Abstract
Oxidative modifications to cellular proteins are critical in mediating redox-sensitive processes such as autophagy, the antioxidant response, and apoptosis. The proteins that become modified by reactive species are often compartmentalized to specific organelles or regions of the cell. Here, we detail protocols for identifying the subcellular protein targets of lipid oxidation and for linking protein modifications with biological responses such as autophagy. Fluorophores such as BODIPY-labeled arachidonic acid or BODIPY-conjugated electrophiles can be paired with organelle-specific probes to identify specific biological processes and signaling pathways activated in response to oxidative stress. In particular, we demonstrate “negative” and “positive” labeling methods using BODIPY-tagged reagents for examining oxidative modifications to protein nucleophiles. The protocol describes the use of these probes in slot immunoblotting, quantitative western blotting, in-gel fluorescence, and confocal microscopy techniques. In particular, the use of the BODIPY fluorophore with organelle- or biological process-specific dyes and chromophores is highlighted. These methods can be used in multiple cell types as well as isolated organelles to interrogate the role of oxidative modifications in regulating biological responses to oxidative stress.
Keywords: lipid peroxidation, BODIPY, electrophile, oxidative stress, autophagy, mitochondria
INTRODUCTION
Lipid peroxidation is a consequence of oxidative stress and can regulate multiple biological processes [1–3]. Extensive peroxidation of phospholipids can result in the loss of function of critical enzymes involved in ion homeostasis, intermediary metabolism, cell repair, and cell death [4–7]. A primary mechanism through which this occurs is via site-specific modification of biomolecules by lipid peroxidation-derived electrophiles [8, 9]. Such modifications have the capacity to promote a state of cellular protection or adaptation, or they can have deleterious effects and promote cell death or otherwise maladaptive changes in cell phenotype [1, 2, 10]. Although modifications to nucleophilic residues of critical signaling proteins, enzymes involved in metabolism, and proteins directing apoptosis have been documented in the literature [8, 11–13], it has nevertheless been difficult to determine how such modifications activate or regulate complex biological processes.
The difficulty in defining changes in biological processes caused by lipid peroxidation, in many cases, is due to methodological limitations. Unambiguous detection and quantification of biologically relevant protein modifications has been particularly difficult for the following reasons: (1) immunological detection of oxidatively modified proteins relies on specific epitopes, which may be biased by surrounding residues; (2) lack of an ability to detect all modifications derived from the broad spectrum of lipid-derived electrophiles formed intracellularly; (3) inadequate sensitivity of detection methods; and (4) inadequate techniques to image the subcellular localization of proteins modified by reactive species. These issues have hindered our ability to understand how the biochemical changes imparted by oxidative protein modifications impact cell physiology.
Fluorophores and chromophores have demonstrated their value in linking biochemistry with changes in multiple cellular processes. Green and red fluorescent proteins, for example, have been used to track protein localization in cell culture experiments [14, 15], and they have played critical roles in tracking cell populations in vivo [16, 17]. Mitochondria-specific probes have also aided in understanding how mitochondrial dysfunction contributes to injury. The sequestration of most, but not all, of these probes is driven by mitochondrial membrane potential, and this property has been exploited to examine how loss of mitochondrial function occurs and relates with cell events such as apoptosis (e.g., [18, 19]). With respect to the field of oxidative stress, several tagged probes have been developed to interrogate the subcellular location of oxidative stress and to quantify free radical production. Several manuscripts and reviews have been written on such probes, and the authors suggest the following for information on measuring and quantifying reactive species for a broad range of applications [20–26].
Probes for measuring lipid peroxidation and the impact of lipid-derived electrophiles have also been described. Boron dipyrromethene (BODIPY) dyes as well as fluorophores with similar characteristics have been useful in identifying conditions that favor lipid peroxidation [27]. BODIPY 581/591 undecanoic acid (C11-BODIPY) has been a particularly valuable tool in quantifying lipid oxidation and anti-oxidant interventions in cultured cells [28, 29]. More recently, BODIPY conjugation to lipid peroxidation substrates such as arachidonic acid or to electrophiles themselves have been useful in detecting and identifying proteins susceptible to modification by reactive lipid species (RLS) and the electrophile response proteome [6, 25]. Qualities that make the BODIPY chromophore particularly useful are its relative non-sensitivity to changes in pH, insensitivity to solvent environment, high photostability relative to other fluorophores, high extinction coefficient, lack of ionic charge, and ease of conjugation to other molecules such as unsaturated fatty acids and reactive compounds such as iodoacetamide (IAM) [25, 30, 31]. BODIPY maintains narrow excitation and emission bandwidths and high fluorescence quantum yield making it especially useful in tandem with other fluorophores and probes, which can be used collectively to understand how biological processes proceed under conditions of oxidative stress.
PRINCIPLES
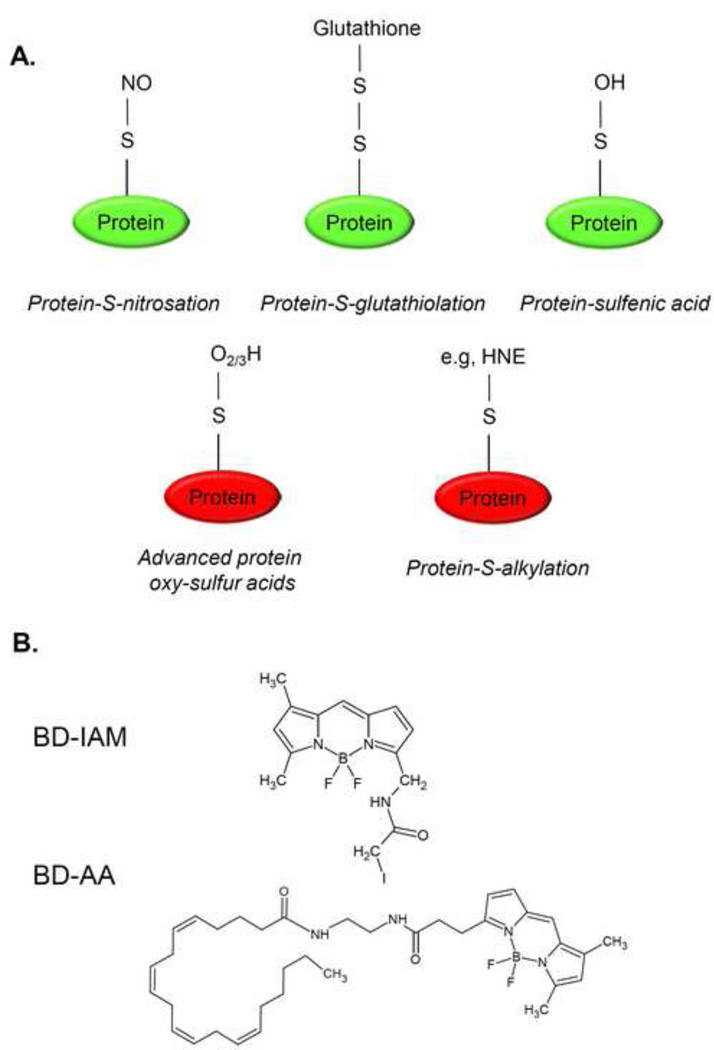
Cysteinyl residues of proteins undergo a variety of modifications. These modifications can have remarkable effects on enzyme activity, and, once formed, they have different stabilities. For example, S-nitrosated, -glutathio(ny)lated, or sulfenic acid-modified proteins are readily reduced back to their unmodified forms; however, advanced oxysulfur acid formation (i.e., sulfinic and sulfonic acids) or addition reactions with electrophilic lipids, such as 4-hydroxynonenal (HNE), result in more stable modifications that can have more prolonged effects on protein function (Fig. 1A). Experimentally, it has been difficult to identify which of these modifications contributes to pathology in tissues and cells under conditions of oxidative stress.
Figure 1. Common oxidative post-translational protein modifications and probes that can be used to interrogate their presence in the proteome.
(A) Simple diagrams showing the readily reversible protein modifications—S-nitrosation, -glutathio(ny)lation, and sulfenic acid—and more stable modifications such as sulfinic and sulfonic acids and Michael adducts with lipid electrophiles. (B) Structures of BD-iodoacetamide (BD-IAM) and BODIPY-labeled arachidonic acid (BD-AA), which can be used in indirect and direct protein thiol labeling strategies, respectively.
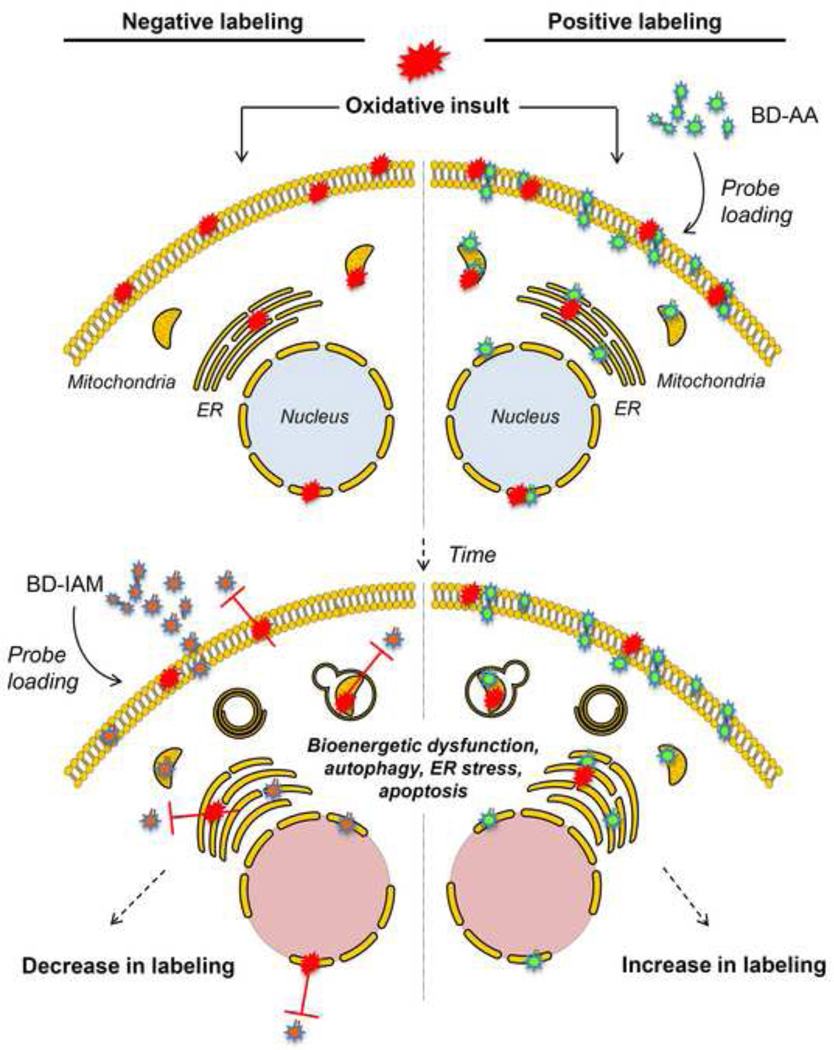
We previously described a protocol to conjugate BODIPY to unsaturated lipids to follow the formation of lipid adducts [25]. In addition, we have developed a separate protocol to examine the reactive thiol proteome using BODIPY conjugated with iodoacetamide [31]. In this protocol, we first show how these two methods can be used in concert with available fluorescent indicators of multiple biological processes, with the goal being to better understand how lipid peroxidation products, in particular, modulate the proteome to induce adaptive or maladaptive responses to oxidative stress. The BODIPY probes shown in Fig. 1B can be used in two different labeling strategies. In the negative labeling strategy, cells are exposed to oxidants or insults that promote oxidative stress (Fig. 2). These exposures often lead to mitochondrial damage [6, 32], activation of the autophagic program [6, 33], endoplasmic reticulum (ER) stress [34], and apoptosis [34, 35]. The proteins that could be involved with each of these processes can then be examined by post-labeling with BODIPY-iodoacetamide (BD-IAM), with the result being a decrease (or negative) labeling in conditions of the oxidative stress compared with control conditions. In the second labeling approach, i.e., positive labeling, cells are pre-loaded with BODIPY-conjugated arachidonic acid (BD-AA) and then subjected to the oxidative insult (Fig. 2). The advantage of this approach is that the lipid peroxidation products will be generated intracellularly and comprised of multiple fluorescently labeled reactive species. Because the BD-AA is uncharged and hydrophobic, it accumulates mostly in hydrophobic compartments and biological membranes, which are primary loci for lipid peroxidation.
