Abstract
Since their description in the late 1990s, human artificial chromosomes (HACs) carrying a functional kinetochore were considered as a promising system for gene delivery and expression with a potential to overcome many problems caused by the use of viral-based gene transfer systems. Indeed, HACs avoid the limited cloning capacity, lack of copy number control and insertional mutagenesis due to integration into host chromosomes that plague viral vectors. Nevertheless, until recently, HACs have not been widely recognized because of uncertainties of their structure and the absence of a unique gene acceptor site. The situation changed a few years ago after engineering of HACs with a single loxP gene adopter site and a defined structure. In this review, we summarize recent progress made in HAC technology and concentrate on details of two of the most advanced HACs, 21HAC generated by truncation of human chromosome 21 and alphoidtetO-HAC generated de novo using a synthetic tetO-alphoid DNA array. Multiple potential applications of the HAC vectors are discussed, specifically the unique features of two of the most advanced HAC cloning systems.
Keywords: Human artificial chromosomes, HAC, Gene expression, Gene delivery vector
Introduction
Genetic manipulation with human cells aiming to complement a gene deficiency or to re-program the cells requires gene expression in a physiologically regulated fashion represents one of the most challenging tasks in modern medicine. It is generally believed that such expression can be achieved only with full-length genes containing all their regulatory sequences, and under conditions where only one copy of the gene is introduced into each gene-deficient cell. The majority of currently used gene delivery systems do not match these requirements. For example, the most advanced adenovirus-, lentivirus-, and retrovirus-derived vectors employ cDNA or ‘minigene’ constructs [1–9] that cannot recapitulate the physiological regulation of endogenous loci. Viral episomal vectors carrying herpes simplex virus type 1 (HSV-1) and Epstein-Barr virus (EBV) amplicons can deliver and express full-length genes up to ~150 kb in size [10, 11]. However, HSV-1 and EBV viral vectors lack strong copy number control. In addition, the use of viral vectors may induce undesired immunological responses and occasional integration of the vector sequences into the host genome, causing insertional mutagenesis and gene silencing [12–16].
Human artificial chromosomes (HACs) represent an alternative system for gene delivery and expression with a potential to overcome many of the problems caused by the use of viral-based gene transfer systems [17–23]. All HACs by definition contain a functional centromere that provides them several advantages over currently used episomal viral vectors for gene function studies and gene therapy applications. Firstly, the presence of a functional centromere enables the long-term stable maintenance of HACs as single copy episomes without integration into the host chromosomes, thereby minimizing such complications as silencing of the therapeutic gene. Secondly, there is no upper size limit to DNA cloned in a HAC: entire genomic loci with all regulatory elements can be used that faithfully mimic the normal pattern of the natural gene expression. Indeed, not only single genes but groups of genes encoding complex pathways can be carried on a single HAC. Thirdly, the HACs can be transferred from one cell to another. Finally, because of the lack of viral sequences, HAC vectors minimize adverse host immunogenic responses and the risk of cellular transformation.
Despite these obvious advantages over viral vectors, until recently, HACs had been used solely for studies of the structure and function of human kinetochores [24–26]. Their use for gene function studies was limited by technical difficulties of gene loading into the HAC and the ill-defined structures of the HACs, although the feasibility of some HACs for gene delivery and gene expression has been demonstrated [17–23, and references therein]. This situation changed several years ago after methods were developed for engineering of HACs with a predefined structure and a unique gene loading site.
In this review, we summarize recent advances in HAC technology leading to the construction of two of the most advanced HAC vectors: the 21HAC based on a truncated human chromosome 21 [27] and the alphoidtetO-HAC based on a de novo kinetochore formed using a synthetic alphoid DNA array [28]. Specific features of the HAC systems are discussed as well as multiple potential applications of the HAC vectors.
Two different types of HACs
Various groups worldwide have developed human artificial chromosomes, and the potential of the HAC technology has been extensively reviewed [17–23]. HACs can be engineered by “top-down” and “bottom-up” (or by de novo formation by centromere seeding) approaches. Both processes generate HACs with a functional centromere that maintain their nuclear location and participate in mitotic and meiotic segregation.
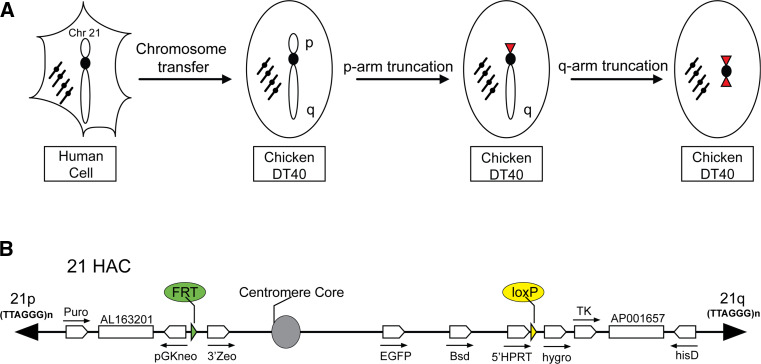
The top-down approach has typically involved a telomere-associated chromosome fragmentation technique in the homologous recombination-proficient chicken DT40 cell line (Fig. 1a). When a vector carrying a cloned telomeric DNA sequence (TTAGGG)n is introduced into cells, the distal portion of the natural chromosome arm is truncated at the vector integration site and a new telomere is formed. As a result, the majority of the sequence of the p- and q- arms can be removed. This approach can generate linear minichromosomes or HACs ranging in size from 0.5 to 10 Mb that retain functional centromeres and are mitotically stable in human and mouse cells. Such HACs have been produced from chromosomes X, Y, 14, and 21 [29–37]. The most advanced top-down HAC in which almost all pericentromeric regions are removed was recently constructed in the Oshimura laboratory by several rounds of the telomere-directed breakage of human chromosome 21 (Fig. 1b) [27].
Fig. 1.
An example of construction of the engineered human artificial chromosome via top-down approach. a The human chromosome 21 was transferred from human cells to recombination-proficient chicken DT40 cells. The HAC was generated by subsequent truncation of the p- and q- arms by targeting constructs in DT40 cells. During arms truncation, a loxP gene loading site was inserted into the HAC in DT40 cells. After the HAC transfer to Chinese hamster ovary (CHO) (hprt −/−) cells, a desired gene can be cloned into the loxP site of the HAC by Cre-loxP-mediated gene insertion. Then, the HAC with a gene of interest can be transferred to desired recipient cells via a micro-cell-mediated transfer technique (MMCT) for gene complementation assays. b A map of the linear 21HAC vector. The TEL/Δq-hisD and TEL/Δp-PGK-Puro constructs were used for chromosome 21 truncation. Two remaining pericentromeric contigs, AP001657 and AL163201, are shown. The 5′HPRT-loxP-Hyg-Tk and pGKneo-FRT-3′Zeo cassettes with loxP and FRT gene loading sites were included in the HAC by homologous recombination. Cre-loxP-mediated gene insertion is accompanied by reconstitution of the HPRT gene. FLP-FRT-mediated gene insertion is accompanied by reconstitution of the function of the Zeo gene. The HAC is marked by the EGFP gene
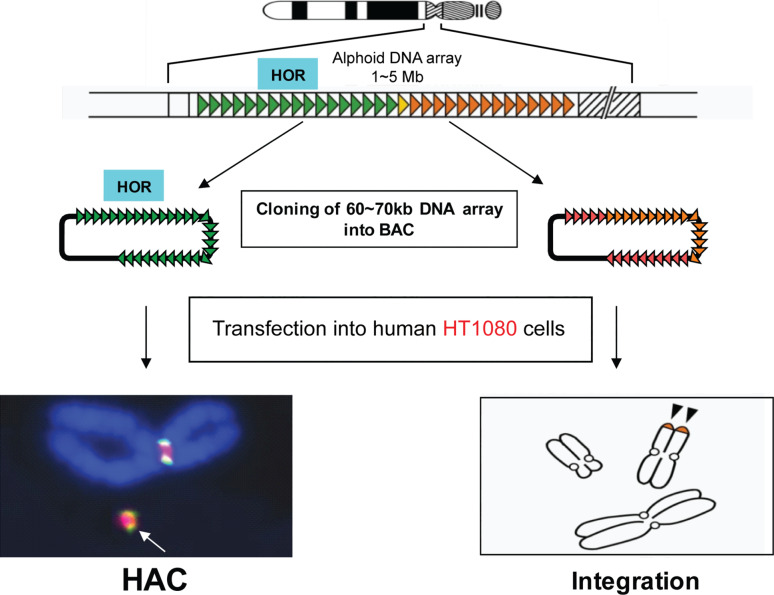
The bottom-up approach of HAC generation, initially developed in human fibrocarcoma HT1080 cells, includes transfection of human cells with either natural high-order repeat (HOR) or synthetic alpha-satellite (alphoid) DNA of 30–200 kb cloned into a circular BAC vector or into a linear YAC vector carrying telomeric sequences (Fig. 2) [38–47]. In all cases, HAC formation (a circular HAC if a BAC is used or a linear HAC if a YAC is used) was accompanied by 20- to 30-fold multimerization of the input DNA. Over the past 14 years, several groups have reported the successful generation of de novo constructed HACs from different substrates [38–47]. The size of those HACs ranges in size from 1 to 10 Mb, and the HACs have been shown to be mitotically stable both in human and rodent cells. However, although protocols for de novo HAC formation have been significantly improved, including the use of a HSV1 amplicon for delivery of alphoid DNA constructs [48], the main limitation—the restriction of HAC formation to a single cell type HT1080—has remained. A recent study has now provided a breakthrough in overcoming this limitation. Masumoto and colleagues [49] discovered that in HT1080 cells the level of H3K9me3 on alphoid DNA is substantially lower than in other human cell lines. Although the critical kinetochore histone, CENP-A can be assembled on transfected alphoid DNA in a variety of cell lines, it remains stable and seeds kinetochore formation only in HT1080 cells. In other types of human cells, heterochromatin enriched by H3K9me3 and HP1 is assembled quickly on the transfected alphoid DNA array preventing CENP-A retention and HAC formation. In a recent technical breakthrough, tethering of histone acetyltransferases (HATs) to the input alphoid DNA arrays breaks this cell-type-specific barrier for de novo CENP-A assembly and allows assembly of other kinetochore proteins leading to HAC formation in a wide range of cell types.
