptor genotype and cerebrospinal fluid homovanillic acid, 5-hydroxyindoleacetic acid and 3-methoxy-4-hydroxy-phenylglycol in alcoholics in Finland and the United States. Acta Psychiatr Scand. 1992;86:351–357. doi: 10.1111/j.1600-0447.1992.tb03279.x. [DOI] [PubMed] [Google Scholar]
- Goldman D, Urbanek M, Guenther D, Robin R, Long JC. Linkage and association of a functional DRD2 variant (Ser311Cys) and DRD2 markers to alcoholism, substance abuse and schizophrenia in Southwestern American Indians. Am J Med Genet B Neuropsychiatr Genet. 1997;74:386–394. doi: 10.1002/(sici)1096-8628(19970725)74:4<386::aid-ajmg9>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Gorwood P, Batel P, Gouya L, Courois F, Feingold J, Ades J. Reappraisal of the association between the DRD2 gene, alcoholism and addiction. Eur Psychiatry. 2000;15:90–96. doi: 10.1016/s0924-9338(00)00207-8. [DOI] [PubMed] [Google Scholar]
- Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry. 2009;66(8):795–800. doi: 10.1016/j.biopsych.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;15;70(6):513–8. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA—a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hesselbrock V, Hesselbrock M. Alcoholism and subtypes of antisocial personality disorder. Alcohol Alcohol. 1994;2(Suppl):479–484. [PubMed] [Google Scholar]
- Hietala J, Pohjalainen T, Heikkila-Kallio U, West C, Salaspuro M, Syvalahti E. Allelic association between D2 but not D1 dopamine receptor gene and alcoholism in Finland. Psychiatr Genet. 1997;7:19–25. doi: 10.1097/00041444-199700710-00003. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Muramatsu T, Murayama M, Hayashida M. Association of structural polymorphism of the dopamine D2receptor gene and alcoholism. Biochem Biophys Res Commun. 1994;204:1199–1205. doi: 10.1006/bbrc.1994.2590. [DOI] [PubMed] [Google Scholar]
- Hill SY, Zezza N, Wipprecht G, Zu J, Neiswanger K. Linkage studies of D2 and D4 receptor genes and alcoholism. Am J Med Genet B Neuropsychiatr Genet. 1999;88:676–685. doi: 10.1002/(sici)1096-8628(19991215)88:6<676::aid-ajmg18>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Arinami T, Akazawa S, Enomoto M, Mitushio H, Fujishiro H, Tada K, Akimoto Y, Mifune H, Shioduka S, Hamaguchi H, Toru M, Shibuya H. Association study between the −141C Ins/Del and TaqI A FAMILY-BASED ASSOCIATION ANALYSES OF ALCOHOL DEPENDENCE PHENOTYPES 1653 polymorphisms of the dopamine D2 receptor gene and alcoholism. Alcohol Clin Exp Res. 1998;22:845–848. [PubMed] [Google Scholar]
- Kaprio J. Twin studies in Finland 2006. Twin Res Hum Genet. 2006;9(6):772–777. doi: 10.1375/183242706779462778. [DOI] [PubMed] [Google Scholar]
- Kaprio J, Pulkkinen L, Rose RJ. Genetic and environmental factors in health-related behaviors: studies on Finnish twins and twin families. Twin Research. 2002;5:358–365. doi: 10.1375/136905202320906101. [DOI] [PubMed] [Google Scholar]
- Kaprio J, Rose RJ, Romanov K, Koskenvuo M. Genetic and environmental determinants of use and abuse of alcohol: The Finnish twin cohort studies. Alcohol & Alcoholism. 1991;(Suppl 1):131–136. [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. JAMA. 1992 Oct 14;268(14):1877–82. [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994 May;151(5):707–15. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. A population-based twin study of lifetime major depression in men and women. Arch Gen Psychiatry. 1999;56(1):39–44. doi: 10.1001/archpsyc.56.1.39. [DOI] [PubMed] [Google Scholar]
- Kendler KSPCA. Genes, Environment, and Psychopathology:Understanding the Causes of Psychiatric and Substance Use Disorders. Guilford Press; New York: 2006. [Google Scholar]
- Kendler KSMJDD, PCA The Relationship between Genetic Influences on Alcohol Dependence and on Patterns of Alcohol consumption. Alcoholism: Clinical and Experimental Research. 2010 doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu R-B, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet. 1998;103:211–227. doi: 10.1007/s004390050809. [DOI] [PubMed] [Google Scholar]
- Konishi T, Calvillo M, Leng A-S, Lin K-M, Wan Y-JY. Polymorphisms of the dopamine D2 receptor, serotonin transporter, and GABA-A receptor B3 subunit genes and alcoholism in Mexican Americans. Alcohol. 2004;32:45–52. doi: 10.1016/j.alcohol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kono Y, Yoneda H, Sakai T, Nonomura Y, Inayama Y, Koh J, Sakai J, Inada Y, Imamichi H, Asaba H. Association between early-onset alcoholism and the dopamine D2 receptor gene. Am J Med Genet B Neuropsychiatr Genet. 1997;74:179–182. doi: 10.1002/(sici)1096-8628(19970418)74:2<179::aid-ajmg13>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Kristenson H, Trell E. Indicators of alcohol consumption: Comparisons between a questionnaire (Mm-MAST), interviews and serum y-Glutamyl transferase (GGT) in a health survey of middle-aged males. British Journal of Addiction. 1982;77:297–304. doi: 10.1111/j.1360-0443.1982.tb02459.x. [DOI] [PubMed] [Google Scholar]