Figure 2. Illustration of negative labeling and positive labeling strategies for detecting oxidative protein modifications.
In negative labeling (left panels), the cells are first exposed to an oxidative insult such as that induced by hydrogen peroxide, hypoxia-reoxygenation, or exposure to lipid peroxidation products. After the desired amount of time, the cells are exposed to BODIPY-labeled iodoacetamide (BD-IAM). A decrease in the extent of labeling, as detected by fluorescence techniques, indicates an increase in oxidative protein modifications. In positive labeling, the cells can be loaded with BODIPY-labeled arachidonic acid (BD-AA) and then exposed to oxidant (or electrophilic) stress. The proteins modified by BD-labeled reactive lipids generated by lipid peroxidation can then be imaged using fluorescence-based strategies For both techniques, microscopy may be used to identify the localization of the protein adducts, and protein fractionation may be used to quantify and identify proteins modified by the reactive species.
Given time with the oxidative stimulus, the labeled probes in both approaches can be followed temporally using fluorescence or confocal microscopy, and the cells can be fractionated to identify products in subcellular compartments. The identification of the proteins in these subproteomes can be facilitated using biochemical separation techniques such as SDS-PAGE, which can be extended to 2D approaches for identifying proteins reactive with oxylipidomes generated endogenously and exogenously. Alternatively, organelles such as the mitochondrion may be isolated and examined using the probes. A major strength of BODIPY probes is their compatibility with multiple organelle-specific probes and oxidative stress dyes, which may be incorporated to examine the localization of proteins modified by lipid peroxidation products and for examining how those modifications relate with cellular processes induced by oxidative stress, such as autophagy.
MATERIALS
Arachidonic Acid (Calbiochem, # 181198)
1-Ethyl-3-[3–dimethylaminopropyl]carbodiimide Hydrochloride (EDC; Pierce Biotechnology, # 22980)
4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl ethylenediamine, hydrochloride (BODIPY FL-EDA; Invitrogen, Product # D2390)
BODIPY® FL C1-IA, N-(4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-yl)Methyl)Iodoacetamide (BODIPY-IAM; Invitrogen, # D6003)
Hoechst 33342 (Enzo Life Sciences, # ENZ-52401)
ER-tracker Blue-White (Invitrogen, # E12353)
Mito-ID (Enzo Life Sciences, # ENZ-51018)
Cell Tracker Blue (Invitrogen, #, C2110)
mCherry-LC3 plasmid
Ethanol, HPLC grade
Acetonitrile, HPLC grade
Methanol, HPLC grade
Chloroform, HPLC grade, preserved with 0.75% ethanol
Acetic Acid, glacial, certified ACS plus grade
Tris base (Fisher, # BP152-1)
Triton X-100 (Sigma-Aldrich, # T-9284)
Hemin chloride (MP biomedical, # 15489-47-1)
4-hydroxynonenal (Cayman Chemical, # 32100)
Recombinant aldose reductase (MP Biomedical, # 199819)
DMSO (Sigma-Aldrich, # D2438)
Nitrocellulose (Biorad, # 162-0112)
35 mm glass bottom confocal petri dish (MatTek, # P35G-0-14-C)
Chambered coverglass, 4-well (Nunc; # 155383)
Amber glass vials, 1.5 ml (SUN SRi, # 200 252)
Caps with rubber septa (SUN SRi, # 500 062)
Borosilicate glass tubes, 16 × 100 mm (Fisher, # 14-961-29)
Bradford protein assay reagent (Biorad, # 500-0006)
Lowry DC protein assay reagent (Biorad,#500-0001)
18 mega-Ω water
Pre-cast 10.5–14% SDS-PAGE Criterion gels (Biorad, # 345-9949)
Anti-rabbit/mouse-Cy5 or Cy3 conjugated antibodies (GE Healthcare, #PA43002 or PA43010V)
INSTRUMENTS
Mass balance with 0.1 mg readability
Pipettes
SDS polyacrylamide gel apparatus
Slot or dot-blotting apparatus
Rocker (tilting shaker)
Vortex mixer
Spectrophotometer
Cuvettes
HPLC system equipped with ultraviolet/visible spectrophotometer and fraction collector.
Preparative Column: Gemini C18 reversed phase column with 10 µm particle size and dimensions of 250 × 21.2 mm. (Phenomenex, # 00G-4436-P0)
Semipreparative Column: Luna C18 reversed phase column with 5 µm particle size and dimensions of 250 × 10 mm. (Phenomenex, # 00G-4041-N0)
Confocal microscopes: (1) Nikon A1 line scanning laser confocal microscope equipped with three solid state lasers to generate 405 nm, 488 nm and 561 nm excitation lines. (2) Leica DMIRBE Microscope with TCS SP Laser Scanning Confocal system 405 nm, 488 nm, and 543 nm lines.
Fluorescent Imager: Typhoon (GE Healthcare) or other fluorescence imaging system suitable for gel scanning and equipped with a laser and filter suitable for BODIPY excitation and detection.
PROTOCOLS
A. Indirect labeling of the thiol proteome sensitive to lipid peroxidation products
This protocol can be used to examine how an oxidative insult or a particular species of reactive lipid affects the thiol proteome. Shown in Fig. 3 is an example of such an experiment. Here, rat aortic smooth muscle cells (RASMCs) were exposed to different concentrations of the lipid peroxidation product 4-hydroxy-2-trans-nonenal (HNE) for 30 min. The HNE-containing medium was then removed and replaced with fresh medium containing BODIPY-iodoacetamide (BD-IAM), which is commercially available and readily implemented into experimental protocols. After the desired time, the BD-IAM containing medium is removed, and the cells are washed to remove any unreacted BDIAM. These cells may then be examined by fluorescence microscopy. Alternatively, the cells may be lysed and the proteins separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteome modified can then be examined by in-gel fluorescence imaging. The thiol proteome modified due to exposure of cells to the reactive species may also be examined by slot blotting techniques.
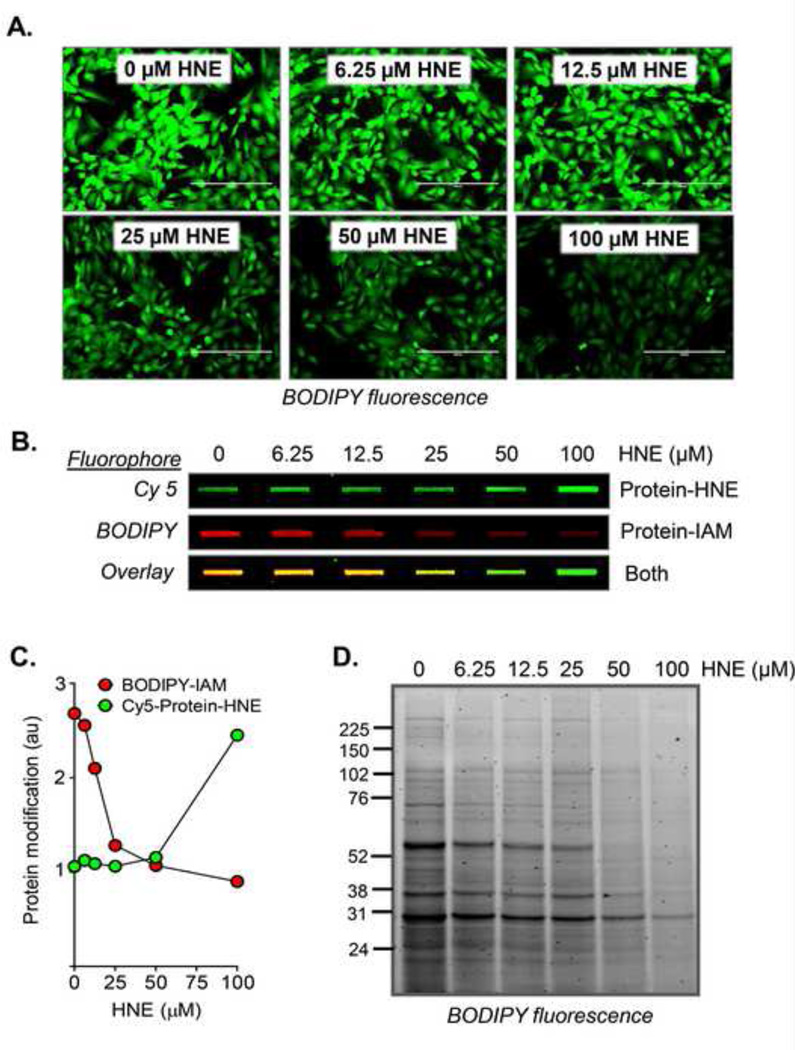
Figure 3. Negative labeling using 4-hydroxynonenal (HNE) as the model stressor.