Fig. 2.
Generation of human artificial chromosomes via bottom-up approach. Each human chromosome contains a centromere consisting of identical (high-order repeats or HOR) and diverged alphoid DNA repeats that form an array of 0.5–5 Mb in size. A HOR DNA array of 30–200 kb in size isolated as a BAC forms a HAC after its transfection into human fibrocarcoma HT1080 cells. HAC formation is accompanied by BAC DNA multimerization up to 1–5 Mb. Highly diverged alphoid DNA arrays lacking CENP-B boxes do not form a HAC. Instead, such arrays randomly integrate into human chromosomes
HACs for correction of gene-deficiencies in recipient human cells
Several studies have demonstrated the use of top-down and de novo generated HACs for delivery and expression of genes in human gene-deficient cell lines. The first reports on insertion of the full-length genes into HACs and subsequent complementation analyses were reported by the Cooke and Larin groups [42, 50], who demonstrated a functional expression of the HPRT gene from the HAC in HPRT-deficient human HT1080 cells. In both studies, the HPRT-containing HACs were circular, stable for several months, and functionally complemented the metabolic deficiency of the host cells. Later on, Okazaki and colleagues generated a HAC containing the GCH1 gene [51], and demonstrated that its expression was induced by interferon-gamma as expected, thus mimicking the regulation of this gene at its natural chromosome location. A comprehensive list of genes loaded on HAC vectors is available in the recent review by Kazuki and Oshimura [22]. Among them, the following genes that are represented as genomic copies that include all regulatory elements: human beta-globin [46], CFTR [52, 53], Factor IX [54], STAT3 [55], TP53 [56], DYS [57], VHL, and NBS1 [58]. However, in almost all cases, the copy number of the gene in the HAC was not precisely controlled either because of the presence of multiple gene accepter sites in the HAC or because the gene was inserted into the HAC during its de novo formation, i.e. the gene in a BAC vector was co-transformed with a BAC containing alphoid DNA into human HT1080 cells. (Remember that vector sequences are typically amplified 20–30 times during de novo HAC formation.)
HACs for animal transgenesis
An increasing number of laboratories around the world employ the mouse as a model of human diseases. Therefore, construction of mice carrying human genes to model specific diseases would be the first step towards future application of the HAC in curing hereditary human diseases. One prerequisite for these studies was the demonstration that HACs are mitotically stable not only in human cells but also in rodent and chicken cells.
Kuroiwa and colleagues were the first to demonstrate introduction of a HAC carrying a large human fragment into mouse ES cells as well as in mice [34]. Specifically, they developed a chromosome-cloning system in which a defined 10-Mb-sized region of the chromosome 22 carrying the IgH genes was transferred into a linear chromosome 14-derived HAC in homologous recombination-proficient chicken DT40 cells. This HAC was stably maintained in mouse ES cells and subsequently in mice. Furthermore, the authors could show functional expression of the genes from the HAC as well as expression of human antibodies in mice. In a later study, the same group of researchers applied a similar technology to cows to produce human immunoglobulin and antigen-specific human polyclonal antibodies from hyperimmunized cattle [59, 60].
Several other groups also succeeded in creation of transgenic mice using de novo constructed HACs carrying human beta-globin [60], GCH1 [51, 61], albumin [62], and CSN2 [63] genes. They demonstrated that the HACs have been transmitted over more than three generations through the mouse germline providing evidence of the meiotic stability of the HACs in vivo. A particularly impressive example is development of mice carrying a HAC expressing the human dystrophin (DYS) gene [57, 64] (discussed in more detail below).
To summarize, the proven availability of HAC vectors to carry certain genes in animals provides an opportunity to develop specific human disease models and also for use in commercial production of therapeutic products. Moreover, human artificial chromosome-based transgenesis can be used to identify genes responsible for recessive phenotypes by complementation or expression of dominant phenotypes. This approach is also applicable to generate reporters for tracking gene expression and to implement subtle changes in regulatory and structural sequences in a near endogenous context.
The most advanced top-down HAC
Two characteristics are needed for HAC vectors to be suitable for gene function studies as well as for human gene and cell therapy: (1) the presence of a single gene loading site for efficient insertion of a gene of interest and (2) complete mapping of the HAC structure.
So far, only a few top-down and bottom-up HACs with a unique gene acceptor site have been developed [36, 65–68]. As mentioned above, the most advanced top-down HACs with a unique gene loading site were constructed by Oshimura’s group [36, 66, 67]. These HACs were generated by telomere-directed breakage of human chromosome 21 in DT40 cells, with the first two HACs, 21ΔqHAC and 21ΔpqHAC, having a size of approximately 10 Mb. Next, a unique gene adaptor (a single loxP sequence—the target sequence for site-specific recombination by Cre recombinase) along with a promoterless Neo gene was introduced into the euchromatic region of the remaining q-arm. As a result, any circular DNA with a loxP site and a promoter can restore the Neo gene expression by Cre-mediated insertion at the loxP site of these HACs. The HACs exhibit a high mitotic stability during their propagation in a variety of host cell types [36, 66, 67]. As a proof of principle, expression of the human erythropoietin (EPO) gene inserted into a loxP site of the 21ΔpqHAC has been reported [69].
Application of these HACs for gene therapy, gene function studies, and animal transgenesis, remained problematic because the exact HAC structure was not determined. Therefore, a novel HAC derivative (21HAC) was developed by a further truncation of the 21ΔqHAC (Fig. 1b) [22]. This novel 5 Mb HAC does not contain any endogenous genes. The final structure of the HAC was determined by a rescue of all remaining p- and q- arm sequences as YACs in yeast using a transformation-associated cloning technique (TAR cloning) followed by sequencing of the TAR-rescued HAC fragments. Such analysis confirmed that no endogenous genes remained in this HAC. Recently, a derivative of the 21HAC was constructed containing a 2.4-Mb genomic fragment encompassing the entire dystrophin gene (DYS) [64]. This HAC was successfully used to correct the genetic deficiency in cells derived from DYS model (mdx) mice and from a human DYS patient [64]. Thus, this recombinant HAC has a potential application in DYS gene therapy.