- Latvala A, Tuulio-Henriksson A, Dick DM, Vuoksimaa E, Viken RJ, Suvisaari J, Kaprio J, Rose RJ. Genetic origins of the association between verbal ability and alcohol dependence symptoms in young adulthood. Psychol Med. 2011 Mar;41(3):641–51. doi: 10.1017/S0033291710001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawford BR, Young RM, Rowell JA, Gibson JN, Feeney GFX, Ritchie TL, Syndulko K, Noble EP. Association of the D2 dopamine receptor A1 allele with alcoholism: medical severity of alcoholism and type of controls. Biol Psychiatry. 1997;41:386–393. doi: 10.1016/S0006-3223(96)00478-7. [DOI] [PubMed] [Google Scholar]
- Lee JF, Lu RB, Ko HC, Chang FM, Yin S-J, Pakstis AJ, Kidd KK. No association between DRD2 locus and alcoholism after controlling the ADH and ALDH genotypes in Chinese Han population. Alcohol Clin Exp Res. 1999;23:592–599. [PubMed] [Google Scholar]
- Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol. 2009 Feb;20(1):1–17. doi: 10.1097/FBP.0b013e3283242f05. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Lobos EA, Todd RD. Association analysis in an evolutionary context: cladistic analysis of the DRD2 locus to test for association with alcoholism. Am J Med Genet B Neuropsychiatr Genet. 1998;81:411–419. [PubMed] [Google Scholar]
- Lu R-B, Ko HC, Chang FM, Castiglione CM, Schoolfield G, Pakstis AJ, Kidd JR, Kidd KK. No association between alcoholism and multiple polymorphisms at the dopamine D2 receptor gene (DRD2) in three distinct Taiwanese populations. Biol Psychiatry. 1996;39:419–429. doi: 10.1016/0006-3223(95)00182-4. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: The Pedigree Disequilibrium Test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita S, Muramatsu T, Murayama M, Nakane J, Higuchi S. Alcoholism, ALDH2*2 allele and the A1 allele of the dopamine D2 receptor gene: an association study. Psychiatry Res. 2001;104:19–26. doi: 10.1016/s0165-1781(01)00290-6. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical Modeling. 5. Box 126 MCV, Richmond, VA 23298: Department of Psychiatry; 1999. [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Kluwer Academic Publishers; Dordrecht: 1992. [Google Scholar]
- Neiswanger K, Hill SY, Kaplan BB. Association and linkage studies of the TAQI A1 allele at the dopamine D2 receptor gene in samples of female and male alcoholics. Am J Med Genet B Neuropsychiatr Genet. 1995;60:267–271. doi: 10.1002/ajmg.1320600402. [DOI] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23(6):540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Noble EP. Addiction and its reward process through polymorphisms of the D2 dopmaine receptor gene: a review. Eur Psychiatry. 2000a;15:79–89. doi: 10.1016/s0924-9338(00)00208-x. [DOI] [PubMed] [Google Scholar]
- Noble EP. The DRD2 gene in psychiatric and neurological disorders and its phenotypes. Pharmacogenetics. 2000b;1:309–333. doi: 10.1517/14622416.1.3.309. [DOI] [PubMed] [Google Scholar]
- Noble EP. D2 dopamine receptor gene in psychiatric and neurologic disorders and its phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2003;116:103–125. doi: 10.1002/ajmg.b.10005. [DOI] [PubMed] [Google Scholar]
- Noble EP, Syndulko K, Fitch RJ, Ritchie T, Bohlman MC, Guth P, Sheridan PJ, Montgomery A, Heinzmann C, Sparkes RS. D2 dopamine receptor TawI A alleles in medically ill alcoholic and nonalcoholic patients. Alcohol Alcohol. 1994;29:729–744. [PubMed] [Google Scholar]
- Noble EP, Zhang X, Ritchie TL, Sparkes RS. Haplotypes at the DRD2 locus and severe alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2000;96:622–631. doi: 10.1002/1096-8628(20001009)96:5<622::aid-ajmg7>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A, Cloninger CR, Zhang Z-H. Functional variant in the DRD2 receptor promoter region and subtypes of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2000;96:407–411. doi: 10.1002/1096-8628(20000612)96:3<407::aid-ajmg32>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A, Todd RD, Devor EJ, O’Malley KL, Suarex BK, Reich T, Cloninger CR. Alcoholism and alleles of the human D2 dopamine receptor locus. Arch Gen Psychiatry. 1991;48:655–663. doi: 10.1001/archpsyc.1991.01810310073013. [DOI] [PubMed] [Google Scholar]