Examples of different fluorescence labeling detection methods: (A) Rat aortic smooth muscle cells (RASMCs) were exposed to HNE (0–100 µM) for 30 min. The HNE-containing medium was then removed and medium containing BD-IAM (15 µM) was added to the cells. After 30 min, the cells were washed and imaged by fluorescence microscopy. (B) The same cells were then lysed, and total protein modification was assessed by slot blotting. Protein-HNE adducts (green) were detected using a rabbit polyclonal protein-HNE antibody; the secondary antibody was a Cy5-labeled anti-rabbit antibody. The BD-IAM modified proteins (red) were detected by fluorescence imaging on the membrane. Shown is the overlay of both modifications. (C) Quantification of protein-HNE and BODIPY-IAM adducts from panel B. (D) The proteins were then separated by SDS-PAGE, and the BODIPY adducts were imaged by in-gel fluorescence imaging. As shown in all of these examples, a loss of BODIPY signal was indicative of increased protein-HNE adducts and possibly secondary oxidative modifications occurring as a result of the electrophilic insult.
In this experiment, the cells treated with HNE showed an apparent concentration-dependent decrease in intracellular BODIPY fluorescence, indicating that the HNE exposure resulted in loss of intracellular free thiols, which would otherwise be reactive with the IAM probe (Fig. 3A). To examine total protein-HNE adducts and BD-IAM modifications simultaneously, the cell lysates were deposited onto nitrocellulose membranes, and they were probed with an anti-protein HNE polyclonal antibody; here, the secondary antibody used was a Cy5-labeled anti-rabbit antibody. Slot-immunoblotting for HNE adducts showed very little change at the lower concentrations of HNE exposure; however, the adducts began to appear when the concentration reached the 50–100 µM range (Fig. 3B). Interestingly, BD-IAM modifications to the proteome began to decrease with even the lowest concentration of HNE (Fig. 3B and C). The proteome affected by the oxidative insult may be further assessed by 1D or 2D protein fractionation. Shown in Fig. 3D is an SDS-PAGE in-gel fluorescence image of the thiol proteome affected by exposure of cells to HNE. Below is a step-by-step protocol for performing this type of experiment. This may be performed with any cell type and with multiple types of oxidative insults.
Procedure
Cells are seeded in 6-well dishes 1–2 days before the experimental protocol is implemented. Seed ~100,000–250,000 cells/well (depending on the proliferation rate of the cell type) in suitable medium such as DMEM containing 10% FBS and 1% penicillin/streptomycin at 37°C, 5% CO2 (if bicarbonate-buffered). The cells should be nearly (>90%) confluent to ensure the greatest yield of protein. If cell cycle changes are of particular interest, serum-starve cells for ~24 h before exposure to reactive species. Note: Analysis by confocal microscopy will require seeding in glass-bottom tissue culture dishes or slides (see Section D).
Wash the cells twice with sterile phosphate-buffered saline (PBS).
Exchange growth medium for DMEM or other suitable medium containing appropriate carbon sources (such as pyruvate, glutamine, glucose). This medium should be phenol red-free (for microscopy), and, in most cases, serum should be excluded to prevent reaction of the reactive species (e.g., electrophiles) with proteins such as albumin.
Add the desired concentration of reactive species or vehicle. In this experiment, ethanol was the vehicle, and a range of 6.25–100 µM HNE was used.
Expose cells to the reactive species for the desired time, e.g., 0.5–4 h, at 37°C. If bicarbonate is the medium-buffering agent, then perform all incubations at 5% CO2.
During incubation, make a stock BD-IAM solution by solubilizing in DMSO.
Determine BD-IAM concentration by diluting the BD-IAM in methanol, measuring the absorbance at 504 nm, and calculating the stock concentration using an extinction coefficient of 76,000 M−1 cm−1. Perform all measurements in low light conditions to avoid any photobleaching of the fluorophore. Keep in low light conditions from this point forward in the protocol.
Dilute BD-IAM to 20 µM in the appropriate volume of phenol-free medium and incubate the cells in this medium for 0.5 h at 37°C.
Wash the cells twice with PBS.
If desired, image cells using a fluorescent or confocal microscope by exciting at 488–505 nm and detecting emission at ~500–550 nm (see section D for more on fluorescence imaging). The excitation and emission maxima for BODIPY FL is shown in Table 1 and the spectra are shown in Fig. 7.
To examine protein modifications by slot/dot-blotting or SDS-PAGE, lyse cells in buffer containing 25 mM HEPES, pH 7.0, 1 mM EDTA or DTPA, 1% NP40, 0.1% SDS, and at least 1 mM N-ethylmaleimide (NEM). NEM is preferred if using a detergent-compatible Lowry assay. Note: It is critical to include either NEM or DTT in the lysis buffer, as this will stop any further reaction of BD-IAM with other cellular nucleophiles. Also, if 2D-IEF-SDS-PAGE will be performed, leave the SDS out of the lysis buffer.
Collect supernatant and perform protein assay.
- For dot or slot blotting, load between 1 and 10 µg of protein into each well of the dot/slot-blot apparatus (generally 1–4 µg is adequate, depending on the sensitivity of the imaging instrument). The general steps for dot/slot blotting are below:
- Cut a piece of nitrocellulose to fit apparatus (or use factory pre-cut nitrocellulose membranes).
- Soak three pre-cut pieces of filter paper and nitrocellulose in 1× Tris-buffered saline (TBS).
- Stack the filter paper and nitrocellulose in the apparatus and seal well. The nitrocellulose should be on top of the filter paper.
- Add 500 µl 1× TBS to each well of the apparatus.
- Pull the TBS through the membrane under vacuum.
- Add equal amounts of protein to each well of the apparatus. Be sure to dilute the protein in at least 400 µl TBS prior to adding to well.
- Pull dry under vacuum.
- Add 600 µl of 1× TBS to each well.
- Pull dry under vacuum.
- Disassemble apparatus and allow nitrocellulose to dry fully in the dark at room temperature.
- Block in appropriate blocking buffer for Western blotting or image directly for BD modifications (see below).
For gel-based analysis, add protein lysates to Laemmli sample buffer (or to 2D sample buffer if isoelectric focusing is desired). Make sure to include enough DTT or mercaptoethanol to quench all free NEM (if NEM is used in lysis buffer). We use up to 100 mM DTT (final concentration) in our sample buffer.
Load 20–40 µg of protein into the wells of an SDS-PAGE gel. Results from Fig. 2D were acquired using a 10.5–14% BioRad Criterion pre-cast SDS polyacrylamide gel.
- Image gels on a Typhoon scanner or other suitable imager.
- For gels in glass plates, first place a single strip of laboratory tape on the edges of the gel plates that contact the platen. This will prevent interference lines that would normally occur at the glass-platen interface.
- Place the gel plate on the platen and set focal plane height at +3 mm. For BODIPY imaging, the glass plates typical of most common electrophoresis equipment work well; low-fluorescence plates are not required unless other fluorophores will be used. The Criterion plastic cassettes, after cleaning with deionized water and drying, may be directly placed on the platen and scanned; again, ensure that the focal scan height is set at +3 mm.
- Alternatively, the gel can be imaged using “platen” for the focal plane setting by removing the gel from the glass or plastic cartridge and directly placing it on the platen on a small amount of deionized water. Avoid bubble formation between the gel and the platen.
- For Typhoon scanning, set the excitation wavelength at 488 nm and use the 520BP40 emission filter. Use at least 200 µm resolution.
Table 1.
Common fluorophores used in cell culture studies and their compatibility with BODIPY-FL dyes.
| Fluorophore† | Staining locale or applications |
Ex (nm) |
Em (nm) |
BODIPY-FL compatibility |
Source | Product # |
|---|---|---|---|---|---|---|
| BODIPY 493/503 | Lipid droplets | 500 | 506 | N | Invitrogen | D3922 |
| BODIPY FL* | - | 503 | 510 | - | Invitrogen | D2390 |
| BODIPY-FL-IA | Free thiols | 507 | 510 | - | Invitrogen | D6003 |
| BODIPY-C11 | Lipid peroxidation sensor | 581 | 591 | N | Invitrogen | D3861 |
| Fluorescein | General stain/Tag | 460 | 515 | N | Sigma | F245-6 |
| Alexa Fluor 488 | General stain/Tag | 495 | 519 | N | Invitrogen | A11001 |
| RFP | Tag | 563 | 582 | Y | - | - |
| mCherry* | Tag | 587 | 610 | Y | - | - |
| GFP | Tag | 493 | 505 | N | - | - |
| YFP | Tag | 529 | 539 | N | - | - |
| CFP | Tag | 458 | 489 | Y | - | - |
| DAPI | Nuclear stain | 350 | 470 | Y | Invitrogen | D21490 |
| Hoechst 33342* | Nuclear stain | 343 | 483 | Y | Invitrogen | H1399 |
| Alexa Fluor 350 phalloidin | Cytoskeleton (actin) | 346 | 446 | Y | Invitrogen | A22281 |
| Alexa Fluor 594 phalloidin | Cytoskeleton (actin) | 593 | 617 | Y | Invitrogen | A12381 |
| Tetramethylrhodamine methyl ester (TMRM) | Mitochondria (ΔΨ-dependent) | 552 | 577 | Y | Invitrogen | T668 |
| TMRM-IA | Mitochondrial thiols (ΔΨ-dependent) | 555 | 580 | Y | Invitrogen | T6006 |
| MitoTracker Deep Red | Mitochondria (ΔΨ-dependent) | 640 | 661 | Y | Invitrogen | M22425 |
| MitoTracker Red | Mitochondria (ΔΨ-dependent) | 578 | 599 | Y | Invitrogen | M7512, M7513 |
| MitoTracker Orange | Mitochondria (ΔΨ-dependent) | 554 | 576 | Y | Invitrogen | M7510, M7511 |
| MitoTracker Green | Mitochondria (ΔΨ-dependent) | 490 | 516 | N | Invitrogen | M7514 |
| Mito-ID Red* | Mitochondria (ΔΨ-independent) | 558 | 690 | Y | Enzo | ENZ51007 |
| Nonyl acridine orange | Mitochondria (ΔΨ-independent) | 495 | 519 | N | Invitrogen | A1372 |
| 3,3'-dihexyloxacarbocyanine iodide | Mitochondria (ΔΨ-dependent) | 482 | 501 | N | Enzo | ENZ52303 |
| CellLight® Golgi-RFP | Golgi apparatus | 555 | 584 | Y | Invitrogen | C10593 |
| Lysotracker Blue DND2 | Lysosomes | 373 | 422 | Y | Invitrogen | L7525 |
| Lysotracker Red DND99 | Lysosomes | 577 | 590 | Invitrogen | L7528 | |
| ER Tracker Blue-White DPX* | Endoplasmic reticulum | 374 | 430 (640) | Y | Invitrogen | E12353 |
| ER Tracker Red | Endoplasmic reticulum | 587 | 615 | Y | Invitrogen | E34250 |
| CellTracker Blue CMAC* | Whole cell | 353 | 466 | Y | Invitrogen | C2110 |
| H2DCFDA | Oxidant sensor | 492 | 517 | N | Invitrogen | D399 |
| Dihydroethidium/hydroethidine** | Oxidant sensor | 518 | 606 | Y,N | Invitrogen | D23107 |
| MitoSox Red** | Oxidant sensor | 396, 510 | 580 | Y,N | Invitrogen | M36008 |
| CellROX Deep Red | Oxidant sensor | 644 | 665 | Y | Invitrogen | C10422 |
| DAF-FM diacetate | Nitric oxide sensor | 495 | 515 | N | Invitrogen | D23844 |
| Monobromobimane | Glutathione sensor | 394 | 490 | Y | Invitrogen | M1378 |
| Cy5 (antibody conjugate)* | Tag | 632 | 699 | Y | GE Healthcare | PA45012 |
| Cy3 (antibody conjugate) | Tag | 534 | 614 | N | GE Healthcare | PA43010V |
indicates fluorophores shown in this paper.