The most advanced de novo constructed HAC
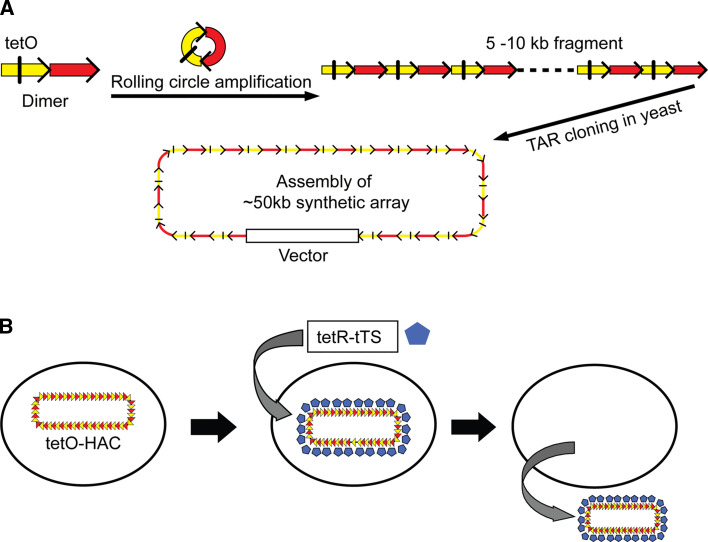
Recently, a new generation HAC, alphoidtetO-HAC, was engineered de novo in HT1080 human cells using a synthetic alphoid DNA array containing tetracycline operator (tetO) sequences embedded into the alphoid DNA [28, 70] (Fig. 3a). One powerful advantage of this HAC is that its centromere can be inactivated by expression of tet-repressor (tetR) fusion proteins (Fig. 3b). Such inactivation results in HAC loss during subsequent cell divisions. This feature of the alphoidtetO-HAC provides a unique possibility to compare the phenotypes of the human cell with and without a functional copy of a rescue transgene, i.e. the phenotypes arising from stable gene expression can be reversed when cells are “cured” of the HAC by inactivating its kinetochore in proliferating cell populations. This provides a control for phenotypic changes attributed to expression of HAC-encoded genes, thereby excluding effects on the endogenous chromosomes following introduction of the HAC.
Fig. 3.
An example of construction of the de novo generated human artificial chromosome via bottom-up approach using a synthetic alphoid DNA array. a Schematic representation of construction of the synthetic tetO-containing DNA tandem repeats array by rolling circle amplification (RCA) in vitro and transformation-associated recombination (TAR) cloning in yeast cells. The first step included amplification of the dimer by RCA up to 5–10 kb in size fragments. One monomer of the dimer is derived from a chromosome 17 alphoid type I 16-mer unit and contains a CENP-B box. The second monomer is a wholly synthetic sequence derived from alohoid DNA consensus, with sequences corresponding to the CENP-B box replaced by a 42-bp tetO motif. The second step included co-transformation of the RCA-amplified fragments into yeast cells along with a vector containing alphoid-specific hooks. End-to-end recombination of alphoid DNA fragments followed by interaction of the recombined fragments with the vector resulted in the rescue of an approximately 50-kb array as a circular molecule in yeast. The targeting vector contains a yeast cassette, HIS3/CEN/ARS (a selectable marker HIS3, a centromere sequence CEN6 from yeast chromosome VI and yeast origin of replication ARSH4) and a mammalian selectable marker (the Blasticidin resistance gene) and a BAC replicon that allows a YAC clone to be transferred into E. coli cells. b The alphoidtetO-HAC loss induced by targeting of the transcriptional silencer (tTS) fused with the tet-repressor (tetR) into the HAC kinetochore. After expression of a chromatin modifier gene (tTS), the HAC is maintained stably when cells are growing in the presence of doxycyclin that prevents binding of tTS to tetO sequences or the HAC is destabilized when cells are growing in the absence of doxycyclin
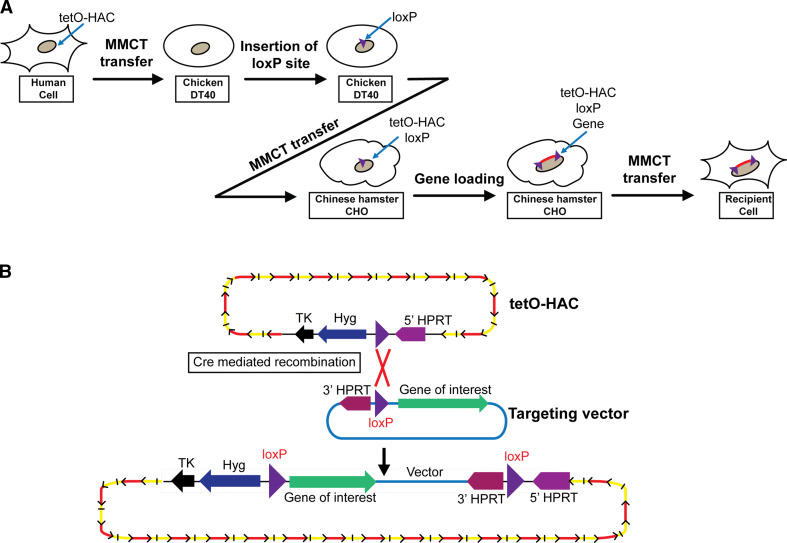
To adopt this HAC for gene delivery and expression studies, a single gene acceptor site (a loxP cassette similar to that described above) was inserted into the alphoidtetO-HAC by homologous recombination in chicken DT40 cells (Fig. 4a) [68]. Next, the HAC was transferred to Chinese hamster ovary (CHO) cells. In these cells, a gene of interest can be easily inserted into the loxP site of the HAC by Cre-mediated recombination (Fig. 4b) and then the HAC can be transferred into different recipient cells. Recently, the capacity of the alphoidtetO-HAC to deliver genomic copies of two human average-size genes, VHL (~25 kb) and NBS1 (~60 kb), and to complement genetic deficiencies in cell lines derived from the patients with deficiencies in either VHL or NBS1, was examined [58]. Functional expression of pVHL and pNBS1 in recipient cells was demonstrated by a set of specific tests based on the known functions of proteins. Importantly, corresponding controls were conducted following targeted and specific elimination (“curing”) of the HAC from the cells following targeted inactivation of its kinetochore. Gene silencing is a major problem limiting the rescue of genetic defects by transgene expression in vertebrate cells. However, no significant changes in the level of expression of pNBS1 and pVHL were detected for more than 6 months after introduction of the alphoidtetO-HAC into patient-derived cell lines. Recently, the alphoidtetO-HAC was physically characterized using a transformation-associated recombination technique (TAR cloning) and Southern blot hybridization analysis [71]. Analysis revealed a specific pattern of the input DNA molecules assembled into a megabase-size array that provides a good control of the HAC integrity during HAC manipulations. Similar patterns had been previously reported for other de novo formed HACs [43, 54, 72, 73]. For therapeutic purposes, it is very important to know whether the HAC structure changes over many months of culture and after multiple transfers from one cell line to another. The epigenetic status of the HAC centromere was analyzed in human HT1080 cells, where the alphoidtetO-HAC was originally formed, and then in the same HT1080 cells after several rounds of MMCT [71]. Immuno-FISH analysis confirmed the assembly of CENP-A chromatin on the HAC sequences in both the starting and final derived cell lines. Chromatin immunoprecipitation (ChIP) analysis also showed a pattern of histone H3 modifications in the final human cell line similar to that of the original alphoidtetO-HAC clone for CENP-A chromatin as well as H3K4me3 (a marker for promoter-proximal transcriptionally active chromatin), H3K4me2 (a marker for open chromatin) and H3K9me3 (a marker for heterochromatin) on alphoidtetO DNA. These results indicate that the alphoidtetO-HAC retained the original histone modification status of its kinetochore after several steps of MMCT. In addition, it was shown that the alphoidtetO-HAC remains circular after several steps of MMCT. Finally, to evaluate the risk of the alphoidtetO-HAC structural rearrangements, the integrity of the alphoidtetO-HAC was checked after its transfer from HT1080 to DT40, from DT40 to CHO, and from CHO into human HT1080 cells. Based on Southern blot hybridization, the majority of clones obtained after MMCT contained a non-rearranged HAC. Thus, once established, the alphoidtetO-HAC remains structurally stable during several months of propagation in host cells and never integrates into the host chromosomes.