- Ponce G, Jimenez-Arriero MA, Rubio G, Hoenicka J, Ampuero I, Ramos JA, Palomo T. The A1 allele of the DRD2 gene (TaqI A polymorphisms) is associated with antisocial personality in a sample of alcohol-dependent patients. Eur Psychiatry. 2003;18:356–360. doi: 10.1016/j.eurpsy.2003.06.006. [DOI] [PubMed] [Google Scholar]
- Reich T. A genomic survey of alcohol dependence and related phenotypes: results from the Collaborative Study on the Genetics of Alcoholism (COGA) Alcohol Clin Exp Res. 1996;20(Suppl 8):133A–137A. doi: 10.1111/j.1530-0277.1996.tb01763.x. [DOI] [PubMed] [Google Scholar]
- Rose RJ. A developmental behavior-genetic perspective on alcoholism risk. Alcohol Health Res World. 1998;22(2):131–143. [PMC free article] [PubMed] [Google Scholar]
- Sander T, Harms H, Podschus J, Finckh U, Nickel B, Rolfs A, Rommelspacher H, Schmidt LG. Dopamine D1, D2, and D3 receptor genes in alcohol dependence. Psychiatr Genet. 1995;5:171–176. doi: 10.1097/00041444-199524000-00004. [DOI] [PubMed] [Google Scholar]
- Sander T, Ladehoff M, Samochowiec J, Finckh U, Rommelspacher H, Schmidt LG. Lack of an allelic association between polymorphisms of the dopamine D2 receptor gene and alcohol dependence in the German population. Alcohol Clin Exp Res. 1999;23:578–581. [PubMed] [Google Scholar]
- SAS Institute. SAS OnlineDoc Version 9.2. SAS Institute, Inc; Cary, NC: 2008. [Google Scholar]
- Schuckit MA, Danko GP, Smith TL, Hesselbrock V, Kramer J, Bucholz KK. A 5-year prospective evaluation of DSM-IV alcohol dependence with and without a physiological component. Alcohol Clin Exp Res. 2003;27:818–825. doi: 10.1097/01.ALC.0000067980.18461.33. [DOI] [PubMed] [Google Scholar]
- Schwab S, Soyka M, Niederecker M, SAckenheil M, Scherer J, Wilderauer DB. Allelic association of human dopamine D2-receptor DNA polymorphism ruled out in 45 alcoholics. Am J Hum Genet. 1991;49(Suppl):203. [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proença C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernández-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Núñez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tönjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 2011 Apr 26;108(17):7119–24. doi: 10.1073/pnas.1017288108. Erratum in: Proc Natl Acad Sci U S A. 2011 May 31;108(22):9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, et al. 2006 [Google Scholar]
- Suarez BK, Parsian A, Hampe CL, Todd RD, Reich T, Cloninger CR. Linkage disequilibria at the D2 dopmaine receptor locus (DRD2) in alcoholics and controls. Genomics. 1994;19:12–20. doi: 10.1006/geno.1994.1005. [DOI] [PubMed] [Google Scholar]
- Thanos PK, Taintor NB, Rivera SN, Umegaki H, Ilkari H, Roth G, Ingram DK, Hitzemann R, Fowler JS, Gatley SJ, Wang G-J, Volkow ND. DRD2 gene transfer into the nucleus accumbens core of the alcohol preferring and nonpreferring rats attenuates alcohol drinking. Alcohol Clin Exp Res. 2004;28:720–728. doi: 10.1097/01.alc.0000125270.30501.08. [DOI] [PubMed] [Google Scholar]
- Turner E, Ewing J, Shilling P, Smith TL, Irwin M, Schuckit M, Kelsoe JR. Lack of association between an RFLP near the D2 dopamine receptor gene and severe alcoholism. Biol Psychiatry. 1992;31:285–290. doi: 10.1016/0006-3223(92)90052-2. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G-J, Swanson JM. Dopamine in drug abuse and addiction: results from imaging studies and treatment implications. Mol Psychiatry. 2004;9:557–569. doi: 10.1038/sj.mp.4001507. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Aragam N, Jian X, Mullersman JE, Liu Y, Pan Y, Waldman ID, Robinson BF, Rhee SH. Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J Neural Transm. 1999;118(9):1293–9. doi: 10.1007/s00702-011-0628-3. [DOI] [PubMed] [Google Scholar]
- Waldman ID, Robinson BF, Rhee SH. A logistic regression extension of the transmission disequilibrium test for continuous traits: application to linkage disequilibrium between alcoholism and the candidate genes DRD2 and ADH3. Genet Epidemiol. 1999;17(Suppl 1):S379–S384. doi: 10.1002/gepi.1370170764. [DOI] [PubMed] [Google Scholar]
- White HR, Labouvie EW. Toward the assessment of adolescent problem drinking. Journal of Studies on Alcohol. 1989;50:30–37. doi: 10.15288/jsa.1989.50.30. [DOI] [PubMed] [Google Scholar]
- Wise RA, Rompre P-P. Brain dopamine and reward. Annu Rev Psychol. 1989;40:191–225. doi: 10.1146/annurev.ps.40.020189.001203. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008 Dec;32(12):2117–27. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]