Dihydroethidium/hydroethidine (DHE)-based dyes, when excited at ~510, may have some overlap with BODIPY FL; however, excitation of the DHE dyes near 510 nm may also report ethidium oxidation products other than the 2-OH-Mito-E+, which is formed by direct reaction of the dye with superoxide. The DHE dyes have another excitation peak at 396 nm that appears to primarily excite 2-OH-Mito-E+ (see Zielonka et al. [24] for further details).
Notes: Peak excitation and emission wavelengths often vary depending on the solvent environment.
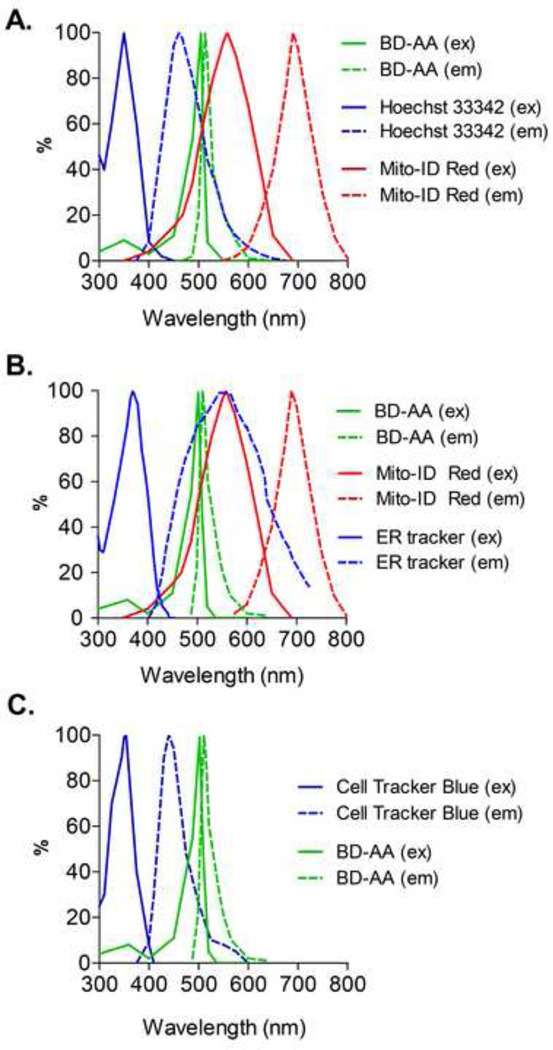
Figure 7. Excitation/emission spectra of tracking dyes compatible with BD-AA.
(A) Spectra for BD-AA, Hoescht 33342 (a nuclear stain), and Mito-ID Red (a non-membrane potential-dependent mitochondrial dye). (B) Spectra for BD-AA, Mito-ID Red and ER tracker Blue-White (an endoplasmic reticulum-specific dye). (C) Spectra for Cell Tracker Blue (a whole cell dye) and BD-AA. In all figures, excitation spectra (ex) are denoted by the solid lines and emission spectra (em) are denoted by the dashed lines.
Although this protocol and the results in Fig. 3 illustrate the ability to identify thiols modified directly by reactive lipid species, some caveats are worth noting. It is likely that the indirect BD-IAM labeling approach would not be useful in detecting modifications to other nucleophilic side chains such as lysine and histidine, both of which can be modified by HNE. Furthermore, the immunological approach used to detect aldehyde-modified proteins directly may not be sensitive enough to distinguish relatively lower levels of protein adducts, or the antibodies may react preferentially with only one or a few types of HNE adduct. These shortcomings could negate convergence between immunological labeling strategies and the indirect labeling strategy shown here using BD-IAM. Nevertheless, this technique does allow one to determine the impact of an electrophile or oxidant on the reactive thiol proteome and demonstrates an ability to combine indirect and direct labeling approaches to more fully elucidate the biological impact of oxidative post-translational protein modifications.
B. Direct labeling of the electrophile response proteome
Synthesis of BODIPY-labeled arachidonic acid (BD-AA)
Synthesis is performed using a carbodiimide-mediated conjugation reaction. After synthesis, the reaction preparations have a mixture of the original unreacted lipid, the tag, the expected product, and the priming compound, EDC. High performance liquid chromatography (HPLC) with UV-Vis detectors is then used to purify the tagged lipid. Following chromatographic separation, product purity is verified using electrospray ionization mass spectrometry (ESI-MS), and the compound is stored in an argon- or nitrogen-purged dark glass vial. For the detailed synthesis and purification protocol, the reader is referred to [25].
Detecting formation of intracellular products of lipid peroxidation products using BD-AA
To directly detect proteins modified by arachidonic acid oxidation products that are formed intracellularly, the BD-AA is incubated with the cells either before or concomitantly with the administration of an oxidizing stimulus. It should be noted that BD-AA will not be incorporated into phospholipids because the carboxylate moiety of AA is removed during derivatization with BODIPY. However, its hydrophobicity would likely sequester it primarily to membranes in the vicinity of lipid oxidation processes. As shown in Fig. 3, the BD-AA fluorescence may be viewed using fluorescence microscopy (Fig. 4A) and the proteins modified by lipid peroxidation products may be separated and visualized by in-gel fluorescence imaging (Fig. 4B and C). The following is a detailed protocol for this type of experiment.
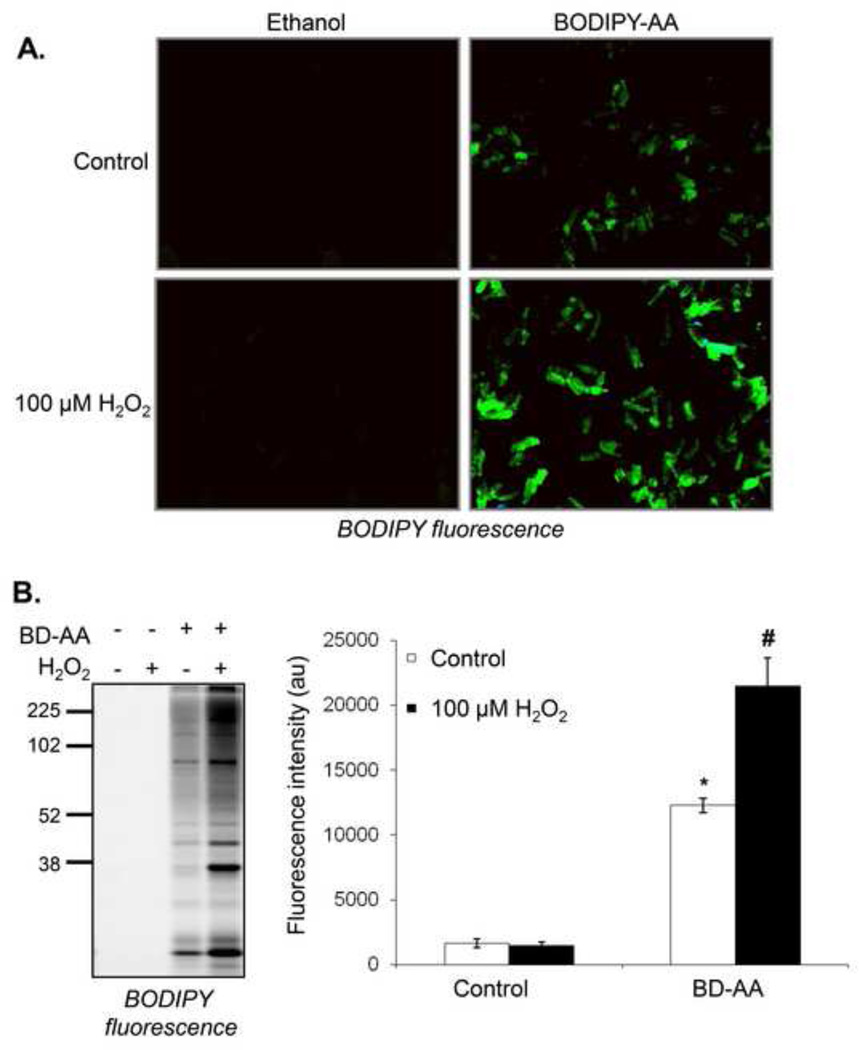
Figure 4. Positive labeling using a hydrogen peroxide and BODIPY-arachidonic acid (BD-AA) lipid peroxidation system.
Fluorescence strategies for detecting protein modifications induced by lipid peroxidation in situ: (A) Adult rat ventricular myocytes (ARVM) were isolated from Sprague-Dawley rats and treated with 10 µM BD-AA or ethanol (vehicle control). The cells were then exposed to vehicle or 100 µM H2O2. Cells were fixed on glass slides and imaged for BODIPY fluorescence. Representative images are shown. (B) The myocyte proteins were then separated by SDS-PAGE, and the BODIPY adducts were imaged by in-gel fluorescence imaging. Shown in panel A are representative images at the 2 h timepoint, and panel B shows proteins modified by the 4 h timepoint. n = 3 per group; *p<0.05 vs. control without BD-AA, #p<0.05 vs. BD-AA alone.
Procedure
Measure the absorbance spectra of the BD-AA stock using a spectrophotometer. Calculate the concentration using an extinction coefficient of 76 mM−1 cm−1 at 504nm.
Dilute BD-AA stock in neat ethanol to obtain a working stock of 5 mM.
Treat cells with BD-AA to a final concentration of 10 µM. Treat vehicle controls with the same volume of ethanol.
Without changing media, add treatment (in this case 100 µM H2O2) and incubate the cells for the desired time. Note: Hydrogen peroxide will work best in cells that contain high levels of heme-containing proteins such as myoglobin. For cell types that do not express myoglobin or adequate levels of heme-containing proteins that facilitate decomposition of unsaturated lipids, hemin at a concentration of ~10–25 µM may be used to induce lipid peroxidation [6, 36, 37].
For imaging using an epifluorescence microscope, cells in Fig. 4A were washed with PBS to remove media containing BD-AA, fixed using paraformaldehyde in PBS, and stored in the dark until imaging. Results typical of this protocol are shown in Fig. 4A. Note: Cells may also be live-imaged.