Fig. 4.
Insertion of a gene loading site into the alphoidtetO-HAC. a The alphoidtetO-HAC was transferred from human HT1080 cells to recombination-proficient chicken DT40 cells. A loxP gene loading site was inserted into the alphoidtetO-HAC by homologous recombination in DT40 cells. The alphoidtetO-HAC with the loxP site was transferred to hamster CHO (hprt −/−) cells to insert the desired gene. The alphoidtetO-HAC with the gene of interest can be transferred to desired recipient cells via microcell-mediated chromosome transfer technique (MMCT). b A scheme of gene loading into the unique loxP gene acceptor site of the alphoidtetO-HAC. A desired gene can be cloned into the HAC by Cre-loxP mediated gene insertion followed by reconstitution of the HPRT gene
Thus, the alphoidtetO-HAC matches all features required for gene function studies. Because its kinetochore may be regulated, this new generation HAC may be even more suitable for gene function studies than the 21HAC.
HAC delivery into recipient cells
The delivery of transgene-loaded HACs to primary human cells is a fundamental challenge for gene function studies as well as for gene therapy applications. However, the efficiency of the HAC transfer into the desired host cells still remains problematic because of their relatively large size.
At present, the preferred method to move the HAC from donor to recipient cells is microcell-mediated chromosome transfer (MMCT), a technique developed more than 30 years ago [74, 75]. However, the method is tedious and only a restricted number of donor cell lines, including hamster CHO and mouse A9 cells, are suitable for microcell delivery [36, 69]. Therefore, to be “mobilized”, a HAC first must be transferred into one of these rodent cell lines. This technically challenging step has been performed for almost all published HAC vectors. The frequency of the HAC transfer from rodent donor cells into recipient cells varied between 10−5 and 10−4. For example, frequencies of 21HAC transfer from CHO cells to human primary fibroblasts [69] and to human stem cells [56, 66, 76, 77] correspond to 1.26 × 10−4 and 4.0 × 10−4, respectively. Though MMCT is thought to have a low risk of truncation or rearrangement of the transferred chromosome, it is not applicable for all types of recipient cells, particularly those whose fusion with microcells is very inefficient. Two different modifications of MMCT have been developed to overcome these problems. In one work a lipid envelope of an inactivated hemagglutinating virus of Japan (HVJ) that has a similar effect on membrane fusion as PEG was utilized for microcell–cell fusion. This increased the transfer efficiency approximately 3–8 times [78]. However, this modified MMCT procedure is toxic for many recipient cells. Recently, a modification of this procedure involving expression of two measles virus envelop proteins, H and F, in the recipient cells has been described [79]. The H (hemagglutinin) and F (fusion) proteins are essential for both virus attachment and membrane fusion. Following this protocol, the frequency of HAC transfer via MMCT was 30–50 times higher for human donor cells that expressed the surface receptor CD46.
For cells that do not form microcells in response to nocadozole treatment, alternative procedures, including the purification of metaphase chromosomes, were investigated. One of those procedures includes isolation of the target chromosome from the host cells by flow sorting [80]. In principle, this procedure could allow chromosome purification to near homogeneity. However, such purification is accompanied by a significant fragmentation of the metaphase chromosomes. Those random fragments then co-sort with the HAC. This may be one reason why to date no publications have reported the successful use of HACs isolated by flow sorting for functional studies.
Recently, Ikeno and colleagues suggested a more gentle method for the isolation of metaphase HACs suitable for their further transfer into recipient cells [81]. They isolated HACs from metaphase cells using a 25 % sucrose cushion centrifugation. Transfer of HACs into human cells was achieved using conventional transfection reagents, which enabled direct transfer of HACs from a variety of donor cells, including those that do not form microcells as required for MMCT. The efficiency of HAC transfer using this technique was comparable to that of MMCT. Unfortunately, no information is yet available concerning HAC integrity when using this protocol.
Thus, safe and efficient procedures for HAC transfer remain an important challenge for the future.
Selective isolation of full-length genes for their loading into the HAC
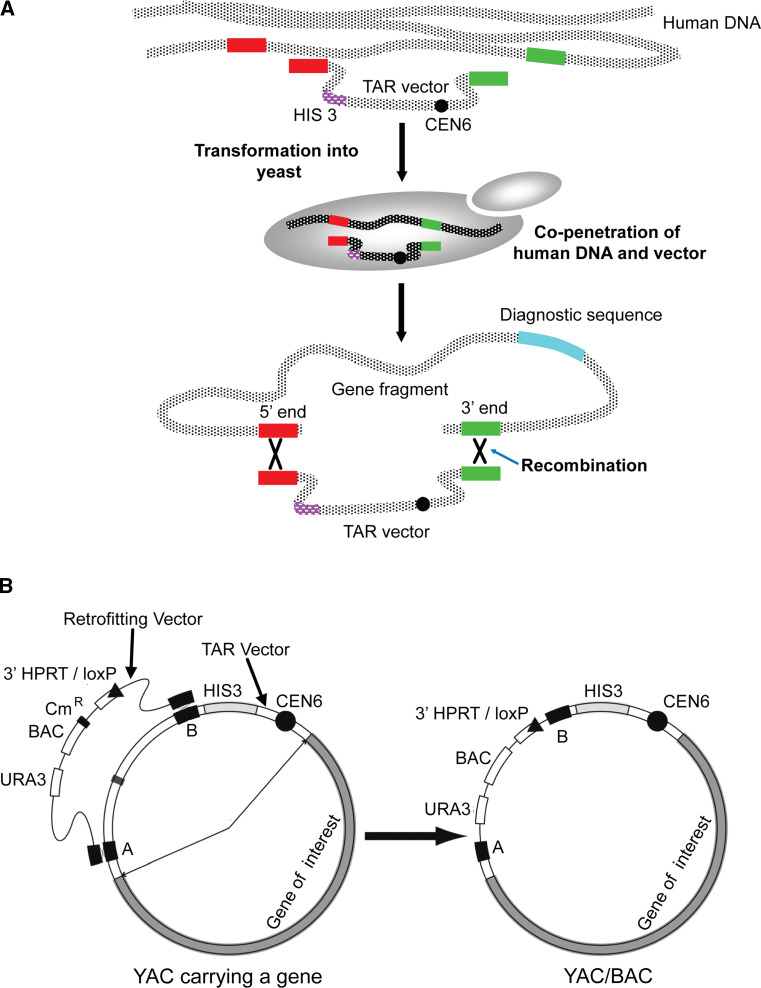
One advantage of HAC technology is the ability to work with full-length genes. More and more publications have appeared describing complex mechanisms regulating gene expression by alternative splicing, alternative promoter-enhancer usage, intronic gene expression, and expression of inhibitory RNAs. At present, the full-length genes including all their regulator elements are available in a BAC or YAC form from BAC and YAC libraraies. However, typically, the BAC/YAC inserts include other genes in addition to the gene of interest, or alternatively the desired gene may be presented only partially with its 5′ or 3′ end deleted during generation of BAC or YAC libraries. A gene cloning technique that is based on transformation-associated recombination (TAR) in the yeast Saccharomyces cerevisiae enables the cloning of defined gene-sized regions of chromosomes [82–84]. The efficient homologous recombination machinery of budding yeast host allows the selective isolation of desired genomic loci from the entire human genome. Recombination between a circularizing TAR cloning vector containing targeting sequences homologous to a region/gene of interest and homologous sequences in the co-transformed human DNA results in rescue of the desired fragment or gene in a circular YAC form (Fig. 5a) that is able to propagate, segregate, and be selected for in yeast cells. The yield of gene-positive clones varies from 1 to 5 %. The entire procedure takes 2–3 weeks to complete once the TAR vector is constructed and genomic DNA is prepared. TAR cloning is a powerful tool to selectively recover chromosome segments and genes up to 200 kb in length from complex genomes.
Fig. 5.