Alternatively, cells can be lysed in cold lysis buffer and separated via non-reducing SDS-PAGE. Due to the fluorescence of the BODIPY tag, protein bands containing lipid-protein adducts can be directly visualized using a Typhoon imager at an excitation wavelength of 488 nm and the 520BP40 emission filter. An example of results using this method is shown in Fig. 4B. The fluorescence intensity of the bands was quantified using ImageQuant TL software. Note: We have found that some of the lipid adducts with proteins are reducible with DTT. It is the experimenter’s option to use reducing conditions for examining the BD-lipid-protein adducts. Also, any free BODIPY label will travel in the dye front. Therefore, it is important to run the dye front completely off of the gel prior to imaging.
Synthesis of an external standard for quantification of protein modifications
The BD-IAM reagent may be used to make an external standard. For this, we have used recombinant aldose reductase (AR) protein. Note: We have noticed that GAPDH precipitates when conjugated with BD-IAM; therefore, it is recommended that BODIPY-FL-SSE, which will react with amines instead of thiols, be used to make a BD-GAPDH [31]. However, AR remains soluble after conjugation with BD-IAM. Other proteins may also be suitable for making a standard using BD-IAM, but these would require additional validation. The steps for making BD-AR are found below:
First, AR (1 mg/ml) is incubated in 100 mM Tris, pH 8.0, containing 100 mM DTT for 30 min at 37°C. During this time, equilibrate a Sephadex G25 (PD-10) column with 50 mM potassium phosphate buffer, pH 7.4.
The fully reduced AR is loaded into the equilibrated PD-10 column to remove DTT. Collect protein fractions (10 drops per fraction) in glass tubes or Eppendorf tubes. When using this PD-10 column, collect a total of 15 fractions. Note: The protein-containing fractions usually elute within fractions 3–9.
In 6 clean, separate tubes, place 100 µl of Bradford reagent. Add 10 µl of your samples from each fraction (i.e., fractions 3–9) to corresponding tubes containing Bradford reagent. The protein should elute sequentially in 2 to 3 tubes. Pool those fractions that turn blue in color.
Measure protein concentration by Lowry, Bradford, or other appropriate assay. Calculate the molar concentration of the protein.
The protein is then reacted with an equimolar amount of BD-IAM for 30 min at 37°C in the dark. During this incubation, equilibrate a PD-10 column with phosphate-buffered saline.
Pass the reaction mixture through the equilibrated PD-10 column to remove unreacted BD-IAM as in steps 1–3 above.
Calculate the protein concentration as in step 4.
Calculate the amount of BODIPY bound to the protein spectrophotometrically at 504 nm (ε = 76000 M–1cm–1).
Test the standard by running in an SDS-PAGE gel under reducing conditions.
An example of the use of the BD-AR standard is shown in Fig. 5. The dynamic range of this standard is ~10 fmoles to 100 pmoles. Calculate the extent of protein modification by dividing the picomoles of BODIPY in each lane (acquired using the standard curve) by the amount of protein (in µg) loaded into the respective lane.
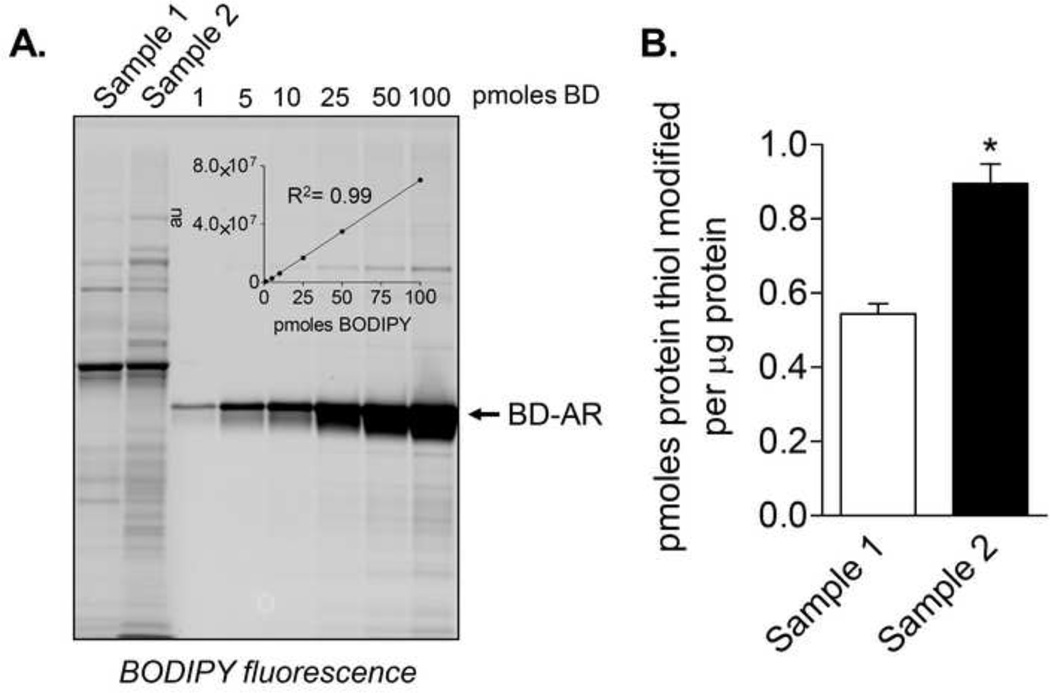
Figure 5. Example of an external standard constructed using BODIPY-iodoacetamide (BD-IAM).
Fluorescence imaging and quantification of samples using an in-gel external standard: (A) A BD-labeled protein standard was constructed by treating pre-reduced recombinant aldose reductase (AR) with BD-IAM. In this example, the usefulness of the standard is shown in context of samples (isolated mitochondria) that were treated with BD-IAM in a medium at pH 5.0 (sample 1) and in a medium at a pH of 7.2 (sample 2). The mitochondria were then lysed and equal amounts of protein were loaded on SDS-PAGE gels. The BD-AR standard was then loaded alongside the sample lanes and used to construct a standard curve (inset). (B) Quantification of BD-IAM adducts using the external standard. n = 3 per group; *p<0.05 vs. sample 1.
C. Detecting and identifying modifications in isolated organelles
The BD-AA and BD-IAM probes may also be used in isolated organelles such as mitochondria to examine protein susceptibility to modification by lipid peroxidation products or protein thiol reactivity. For this, the organelle should be intact after isolation and exposed to the BD-AA or BD-IAM under the desired conditions (e.g., Fig. 6A). Here, mitochondria were isolated from mouse hearts and loaded with BD-AA. The mitochondria were then placed under conditions of oxidative stress. For example, mitochondria incubated with hemin show an increase in lipid peroxidation-derived protein modifications compared with control mitochondria treated with BD-AA alone (Fig. 6B and C). Antioxidant interventions may also be used to determine specificity of the response; e.g., BHT was included as an antioxidant control in the hemin group (Fig. 6B and C). The lysates may also be used in 2D approaches for identifying proteins modified by reactive lipid species (Fig. 6B, lower panel). A general procedure for isolating mitochondria from heart or liver and examining targets of lipid peroxidation is found below.
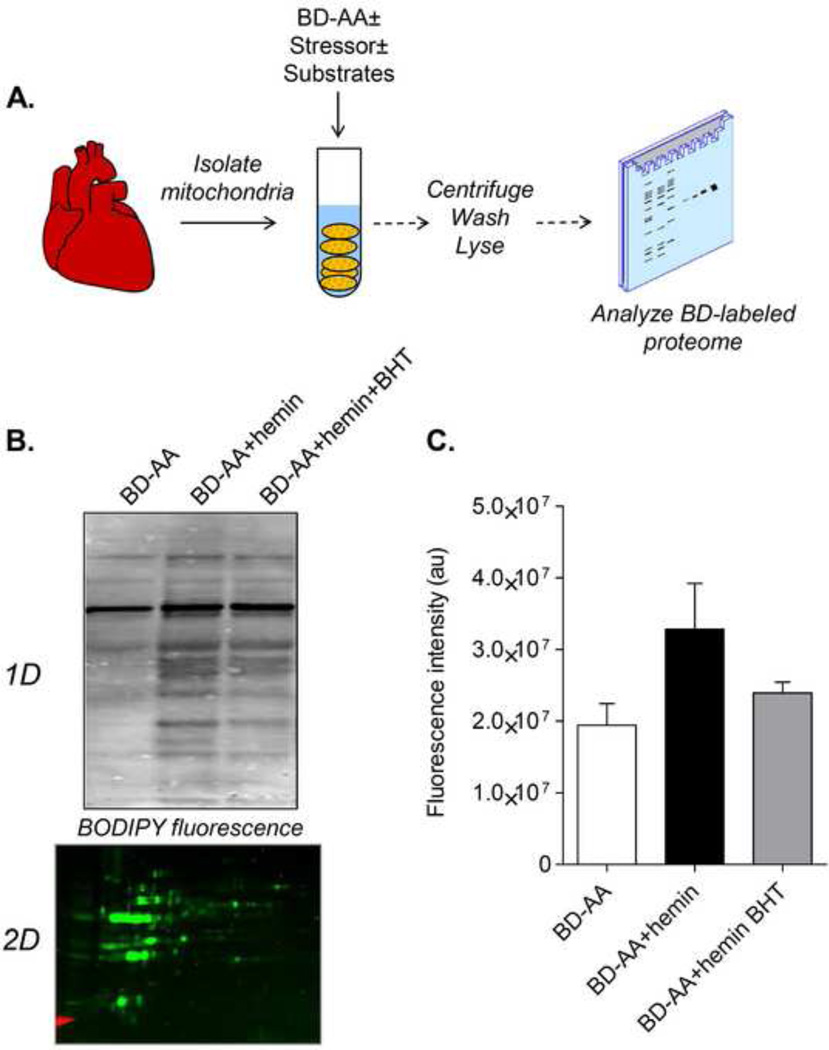
Figure 6. A positive labeling strategy to identify protein targets of lipid peroxidation in isolated organelles.
Example of the use of fluorescence imaging to detect mitochondrial proteins modified by oxidized products of BD-AA: (A) Illustration of the procedure for examining mitochondrial protein targets of lipid peroxidation: Isolated, intact mitochondria derived from rodent hearts may be treated with BD-AA in the absence or presence of oxidative stressors. Additionally, the mitochondria may be placed in different respiratory conditions. The mitochondria are then lysed and the protein targets can be examined using in-gel fluorescence imaging. (B) Isolated mitochondria were treated with BD-AA in the absence or presence of hemin. Lipid peroxidation processes were inhibited in the hemin group using the antioxidant butylated hydroxytoluene (BHT). The image in the lower panel shows that such modifications may also be detected in 2D proteomic strategies. (C) Quantification of the groups in panel B. n = 3 per group.