A scheme of selective gene isolation from human genomic DNA by transformation-associated recombination (TAR-cloning) in the yeast S. cerevisiae. a A high-molecular weight human genomic DNA prepared in agarose plugs or in solution is co-transformed along with the TAR vector into yeast spheroplasts. The TAR vector contains a yeast selectable marker HIS3 for selection of yeast transformants on His-minus medium, a yeast centromere for proper propagation of the vector during cell division and two targeting sequences (hooks) homologous to the 5′ and 3′ ends of the gene of interest. After penetration, the targeting hooks of the vector recombine with the homologous sequences of the gene resulting in a rescue of the desired gene as a circular molecule (YAC) in yeast cells. The gene-positive yeast transformants are selected by PCR using diagnostic primers specific for a gene of interest. b Retrofitting of the circular gene-containing YAC into a BAC/YAC. The retrofitting vector contains a yeast selectable marker URA3, a BAC replicon for propagation in bacterial cells, a chloramphenicol gene (Cm), a loxP gene targeting cassette and two targeting sequences homologous to the TAR vector sequence. Transformation of the linearized retrofitting vector into yeast cells containing the YAC with the gene followed by selection on Ura-minus medium results in a rescue of circular molecules in a YAC/BAC form. The YAC/BAC can be transferred from yeast into E. coli cells by electroporation for further BAC DNA isolation
To prepare a YAC TAR isolate containing a gene of interest for its loading into a HAC, the YAC is converted into a BAC form containing a loxP cassette. Conversion of the YAC into a BAC is advantageous because purification of circular DNA molecules is much easier from E. coli than from yeast cells. For this purpose, a yeast–bacteria–mammalian cell shuttle vector is used to retrofit YAC gene-containing isolates into YAC/BACs (Fig. 5b) [58]. The retrofitting vector contains a 3′ HPRT-loxP-eGFP cassette, allowing gene loading into a unique loxP site of the HAC in CHO cells and selection of these events. An appropriate BAC gene-containing construct and a Cre-recombinase expression vector are co-transfected into HPRT-minus CHO cells carrying the HAC and HPRT-positive colonies are selected on HAT medium. Insertion of genes into the HAC is confirmed by PCR using specific primers that detect reconstitution of the HPRT gene (Fig. 4b).
So far, two HAC vectors, a derivative of the truncated chromosome 21 and alphoidtetO-HAC, containing unique acceptor loxP sites have served as a platform for the reproducible expression of TAR-isolated full-length genes, VHL, NBS1, and HPRT [58, 85]. The entire copies of these genes were isolated by TAR cloning, retrofitted with the donor loxP site and loaded onto the HAC vectors by the Cre/loxP system. It was later shown that these HACs complement the genetic defects in patient-derived human cells. Thus, the combination of gene cloning by TAR and HAC vector technology represents a powerful tool for functional genomic studies and potentially for gene therapy.
HAC applications and perspectives
Two recently constructed HAC vectors, the 21HAC and the alphoidtetO-HAC, open new opportunities for HAC-based technology, as they meet an important requirement for safe and controlled gene transfer experiments, i.e. their physical structure is determined. Efficient and accurate gene loading and the subsequent transfer and stable transgene expression in patient-derived human cells as well as in mice have been demonstrated for those HACs [58, 64]. These proof-of concept studies are the first steps towards exploiting HAC potential for further gene transfer and expression studies.
HACs for gene function studies and treatment of monogenic diseases
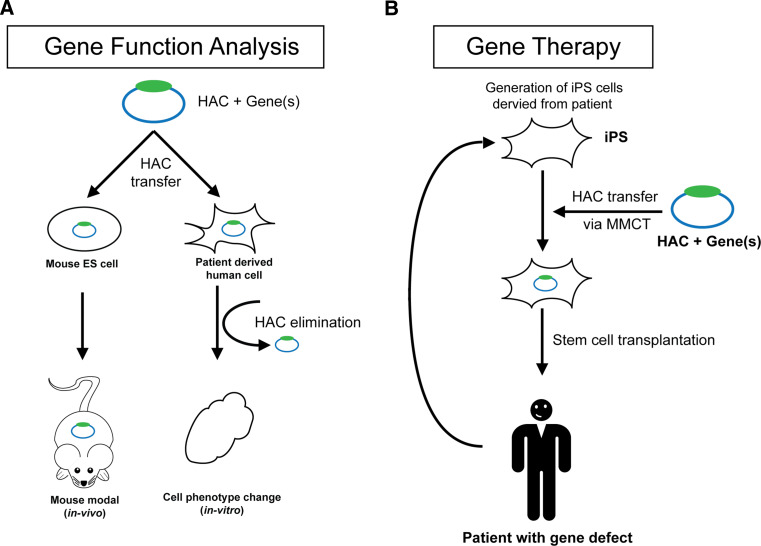
Until now, the functions of novel genes have been deduced mostly based on experiments using viral transfection or integrative transfection by BAC or YAC vectors, where often the gene cannot be expressed at a physiological level because the gene copy number is not regulated and a BAC/YAC transgene randomly integrates into the host genome. Both the above HACs provide a way to overcome this problem [22, 58]. The alphoidtetO-HAC, having a conditional centromere, also provides a way to control for the phenotypic changes attributed to expression of HAC-encoded genes [58]. Construction and physical characterization of these HACs is an initial step towards their future application for treatment of monogenic diseases.
Loading of several genes or entire locus into HACs
For situations in which several genes need to be inserted into the HAC, a multi-integrase HAC vector containing five recombination sites (ΦC31 attP, R4 attP, TP901-1 attP, Bxb1 attP, and FRT) was designed [86]. This 21HAC derivative has several advanced functions, including gene integration in the precise loci, thus avoiding genomic position effects. This vector is also transferable to other cell lines and is capable of accepting genes of interest in those environments. A HAC vector containing multiple integrase sites is also suitable for assembly of a gene which size is bigger than 300 kb from several DNA fragments. It is worth noting that a direct loading of large-size genes into the HAC is very inefficient due to random linearization of large circular DNA molecules during DNA transfection procedure.
A HAC-based iPS protocol meets safety and efficacy requirements
Use of iPS cells [87–90] is in part impeded by the low efficiency of iPS formation using established viral-based iPS protocols and by artifacts caused by vector integration into the host genome. Such integration events greatly increase the risk of tumorigenicity. These problems may potentially be overcome using a new generation of HACs as vectors for the iPS cassette.
Recently, Oshimura and colleagues demonstrated the use of the 21HAC vector for reprogramming mouse embryonic fibroblasts (MEFs) into induced iPS cells [91]. They constructed a HAC vector carrying four reprogramming factors, Oct3/4, Sox2, Klf4, and c-Myc, that efficiently reprogrammed MEFs. Global gene expression patterns showed that the HAC, unlike other vectors, generated relatively uniform iPS cells at a level comparable to that observed with established retrovirus-based iPS protocols. Next, they established HAC-free iPS cells by isolating cells that spontaneously lost the HAC. Analyses of pluripotent markers, teratomas and chimeras, confirmed that these HAC-free iPS cells were pluripotent.
A cell-reprogramming construct containing four transcriptional factors planned to be inserted into the alphoidtetO-HAC. Because the HAC along with the iPS cassette can be efficiently eliminated from the cells after reprogramming by inactivation of the HAC centromere, it provides an advantage over the 21HAC when elimination of the HAC is desired. Previously, elimination of the alphoidtetO-HAC was induced by transfecting cells with a centromere-inactivating module (tetR-tTS) [28, 58]. This procedure could potentially be mutagenic. To refine the alphoidtetO-HAC derivative carrying the cell-reprogramming cassette and avoid the transfection step, a module controlling activity of the HAC centromere was inserted into the alphoidtetO-HAC itself. Our recent data showed that one copy of the tetR-tTS module that is regulated by doxycycline is enough to conditionally inactivate the HAC kinetochore (unpublished data). Thus, derivatives of the alphoidtetO-HAC with conditional centromeres represent a very promising system to develop a new iPS protocol that meets safety and efficacy requirements. HAC-induced stem cells might provide a great potential to address a wide spectrum of monogenic diseases (Fig. 6).
Fig. 6.