Procedure
After anesthetization, isolate the heart or liver and immediately place it into 10 mL of cold buffer A (containing 220 mM mannitol, 70 mM sucrose, 5 mM MOPS, 1 mM EGTA, adjust pH to 7.4 with KOH) in a 15 ml conical tube.
Wash the heart 5× with 10 ml of cold buffer A (until blood is mostly gone). If using Langendorff-perfused hearts, no washes are required.
Weigh the heart and then chop finely with scissors in 10 ml of Buffer A containing 0.2% defatted BSA and protease inhibitor cocktail (optional). Note: use 10 ml buffer to 1 g of tissue.
Homogenize tissue with a tissue tearer, or mince the tissue on ice with a razor blade and use a Teflon-coated Glas-Col homogenizer to homogenize tissue.
Transfer the suspension into a centrifuge tube.
Centrifuge at 500–1000×g for 10 min.
Collect supernatant. Optional: The supernatant may be passed through cheesecloth to filter out any large particles that may dislodge from the pellet.
Centrifuge the filtered supernatant at 10,000×g for 10 min.
Discard the supernatant (or keep as the crude cytosolic fraction if desired).
Resuspend the mitochondrial pellet in 5 ml of ice cold buffer A (without BSA).
Centrifuge once again at 10,000×g for 10 min.
Resuspend the pellet in 0.5–1 ml of either buffer A, respiration buffer, or buffer appropriate for the planned assay or experiment. Typical respiration buffer is: 120 mM KCl, 25 mM sucrose, 10 mM HEPES, 1 mM MgCl2 and 5 mM KH2PO4, pH to 7.2 with KOH.
If assaying respiration, make 0.5 M stocks of glutamate-malate (or pyruvate/malate) and/or succinate, and 100 mM ADP preparations. Remember to always use KOH to pH to 7.2.
Note: If a highly pure population of mitochondria is desired, resuspend mitochondria in 5 ml of Buffer A containing 19% Percoll after step 11. Centrifuge at 14,000×g for 10 min, followed by resuspension of the pellet in the desired buffer.
Measure protein concentration of isolated mitochondria suspension by first lysing 5 µl of mitochondrial suspension in 45 µl of 10% SDS. Use the Lowry DC protein assay reagent to determine protein concentration.
Preload a 1 mg/ml solution of intact mitochondria with 20–40 µM BD-AA and incubate at room temperature for 5 min.
Add oxidizing stimulus, vortex briefly, and place at 37°C for the desired time. If using energized mitochondrial preparations (i.e., mitochondria with substrate present), leave the tube tops open and vortex occasionally to help prevent depletion of oxygen in the medium. If assaying under state 3 or uncoupled conditions, decrease the amount of mitochondrial protein to 0.25 mg/ml or less per incubation.
Centrifuge mitochondria at 13,000×g for 5 min. Aspirate medium and freeze pellet or lyse immediately.
Lyse using appropriate buffer containing detergent. A simple lysis buffer that is compatible with both 1D and 2D electrophoresis is 10 mM HEPES (or Tris), pH 7.0, containing 1% NP-40, 0.5% deoxcholate, 1 mM EDTA, protease and phosphatase inhibitor cocktail, and 1 mM NEM. Allow lysates to incubate on ice for 1 h followed by sonication (3× for 10 sec each) on ice.
Centrifuge lysates at 13,000×g for 10 min and transfer supernatants to fresh tubes.
Perform protein assay on lysates and analyze BD-modified proteins as described in the above sections.
D. Detecting the localization of BD-AA and its derived oxidation products
One of the major strengths of the BODIPY fluorophore is its narrow excitation and emission bandwidths, which allow for it to be used with multiple other fluorophores. Of particular interest are the sites at which lipid-derived reactive species are generated and where they localize under conditions of oxidative stress. Hence, colocalization of BD-AA with specific organelles can be used to indicate primary targets of lipid oxidation products and identify biological processes regulation by oxidized lipids. Here we present a protocol for using BD-AA and BD-IAM in confocal microscopy applications. See Table 1 for a more extensive list of the compatible fluorophores and probes that may be used with BD-AA.
Shown in Fig. 7 are the excitation-emission spectra of BD-AA and Hoescht 33342 (a nuclear dye), Mito-ID Red [a non-membrane potential (ΔΨ)-dependent mitochondrial dye], ER Tracker Blue-White (an endoplasmic reticulum-specific dye) and Cell Tracker Blue (a dye that stains the entire cell). Prior to each experiment, the excitation and emission spectra should be examined to determine if the stain of choice is compatible with BODIPY. Applications and tools for spectral comparisons are available from multiple vendors and sites. For example, Molecular Probes (SpectraViewer at http://www.invitrogen.com/site/us/en/home/support/Research-Tools/Fluorescence-SpectraViewer.html or the iPad App), Heliophor, and Zeiss offer guidance for initial screening of compatible dyes.
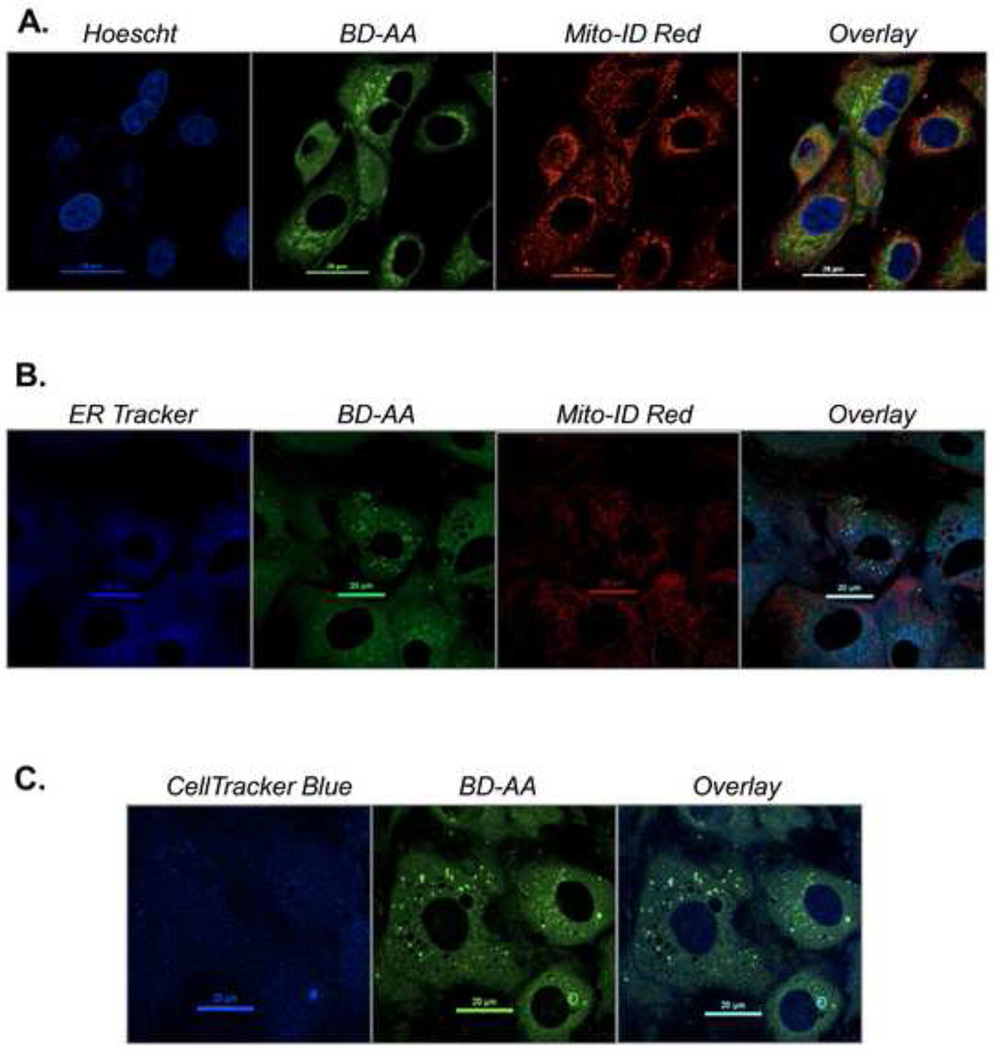
Shown in Fig. 8 are the confocal images of the above-mentioned dyes used in concert in unstressed cells. Most tracking and organelle dyes minimally affect cell viability, and they can be used for prolonged periods or for time-lapse imaging. Mito-ID Red (shown in Fig. 8A and B) is a good choice for examining mitochondrial colocalization since its localization should not be affected by changes in respiratory function. Should the confocal microscope not have differential interference contrast (DIC) capabilities to show clear boundaries of the cell, a total cell dye such as CytoTracker Blue may be used (Fig. 8C).
Figure 8. Using compatible dyes for identifying the localization of BD-AA by confocal microscopy.
Confocal fluorescence images of unstressed rat aortic smooth muscle cells in which BD-AA is shown in concert with tracking dyes. (A) Hoechst 33342, BD-AA, and Mito-ID stains; (B) ER Tracker Blue-White, BD-AA, and Mito-ID; and (C) Cell Tracker Blue and BD-AA. Table 2 shows the confocal settings and conditions for these images. Additional compatible dyes are found in Table 1.