Schematic diagram of application of the HAC vector system for gene functional analyses and for the treatment of genetic disorders. a A HAC along with a gene(s) of interest is transferred to desired recipient cells (for example, mouse ES cells or patient-derived human cells deficient for this gene). The HAC containing a gene can be utilized for functional analyses in vitro and in vivo, including a humanized mouse model. For alphoidtetO-HAC, the phenotypes arising from stable gene expression from the HAC can be reversed when the HAC is eliminated from the cells by inactivating its kinetochore that provides a control for phenotypic changes attributed to expression of HAC-encoded genes. b HAC for gene therapy. A HAC containing a therapeutic gene(s) can be utilized for the treatment of patients with genetic disorders. As a first step, pluripotent stem cells are produced from patient fibroblasts using one of the currently available protocols. Then, a HAC carrying a therapeutic gene is introduced into iPS cells from CHO cells using MMCT. A final step includes transplantation of stem cells into the patient
HACs for production of human antibodies and proteins
Antigen-specific human polyclonal antibodies, produced by hyperimmunization, have many potential used in treating a variety of human diseases. Kuroiwa and colleagues [59, 92] demonstrated the feasibility of HAC vectors for production of antigen-specific human polyclonal antibodies from hyperimmunized cattle. The entire human heavy- and light-chain immunoglobulin loci were inserted into a HAC vector generated by truncation of chromosome 14. This HAC vector was introduced into bovine primary fibroblasts, which were rejuvenated and expanded by producing cloned fetuses. Cloned fetal cells were selected and recloned to produce healthy, transchromosomic (Tc) calves. These Tc cattle containing human immunoglobulin genes were shown to produce novel human antibodoes. These results demonstrate the feasibility of HAC vectors for production of large quantities of human polyclonal antibodies. A similar approach could potentially be used for production of proteins that cannot be produced in bacterial and yeast cells.
HACs as an evaluation system for bioactive substances
Another novel application of HACs recently demonstrated by Takahashi and colleagues [93] was development of an evaluation system for bioactive substances widely used in daily food and supplements. Specifically, the authors constructed a HAC vector to evaluate osteogenic activity in food supplements. The osteocalcin gene along with the green fluorescence protein (GFP) regulated by an osteocalcin gene promoter was inserted into a unique acceptor site of the 21HAC. The bioactivity of different fish oil extracts was evaluated by comparison of fluorescence intensity in response to the content of vitamin D3 in oils. Using a HAC-based evaluation system for bioactive substances, the authors could demonstrate osteogenic activity in some fish oils. A similar approach may be applied to evaluate activity of other bioactive compounds.
HACs for screening the drugs affecting chromosome stability
Many cancer cells exhibit an elevated rate of chromosome mis-segregation referred to as chromosome instability or CIN. The altered chromosome number and large-scale changes in gene expression resulting from CIN have been hypothesized to be a major driver of cancer progression, drug resistance, and relapse. While CIN can act as a driver of cancer genome evolution and tumor progression, recent findings point to the existence of a threshold level beyond which CIN becomes a barrier to tumor growth [94, 95]. Therefore, new drugs that further increase the level of CIN in cancer cells may be a useful adjunct to current cancer therapy.
Because the HAC is a non-essential 47th chromosome, and is several orders less stable than natural chromosomes, it may be useful as a sensitive screening tool for identification of drugs affecting CIN. For this purpose, a HAC marked by a color marker (GFP or RFP) would be specifically useful. One such derivative of the alphoidtetO-HAC containing the GFP gene, has been recently exploited in our laboratory to evaluate the effect of anti-microtubule drugs on chromosome stability using a high throughput fluorescence-based assay (unpublished data). This system will be further developed as a screening tool for future identification of novel therapeutic agents that increase CIN in cancer cells and promote lethal aneuploidy.
HACs for optimization of drug treatment
HACs may also be used as a pre-screen tool to study dosage effects of different anti-cancer drugs on oncogenic alleles of certain genes [for example, optimization of treatment of NSCLC (lung cancer) patients with inhibitors of the tyrosine kinases EGFR and HER2]. Certain mutations in EGFR and HER2 genes lead to lung cancer [96]. A set of isogenic cell lines containing the HAC with different numbers of EGFR or HER2 genes or expressing different alleles of these genes is being constructed to test cell response and toxicity to anti-cancer drugs such as iressa or erlotinin. Similar experiments may be performed to optimize treatment conditions for other drugs used for cancer therapy.
HACs for identification of new genes controlling chromosome segregation
The spectrum of mutations that cause CIN is only partly known, and it is not possible to predict a priori all pathways whose disruption might lead to CIN. Recently, Hieter and colleagues have completed two genome-wide chromosome instability screens in yeast S. cerevisiae and identified 257 genes whose over-expression decreases genome stability [97]. Based on the sequences of orthologs, they created a list of human CIN candidate genes, which are cross-referenced to the published somatic mutation databases. Interestingly, over 50 % of these genes are amplified and/or over-expressed in breast cancer. To assess the cross-species relevance of these dosage CIN genes, a HAC-GFP assay can be used to examine if over-expression of the candidate genes results in chromosome instability in human cells. This approach could potentially result in identification of new genes controlling chromosome segregation in human cells. It could also help to better understand the CIN phenotype and its role in cancer.
HACs to study human kinetochore structure and assembly
Human artificial chromosomes mimic endogenous chromosomes in their kinetochore structure and mitotic behavior and therefore represent a powerful system for functional studies of the human kinetochore. The alphoidtetO-HAC specifically is applicable for kinetochore studies because it contains ~6,000 copies of the tetracycline operator (tetO) sequence. The tetO sites are recognized by the tetR (tetracycline repressor) protein, thereby allowing a specific and conditional tethering of a wide range of tetR fusion proteins to the tetO-containing alphoid array. Targeting of different chromatin modifiers into the HAC kinetochore has already shed light on centrochromatin and its impact into kinetochore structure and function in human cells [28, 49, 98–100]. More specifically, tethering of different tetR-fusion proteins is allowing us to understand the requirements for individual histone modifications necessary for formation and maintenance of kinetochore structure. A recently published review summarizes the results of human kinetochore analyses using alphoidtetO-HAC [101].
Concluding remarks
HACs potentially represent new promising vectors with multiple applications. HACs are already being used for gene function studies in functional genomics, since HAC-based vector systems lack many of the problems that plague viral vector systems. So far, the potential utility of HACs to correct genetic defects has been demonstrated in cultured cells and animal models. However, any application of HAC vectors for gene therapeutics will require many additional studies. Present research should focus on the efficiency and safety of delivery of HAC vectors, not only to cells cultured in vitro but also in animal models. In addition, optimization of HAC transfer protocols is a priority. Protocols were developed three decades ago for the transfer of entire chromosomes, but have not been improved since due to the absence of a specific demand. Additional studies will include the systematic analysis of mitotic HAC stability and gene expression from the HAC in different types of non-transformed human cells. It will also be important to determine the effect, if any, of the extra chromosome on the replication and segregation of the endogenous chromosomes.
Clearly, there is a long way to go before the use of HACs as vectors for treating genetic disease will be a reality. However, the potential is there for vectors that can express entire complex signaling pathways under their normal physiological regulation. This is an exciting area for scientific exploration with great potential benefits.
Acknowledgments
This study was supported by the Intramural Research Program of the NIH NCI Center for Cancer Research (V.L.). This study was also supported by a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (H.M.) and by The Wellcome Trust, of which W.C.E. is a Principal Research Fellow (grant number 073915). The Wellcome Trust Centre for Cell Biology is supported by core grant numbers 077707 and 092076.
References
- 1.Lufino MM, Edser PA, Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: episomal maintenance, vector delivery, and transgene expression. Mol Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- 2.Epstein AL. Progress and prospects: biological properties and technological advances of herpes simplex virus type 1-based amplicon vectors. Gene Ther. 2009;16:709–715. doi: 10.1038/gt.2009.42. [DOI] [PubMed] [Google Scholar]
- 3.Mingozzi F, Katherine A, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Genet Rev. 2011;12:341–356. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 4.Maier P, von Kalle C, Laufs S. Retroviral vectors for gene therapy. Future Microbiol. 2010;5:1507–1523. doi: 10.2217/fmb.10.100. [DOI] [PubMed] [Google Scholar]
- 5.Mátrai J, Chuah MK, VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz CJ, Mühlebach MD, Cichutek K. Lentiviral vectors with measles virus glycoproteins—dream team for gene transfer? Trends Biotechnol. 2009;27:259–265. doi: 10.1016/j.tibtech.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Banasik MB, McCray PB., Jr Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- 8.Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 10.Lufino MM, Edser PA, Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: episomal maintenance, vector delivery, and transgene expression. Mol Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- 11.Hibbitt OC, Wade-Martins R. Delivery of large genomic DNA inserts >100 kb using HSV-1 amplicons. Curr Gene Ther. 2006;6:325–336. doi: 10.2174/156652306777592054. [DOI] [PubMed] [Google Scholar]
- 12.Li CM, Park JH, Simonaro CM, et al. Insertional mutagenesis of the mouse acid ceramidase gene leads to early embryonic lethality in homozygotes and progressive lipid storage disease in heterozygotes. Genomics. 2002;79:218–224. doi: 10.1006/geno.2002.6686. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Düllmann J, Schiedlmeier B, Schmidt M, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 14.Raper SE, Chirmule N, Lee FS, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Odom GL, Gregorevic P, et al. Viral-mediated gene therapy for the muscular dystrophies: successes, limitations and recent advances. Biochim Biophys Acta. 2007;1772:243–262. doi: 10.1016/j.bbadis.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saffery R, Choo KH. Strategies for engineering human chromosomes with therapeutic potential. J Gene Med. 2002;4:5–13. doi: 10.1002/jgm.236. [DOI] [PubMed] [Google Scholar]
- 18.Basu J, Willard HF. Human artificial chromosomes: potential applications and clinical considerations. Pediatr Clin North Am. 2006;53:843–53. doi: 10.1016/j.pcl.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Monaco ZL, Moralli D. Progress in artificial chromosome technology. Biochem Soc Trans. 2006;34(Pt 2):324–327. doi: 10.1042/BST20060324. [DOI] [PubMed] [Google Scholar]
- 20.Ren X, Tahimic CG, Katoh M, et al. Human artificial chromosome vectors meet stem cells: new prospects for gene delivery. Stem Cell Rev. 2006;2(1):43–50. doi: 10.1007/s12015-006-0008-9. [DOI] [PubMed] [Google Scholar]
- 21.Oshimura M, Katoh M. Transfer of human artificial chromosome vectors into stem cells. Reprod Biomed. 2008;16(1):57–69. doi: 10.1016/S1472-6483(10)60557-3. [DOI] [PubMed] [Google Scholar]
- 22.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther. 2011;19(9):1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeno M, Suzuki N. Construction and use of a bottom-up HAC vector for transgene expression. Methods Mol Biol. 2011;738:101–110. doi: 10.1007/978-1-61779-099-7_7. [DOI] [PubMed] [Google Scholar]
- 24.Rudd MK, Mays RW, Schwartz S, et al. Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol Cell Biol. 2003;23(21):7689–7697. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimes BR, Babcock J, Rudd MK, et al. Assembly and characterization of heterochromatin and euchromatin on human artificial chromosomes. Genome Biol. 2004;5(11):R89. doi: 10.1186/gb-2004-5-11-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13(8):745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- 27.Kazuki Y, Hoshiya H, Takiguchi M, et al. Refined human artificial chromosome vector for gene therapy and animal transgenesis. Gene Ther. 2011;18:384–393. doi: 10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano M, Cardinale S, Noskov VN, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farr CJ, Stevanovic M, Thomson EJ, et al. Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat Genet. 1992;2:275–282. doi: 10.1038/ng1292-275. [DOI] [PubMed] [Google Scholar]
- 30.Brown KE, Barnett MA, Burgtorf C, et al. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- 31.Heller R, Brown KE, Burgtorf C, et al. Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc Natl Acad Sci USA. 1996;93:7125–7130. doi: 10.1073/pnas.93.14.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills W, Critcher R, Lee C, et al. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum Mol Genet. 1999;8:751–761. doi: 10.1093/hmg/8.5.751. [DOI] [PubMed] [Google Scholar]
- 33.Yang JW, Pendon C, Yang J, et al. Human mini-chromosomes with minimal centromeres. Hum Mol Genet. 2000;9:1891–1902. doi: 10.1093/hmg/9.12.1891. [DOI] [PubMed] [Google Scholar]
- 34.Kuroiwa Y, Tomizuka K, Shinohara T, et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat Biotechnol. 2000;18:1086–1090. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- 35.Shen MH, Mee PJ, Nichols J, et al. A structurally defined mini-chromosome vector for the mouse germ line. Curr Biol. 2000;10:31–34. doi: 10.1016/S0960-9822(99)00261-4. [DOI] [PubMed] [Google Scholar]
- 36.Katoh M, Ayabe F, Norikane S, et al. Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun. 2004;321:280–290. doi: 10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- 37.Kakeda M, Nagata K, Osawa K, et al. A new chromosome 14-based human artificial chromosome (HAC) vector system for efficient transgene expression in human primary cells. Biochem Biophys Res Commun. 2011;415(3):439–444. doi: 10.1016/j.bbrc.2011.10.088. [DOI] [PubMed] [Google Scholar]
- 38.Harrington JJ, Van Bokkelen G, Mays RW, et al. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 39.Ikeno M, Grimes B, Okazaki T, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 40.Guiducci C, Ascenzioni F, Auriche C, et al. Use of a human minichromosome as a cloning and expression vector for mammalian cells. Hum Mol Genet. 1999;8:1417–1424. doi: 10.1093/hmg/8.8.1417. [DOI] [PubMed] [Google Scholar]
- 41.Ebersole TA, Ross A, Clark E, et al. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 42.Grimes BR, Schindelhauer D, McGill NI, et al. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2001;2:910–914. doi: 10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mejía JE, Alazami A, Willmott A, et al. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79:297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 44.Kouprina N, Ebersole T, Koriabine M, et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu J, Stromberg G, Compitello G. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 2005;33:587–596. doi: 10.1093/nar/gki207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basu J, Compitello G, Stromberg G, et al. Efficient assembly of de novo human artificial chromosomes from large genomic loci. BMC Biotechnol. 2005;5:21. doi: 10.1186/1472-6750-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotzamanis G, Cheung W, Abdulrazzak H, et al. Construction of human artificial chromosome vectors by recombineering. Gene. 2005;351:29–38. doi: 10.1016/j.gene.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Moralli D, Simpson KM, Wade-Martins R, et al. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep. 2006;7:911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohzeki JI, Bergmann JH, Kouprina N, et al. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mejía JE, Willmott A, Levy E. Functional complementation of a genetic deficiency with human artificial chromosomes. Am J Hum Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikeno M, Inagaki H, Nagata K, et al. Generation of human artificial chromosomes expressing naturally controlled guanosine triphosphate cyclohydrolase I gene. Genes Cells. 2002;7:1021–1032. doi: 10.1046/j.1365-2443.2002.00580.x. [DOI] [PubMed] [Google Scholar]
- 52.Auriche C, Carpani D, Conese M, et al. Functional human CFTR produced by a stable minichromosome. EMBO Rep. 2002;3:862–868. doi: 10.1093/embo-reports/kvf174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rocchi L, Braz C, Cattani S, et al. Escherichia coli-cloned CFTR loci relevant for human artificial chromosome therapy. Hum Gene Ther. 2010;21:1077–1092. doi: 10.1089/hum.2009.225. [DOI] [PubMed] [Google Scholar]
- 54.Breman AM, Steiner CM, Slee RB, et al. Input DNA ratio determines copy number of the 33 kb Factor IX gene on de novo human artificial chromosomes. Mol Ther. 2008;16:315–323. doi: 10.1038/sj.mt.6300361. [DOI] [PubMed] [Google Scholar]