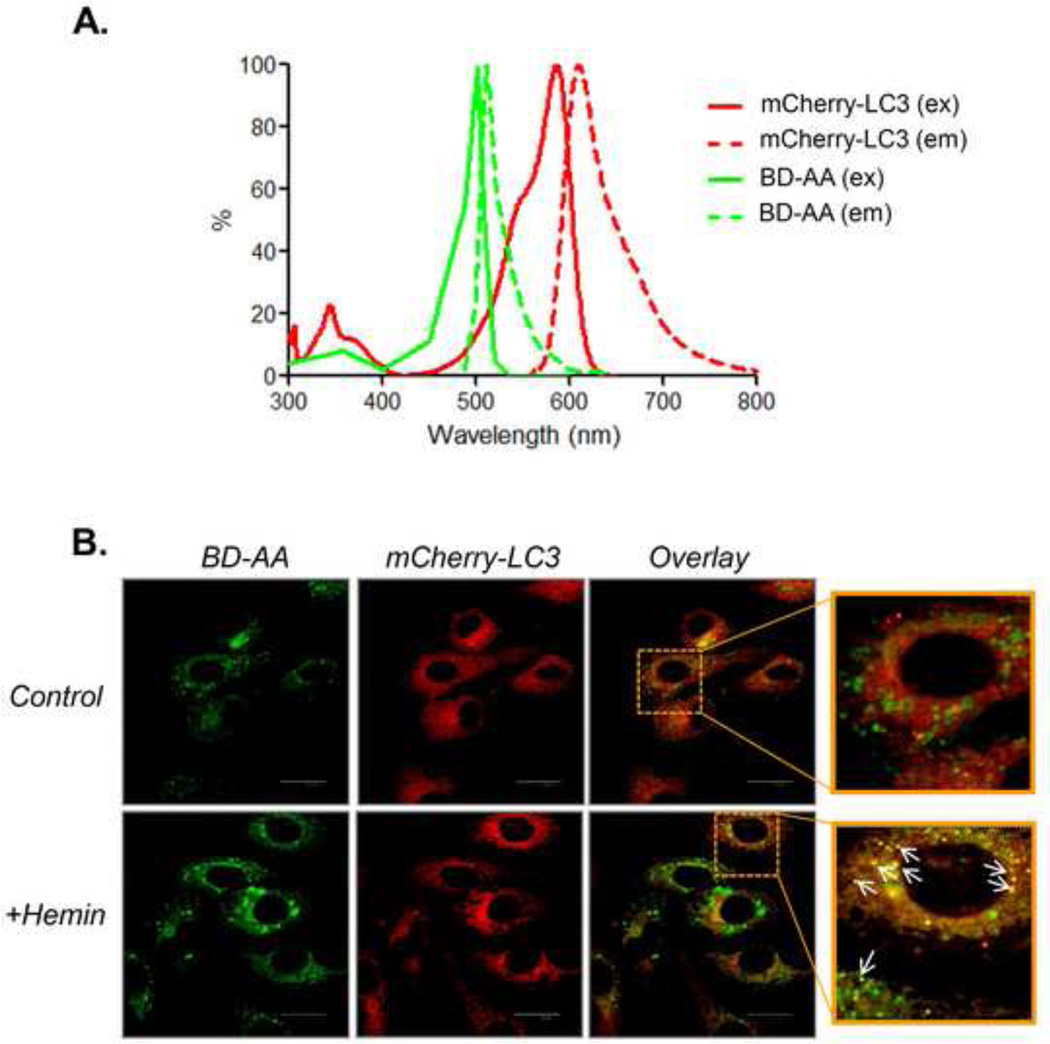
In some experiments, it is useful to transfect cells with reporters such as fluorescently labeled LC3, which forms puncta during times of increased autophagy [38]. The cells can then be stressed with an oxidant or a condition that produces oxidative stress, and the localization of BD-AA can be assessed with respect to fluorescently labeled autophagosomal structures. Shown in Fig. 9 is an example of such an experiment. Here, endothelial cells were transfected with mCherry-LC3 and loaded with BD-AA after 48 h. The spectra of each of these fluorophores are shown in Fig. 9A. The cells were then exposed to vehicle or hemin for 4 h. The localization of the BD-AA with respect to mCherry-LC3 was then assessed by confocal microscopy (Fig. 9B). Hemin has been shown to induce autophagy, which was validated by separate biochemical approaches [6], and BD-AA localized to the puncta. These data and supporting data from similar experiments (see [6, 33] for details) suggest that oxidative stress increases autophagy, which serves to degrade proteins modified by lipid peroxidation products. Below is a detailed protocol for the detection and monitoring of BD-AA under basal and oxidant-stressed conditions, including co-labeling with the aforementioned tracking dyes.
Figure 9. Example of the use of BD-labeled probes for examining the role of lipid peroxidation and oxidative stress in biological processes such as autophagy.
(A) Excitation and emission spectra of mCherry and BD-AA. (B) Representative confocal images of live cells: Cultured bovine aortic endothelial cells were transfected with mCherry-LC3 48 h prior to labeling with BD-AA. The cells were then exposed to vehicle (DMSO) or hemin (25 µM) for 3 h. The confocal images were acquired after replacing the medium with BD- and hemin-free medium. Arrows indicate apparent co-localization of BD-AA products with mCherry-LC3 puncta. Shown in Table 2 are the confocal settings used for acquiring these images.
Procedure
The user should have access to a laser confocal microscope with at least two solid state lasers that generate substantially different excitation lines. In our studies, we use either a line scanning Nikon A1 system equipped with a 60× oil immersion objective mounted on TE-2000E2 inverted microscope as described previously [39, 40] or a Leica DMIRBE Microscope with similar capabilities [6].
Cells should be seeded at ~100,000–250,000 cells per dish (depending on expected proliferation rate) for 1–2 days on 35-mm glass-bottom dishes or on glass coverslips.
- Cells should then be incubated in phenol-free DMEM with 0.5–10 µM BD-AA at 37°C for 2 h before addition of any other dyes.
- For mCherry-LC3 imaging, cells can be seeded per step 1, then at ~75% confluence, they should be transfected with the desired plasmid.
- Carry out transfection per manufacturer’s instructions or in a self-standardized protocol. For the results shown in Fig. 9, the transfection was carried out with 2 µg of plasmid DNA, which was incubated in 150 µl of serum-free DMEM for 5 min at room temperature. The diluted plasmid DNA and lipofectamine were then mixed gently and incubated for 20 min at room temperature. The DNA-lipofectamine mixture was added to cells, and, after 6 h, the medium was removed and replaced with DMEM containing 10% FBS. Transfection efficiency was determined by fluorescence microscopy the following day. The cells were then trypsinized and plated at 5 × 104 cells/well of a 4-well chamber slide. Confluence was ~70% at the time of imaging. Note: Optimization of transfection conditions will be necessary depending on cell type and desired confluence.
- Image cells by confocal microscopy. If desired, the experimenter may then determine the effects of oxidative stress on subcellular localization of lipid peroxidation products. An example of this is shown in Fig. 9B. Here, bovine aortic endothelial cells (BAECs) expressing mCherry-LC3 were incubated with 10 µM BD-AA. The cells were then exposed to vehicle (DMSO) or 25 µM hemin for 3 h prior to imaging. Note transfection efficiency should be optimized for cell type, confluence and DNA-transfection reagent ratios.
For organelle (Mito ID, Hoechst or ER tracker) and cell tracker dyes, add directly into medium with BD-AA for the final 15–30 min of the incubation period. Agitate diluent into medium to evenly distribute dye.
The concentrations of tracking dyes used in Fig. 8 are as follows: ER-tracker was diluted to 500 nM, Cell Tracker Blue was used at 1 µM, Mito ID was used at 1:10000 (from manufacturer’s stock), and Hoechst 33324 was used at final concentration of 1 µg/ml.
Cells were washed 2× in sterile PBS.
Post imaging, all conditions were adjusted equally for image quality, brightness and scaled for comparative analysis. Note: optimization of dye tracker concentration, labeling times and other parameters such as buffer pH and media conditions may be required to improve dye compatibility and image quality.
See Table 2 for the details pertaining to each probe/dye and the specific conditions for excitation and emission data collection. For most confocal experiments, images were natively averaged at 2×, laser offset was maintained at 0, image size was 1024, pinhole size was 36.1 µm, channel series was ON, and pixel dwell time was 1 µs.
Table 2.
Details of confocal microscopy protocol for each dye or labeling component that was combined with BD-AA.
| Probe/Dye | Excitation line (nm) |
Emission-BP (nm) |
Power | HV |
|---|---|---|---|---|
| BD-AA | 488 | 525/50 | 0.3 | 100 |
| mCherry-LC3 | 543 | 630/60 | 4 | 90 |
| Hoechst | 405 | 450/50 | 2.5 | 100 |
| ER Tracker Blue-White | 405 | 450/50 | 2.5 | 100 |
| Mito ID Red | 561 | 700/75 | 4 | 100 |
| Cell Tracker Blue | 405 | 450/50 | 5 | 100 |
HV=photomultiplier tube voltage, BP=Band pass
E. Calculations and Expected Results
To determine differences in protein modifications (such as that induced by HNE or BD-IAM) following imaging, we utilized Image Quant TL (Total Lab GE Healthcare) and centered a single band for the entire lane intensity measurements in arbitrary units (au). Background subtraction is of critical importance, in some cases manual baseline background subtraction may be optimal depending on intensity of BODIPY bands, lower intensity bands should be manually scrutinized for optimal background removal.
F. Caveats and other considerations
This protocol describes the use of BD-labeled probes for the detection of proteins modified by reactive lipids species and to determine which reactive protein thiols are most readily oxidized. However, it may be useful, depending on the experiment or intent, to use other types of probes to interrogate the thiol proteome. For instance, a stronger electrophile, such as BODIPY-labeled NEM may be used to label a wider range of protein thiols and identify changes that IAM may miss [31]. In addition, these protocols could easily be adapted and combined with other fluorophore-labeled probes and protocols to provide further insight into and to quantify potentially important protein thiol modifications. For example, it is possible to extend this protocol not only to detect the overall thiol proteome that is modified but also to begin to discern if those proteins that are S-nitrosated, -glutathiolated, or -oxidized. Disulfide-forming probes or probes similar to those used in protein spin labeling studies such as biotinylated glutathione, methyl methanethiosulfonate, or N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)propionamide may be used alone or in concert to detect specific protein modifications [31, 41, 42]. Other specific derivatization agents, e.g., dimedone or dimedone-like compounds [43], or reductants, e.g., ascorbate [44], could be used to detect and discriminate between reversible posttranslational protein modifications such as S-nitrosation and sulfenylation.
A long-standing problem in understanding how oxidative modifications impact protein, cell, and tissue function is the determination of the extent of modification required to elicit a biologically relevant effect. For example, it is possible that 10% of the total pool of a certain enzyme is inhibited by a thiol modification; yet, this may not have a sizeable effect on pathway flux or cell function. Such a modification would readily be detected in positive labeling strategies, where the detection signal would be well above background. The concern then is that the positive labeling approach could lead an investigator to misinterpret data and place undue importance on a particular modification to a protein or enzyme. In this respect, the indirect labeling approach is advantageous because it can be used to determine the extent of protein thiol modification on a particular protein or subproteome. The indirect approach, however, does require that a fairly substantial proportion of a protein or subproteome is modified to detect a robust difference between groups. Using both indirect and direct approaches can maximize the ability to determine the significance of a particular modification to protein and cell function.
The use of organelle tracker dyes and fluorescent probes should be tested over the range of different stimuli concentrations, time points, temperatures and incubation periods to characterize potential effects attributed to the probe alone compared with those from experimental stimuli. It is particularly important in confocal experiments to control for artifact autofluorescence of the cells themselves. Therefore, in every experiment, it is important to include unstained cells as controls. Moreover, some oxidant stimuli such as hemin are excitable and have fluorescent properties, and these should be included as control samples as well. Empirical determination of the dose of hemin generating minimal auto-fluorescence or spectral interference with BODIPY or other labeling fluorophores should be conducted before integrating into final protocols.
In the above protocols, we generally use 10–20 µM BD-AA or BD-IAM to determine labeling and adduct formation in vitro or in situ. One consideration would be to determine a dose curve of BODIPY for each condition especially when considering new electrophilic target proteomes or protocol development. For confocal experiments, it is prudent to optimize the BODIPY probe concentrations and minimize potential artifact labeling in the cell type of interest and (if desired) the oxidant stimulus.
Lastly, it is important to discuss the reliability of colocalization. The word “colocalization” is used to describe the existence of two or more different molecules in a very close spatial position in the cell [45]. So how close must the two be to truly be colocalized? The phenomenon behind this is quite possibly one of the most misrepresented areas of current basic research, with the most common misconception being that colocalization of antigens equals sharing of their functional characteristics [46]. Therefore, caution and additional biochemical methods are generally required to ensure that the conclusions taken from confocal colocalization studies are accurate. Should more quantitative colocalization studies be required, additional steps should be taken to calculate colocalization coefficients and to perform reliable quantitative colocalization analysis using appropriate software such as CoLocalizer Pro [47].
Highlights.
A method was developed to quantify oxidative modifications using fluorescent tags.
BODIPY-electrophile conjugates track endogenous protein thiol modifications.
Fluorescent lipid peroxidation substrates were created to identify redox-sensitive proteomes.
Organelle-specific proteomes reactive with lipid species can be examined by confocal microscopy.
BODIPY-arachidonic acid can be used to examine autophagic responses to oxidative stress.
ACKNOWLEDGMENTS
The authors acknowledge funding from the following sources: BGH was supported by a grant from the NIH-NCRR (P20 RR024489), VDU was supported by NIH Grants (ES10167, AA13395, DK 75865 and HL109785), JZ was supported by NIH NS064090 and a VA merit award, and LD-I was supported by NIH HL097176 and HL109785.
Abbreviations
- AA
arachidonic acid
- ACM
adult cardiac myocytes
- AR
aldose reductase
- BD or BODIPY FL-EDA, 4
4-difluoro-4-bora-3a,4a-diaza-s-indacene-3-propionyl ethylenediamine hydrochloride
- EDC
1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride
- BAEC
bovine aortic endothelial cells
- BD-AA
BODIPY-arachidonic acid
- BD-IAM
BODIPY-iodoacetamide
- BHT
butylated hydroxytoluene
- BSA
bovine serum albumin
- DIC
differential interference contrast
- DTPA
diethylenetriaminepentaacetate
- DTT
dithiothreitol
- EDTA
ethylenetriaminepentaacetate
- ER
endoplasmic reticulum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HNE
4-hydroxy-2-trans-nonenal
- IAM
iodoacetamide
- NEM
N-ethylmaleimide
- PBS
phosphate-buffered saline
- PD-10
Sephadex G25 PD-10 column
- RASMC
rat aortic endothelial cells
- ROS
reactive oxygen species
- RLS
reactive lipid species
- TBS
Tris-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Higdon A, Landar A, Barnes S, Darley-Usmar VM. The Electrophile Responsive Proteome: Integrating Proteomics and Lipidomics with Cellular Function. Antioxidants & redox signaling. 2012 doi: 10.1089/ars.2012.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higdon A, Diers AR, Oh JY, Landar A, Darley-Usmar VM. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. The Biochemical journal. 2012;442:453–464. doi: 10.1042/BJ20111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickinson DA, Darley-Usmar VM, Landar A. The covalent advantage: a new paradigm for cell signaling by thiol reactive lipid oxidation products. In: Dalle-Donne I, Scalone A, Butterfield DA, editors. Redox Proteomics: from Protein Modifications to Cellular Dysfunction and Diseases. Indianapolis: John Wiley & Sons, Inc.; 2006. pp. 345–367. [Google Scholar]
- 4.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 5.Stark G. Functional consequences of oxidative membrane damage. J Membr Biol. 2005;205:1–16. doi: 10.1007/s00232-005-0753-8. [DOI] [PubMed] [Google Scholar]
- 6.Higdon AN, Benavides GA, Chacko BK, Ouyang X, Johnson MS, Landar A, Zhang J, Darley-Usmar VM. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Heart Circ Physiol. 2012;302:H1394–H1409. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landar A, Shiva S, Levonen AL, Oh JY, Zaragoza C, Johnson MS, Darley-Usmar VM. Induction of the permeability transition and cytochrome c release by 15-deoxy-Delta12,14-prostaglandin J2 in mitochondria. Biochem J. 2006;394:185–195. doi: 10.1042/BJ20051259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diers AR, Higdon AN, Ricart KC, Johnson MS, Agarwal A, Kalyanaraman B, Landar A, Darley-Usmar VM. Mitochondrial targeting of the electrophilic lipid 15-deoxy-Delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. The Biochemical journal. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueraud F, Atalay M, Bresgen N, Cipak A, Eckl PM, Huc L, Jouanin I, Siems W, Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010;44:1098–1124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh JY, Giles N, Landar A, Darley-Usmar V. Accumulation of 15-deoxy-delta(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem J. 2008;411:297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond SP, Allen TD. From live-cell imaging to scanning electron microscopy (SEM): the use of green fluorescent protein (GFP) as a common label. Methods Cell Biol. 2008;88:97–108. doi: 10.1016/S0091-679X(08)00406-8. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman RM, Yang M. Subcellular imaging in the live mouse. Nat Protoc. 2006;1:775–782. doi: 10.1038/nprot.2006.109. [DOI] [PubMed] [Google Scholar]
- 16.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 17.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Grieshaber S, Mathews CE. Methods to assess beta cell death mediated by cytotoxic T lymphocytes. J Vis Exp. 2011 doi: 10.3791/2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marban E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson M, Kurz T, Brunk UT, Nilsson SE, Frennesson CI. What does the commonly used DCF test for oxidative stress really show? Biochem J. 2010;428:183–190. doi: 10.1042/BJ20100208. [DOI] [PubMed] [Google Scholar]
- 21.Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, Kalyanaraman B. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem. 2012;287:2984–2995. doi: 10.1074/jbc.M111.309062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielonka J, Sikora A, Joseph J, Kalyanaraman B. Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J Biol Chem. 2010;285:14210–14216. doi: 10.1074/jbc.M110.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higdon AN, Dranka BP, Hill BG, Oh JY, Johnson MS, Landar A, Darley-Usmar VM. Methods for imaging and detecting modification of proteins by reactive lipid species. Free Radic Biol Med. 2009;47:201–212. doi: 10.1016/j.freeradbiomed.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landar A, Oh JY, Giles NM, Isom A, Kirk M, Barnes S, Darley-Usmar VM. A sensitive method for the quantitative measurement of protein thiol modification in response to oxidative stress. Free Radic Biol Med. 2006;40:459–468. doi: 10.1016/j.freeradbiomed.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 27.Itoh N, Cao J, Chen ZH, Yoshida Y, Niki E. Advantages and limitation of BODIPY as a probe for the evaluation of lipid peroxidation and its inhibition by antioxidants in plasma. Bioorg Med Chem Lett. 2007;17:2059–2063. doi: 10.1016/j.bmcl.2007.01.080. [DOI] [PubMed] [Google Scholar]
- 28.Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic Biol Med. 2002;33:473–490. doi: 10.1016/s0891-5849(02)00848-1. [DOI] [PubMed] [Google Scholar]
- 29.Drummen GP, Gadella BM, Post JA, Brouwers JF. Mass spectrometric characterization of the oxidation of the fluorescent lipid peroxidation reporter molecule C11-BODIPY(581/591) Free Radic Biol Med. 2004;36:1635–1644. doi: 10.1016/j.freeradbiomed.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Benniston AC, Copley G. Lighting the way ahead with boron dipyrromethene (Bodipy) dyes. Phys Chem Chem Phys. 2009;11:4124–4131. doi: 10.1039/b901383k. [DOI] [PubMed] [Google Scholar]
- 31.Hill BG, Reily C, Oh JY, Johnson MS, Landar A. Methods for the determination and quantification of the reactive thiol proteome. Free Radic Biol Med. 2009;47:675–683. doi: 10.1016/j.freeradbiomed.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill BG, Haberzettl P, Ahmed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 34.Vladykovskaya E, Sithu SD, Haberzettl P, Wickramasinghe NS, Merchant ML, Hill BG, McCracken J, Agarwal A, Dougherty S, Gordon SA, Schuschke DA, Barski OA, O'Toole T, D'Souza SE, Bhatnagar A, Srivastava S. The lipid peroxidation product-4-hydroxy-trans-2-nonenal causes endothelial activation by inducing endoplasmic reticulum stress. J Biol Chem. 2012 doi: 10.1074/jbc.M111.320416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodur C, Kutuk O, Tezil T, Basaga H. Inactivation of Bcl-2 through IkappaB kinase (IKK)-dependent phosphorylation mediates apoptosis upon exposure to 4-hydroxynonenal (HNE) J Cell Physiol. 2012 doi: 10.1002/jcp.24057. [DOI] [PubMed] [Google Scholar]
- 36.Patel RP, Svistunenko DA, Darley-Usmar VM, Symons MC, Wilson MT. Redox cycling of human methaemoglobin by H2O2 yields persistent ferryl iron and protein based radicals. Free Radic Res. 1996;25:117–123. doi: 10.3109/10715769609149916. [DOI] [PubMed] [Google Scholar]
- 37.Moore KP, Holt SG, Patel RP, Svistunenko DA, Zackert W, Goodier D, Reeder BJ, Clozel M, Anand R, Cooper CE, Morrow JD, Wilson MT, Darley-Usmar V, Roberts LJ., 2nd A causative role for redox cycling of myoglobin and its inhibition by alkalinization in the pathogenesis and treatment of rhabdomyolysis-induced renal failure. J Biol Chem. 1998;273:31731–31737. doi: 10.1074/jbc.273.48.31731. [DOI] [PubMed] [Google Scholar]
- 38.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 39.Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM, Long BW, Prabhu SD, Xuan YT, Jones SP. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci U S A. 2010;107:17797–17802. doi: 10.1073/pnas.1001907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Facundo HT, Brainard RE, Watson LJ, Ngoh GA, Hamid T, Prabhu SD, Jones SP. O-GlcNAc Signaling is Essential for NFAT-Mediated Transcriptional Reprogramming During Cardiomyocyte Hypertrophy. Am J Physiol Heart Circ Physiol. 2012 doi: 10.1152/ajpheart.00775.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eaton P. Protein thiol oxidation in health and disease: techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radic Biol Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 42.Ying J, Clavreul N, Sethuraman M, Adachi T, Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charles RL, Schroder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001 doi: 10.1126/stke.2001.86.pl1. pl1. [DOI] [PubMed] [Google Scholar]
- 45.Smallcombe A. Multicolor imaging: the important question of colocalization. Biotechniques. 2001;30:1240–1242. 1244–1246. doi: 10.2144/01306bt01. [DOI] [PubMed] [Google Scholar]
- 46.North AJ. Seeing is believing? A beginners' guide to practical pitfalls in image acquisition. J Cell Biol. 2006;172:9–18. doi: 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinchuk V, Grossenbacher-Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr Protoc Cell Biol. 2011 doi: 10.1002/0471143030.cb0419s52. Chapter 4, Unit4 19. [DOI] [PubMed] [Google Scholar]