- 55.Yamada H, Li YC, Nishikawa M, et al. Introduction of a CD40L genomic fragment via a human artificial chromosome vector permits cell-type-specific gene expression and induces immunoglobulin secretion. J Hum Genet. 2008;53:447–453. doi: 10.1007/s10038-008-0268-0. [DOI] [PubMed] [Google Scholar]
- 56.Kazuki Y, Hoshiya H, Kai Y, et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther. 2008;15(8):617–624. doi: 10.1038/sj.gt.3303091. [DOI] [PubMed] [Google Scholar]
- 57.Hoshiya H, Kazuki Y, Abe S, et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol Ther. 2009;17:309–317. doi: 10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JH, Kononenko A, Erliandri I, et al. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc Natl Acad Sci USA. 2011;108(50):20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuroiwa Y, Kasinathan P, Choi YJ, et al. Cloned transchromosomic calves producing human immunoglobulin. Nat Biotechnol. 2002;20:889–894. doi: 10.1038/nbt727. [DOI] [PubMed] [Google Scholar]
- 60.Kuroiwa Y, Kasinathan P, Sathiyaseelan T, et al. Antigen-specific human polyclonal antibodies from hyperimmunized cattle. Nat Biotechnol. 2009;27:173–181. doi: 10.1038/nbt.1521. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki N, Nishii K, Okazaki T, et al. Human artificial chromosomes constructed using the bottom-up strategy are stably maintained in mitosis and efficiently transmissible to progeny mice. J Biol Chem. 2006;281:26615–26623. doi: 10.1074/jbc.M603053200. [DOI] [PubMed] [Google Scholar]
- 62.Ito M, Ikeno M, Nagata H, et al. Treatment of nonalbumin rats by transplantation of immortalized hepatocytes using artificial human chromosome. Transplant Proc. 2009;41:422–424. doi: 10.1016/j.transproceed.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 63.Voet T, Schoenmakers E, Carpentier S, et al. Controlled transgene dosage and PAC-mediated transgenesis in mice using a chromosomal vector. Genomics. 2003;82(6):596–605. doi: 10.1016/S0888-7543(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 64.Kazuki Y, Hiratsuka M, Takiguchi M, et al. Complete genetic correction of iPSs cells from Duchenne muscular dystrophy. Mol Ther. 2010;18:386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dafhnis-Calas F, Xu Z, Haines S, et al. Iterative in vivo assembly of large and complex transgenes by combining the activities of phiC31 integrase and Cre recombinase. Nucleic Acids Res. 2005;33(22):e189. doi: 10.1093/nar/gni192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren X, Katoh M, Hoshiya H, et al. A novel human artificial chromosome vector provides effective cell lineage-specific transgene expression in human mesenchymal stem cells. Stem Cells. 2005;23:1608–1616. doi: 10.1634/stemcells.2005-0021. [DOI] [PubMed] [Google Scholar]
- 67.Kazuki Y, Hoshiya H, Kai Y, et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther. 2008;15:617–624. doi: 10.1038/sj.gt.3303091. [DOI] [PubMed] [Google Scholar]
- 68.Iida Y, Kim JH, Kazuki Y, et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kakeda M, Hiratsuka M, Nagata K, et al. Human artificial chromosome (HAC) vector provides long-term therapeutic transgene expression in normal human primary fibroblasts. Gene Ther. 2005;12:852–856. doi: 10.1038/sj.gt.3302483. [DOI] [PubMed] [Google Scholar]
- 70.Ebersole T, Okamoto Y, Noskov VN, et al. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucl Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kouprina N, Samoshkin A, Erliandri I et al (2012) Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ASC Synthetic Biol (in press) [DOI] [PMC free article] [PubMed]
- 72.Alazami AM, Mejía JE, Monaco ZL. Human artificial chromosomes containing chromosome 17 alphoid DNA maintain an active centromere in murine cells but are not stable. Genomics. 2004;835:844–851. doi: 10.1016/j.ygeno.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 73.Moralli D, Chan DY, Jefferson A. HAC stability in murine cells is influenced by nuclear localization and chromatin organization. BMC Cell Biol. 2009;10:18. doi: 10.1186/1471-2121-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fournier RE, Ruddle FH. Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc Natl Acad Sci USA. 1977;74:319–323. doi: 10.1073/pnas.74.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koi M, Shimizu M, Morita H, et al. Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn J Cancer Res. 1989;80:413–418. doi: 10.1111/j.1349-7006.1989.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamada H, Kunisato A, Kawahara M, et al. Exogenous gene expression and growth regulation of hematopoietic cells via a novel human artificial chromosome. J Hum Genet. 2006;51:147–150. doi: 10.1007/s10038-005-0334-9. [DOI] [PubMed] [Google Scholar]
- 77.Kinoshita Y, Kamitani H, Mamun MH, et al. A gene delivery system with a human artificial chromosome vector based on migration of mesenchymal stem cells towards human glioblastoma HTB14 cells. Neurol Res. 2010;32:429–437. doi: 10.1179/174313209X455718. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi S, Ren X, Katoh M, et al. A new method of microcell-mediated transfer of human artificial chromosome using a hemagglutinating virus of Japan envelope. Chromosoma Sci. 2006;9:65–73. [Google Scholar]
- 79.Katoh M, Kazuki Y, Kazuki K. Exploitation of the interaction of measles virus fusogenic envelope proteins with the surface receptor CD46 on human cells for microcell-mediated chromosome transfer. BMC Biotechnol. 2010;10:37. doi: 10.1186/1472-6750-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Jong G, Telenius A, Vanderbyl S, et al. Efficient in vitro transfer of a 60-Mb mammalian artificial chromosome into murine and hamster cells using cationic lipids and dendrimers. Chromosome Res. 2001;9(6):475–485. doi: 10.1023/A:1011680529073. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki N, Itou T, Hasegawa Y, Okazaki T, et al. Cell to cell transfer of the chromatin-packaged human beta-globin gene cluster. Nucleic Acids Res. 2010;38(5):e33. doi: 10.1093/nar/gkp1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Larionov V, Kouprina N, Graves J, et al. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci USA. 1996;93(1):491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kouprina N, Larionov V. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat Rev Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 84.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes bytransformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protocols. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 85.Ayabe F, Katoh M, Inoue T, et al. A novel expression system for genomic DNA loci using a human artificial chromosome vector with transformation-associated recombination cloning. J Hum Genet. 2005;50:592–599. doi: 10.1007/s10038-005-0300-6. [DOI] [PubMed] [Google Scholar]
- 86.Yamaguchi S, Kazuki Y, Nakayama Y, et al. A method for producing transgenic cells using a multi-integrase system on a human artificial chromosome vector. PLoS ONE. 2011;6:e17267. doi: 10.1371/journal.pone.0017267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanatsu-Shinohara M, Inoue K, Lee J, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 89.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 90.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 91.Hiratsuka M, Uno N, Ueda K, et al. Integration-free iPS cells engineered using human artificial chromosome vectors. PLoS ONE. 2011;6(10):e25961. doi: 10.1371/journal.pone.0025961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuroiwa Y, Kasinathan P, Choi YJ, et al. Cloned transchromosomic calves producing human immunoglobulin. Nat Biotechnol. 2002;20:889–894. doi: 10.1038/nbt727. [DOI] [PubMed] [Google Scholar]
- 93.Takahashi Y, Tsuji S, Kazuki Y, et al. Development of evaluation system for bioactive substances using human artificial chromosome-mediated osteocalcin gene expression. J Biochem. 2010;148:29–34. doi: 10.1093/jb/mvq030. [DOI] [PubMed] [Google Scholar]
- 94.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106(45):19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Colombo R, Moll J. Destabilizing aneuploidy by targeting cell cycle and mitotic checkpoint proteins in cancer cells. Curr Drug Targets. 2010;11(10):1325–1335. doi: 10.2174/1389450111007011325. [DOI] [PubMed] [Google Scholar]
- 96.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 97.Stirling PC, Bloom MS, Solanki-Patil T, et al. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011;7(4):e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cardinale S, Bergmann JH, Kelly D, et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20(19):4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bergmann JH, Rodríguez MG, Martins NM, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30(2):328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bergmann JH, Jakubsche JN, Martins NM, et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125(Pt 2):411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bergman JH, Martins NMC, Larionov V, et al. HACking the centromere chromatin code: in sights from human artificial chromosomes. Chromosome Res. 2012;20:505–519. doi: 10.1007/s10577-012-9293-0. [DOI] [PubMed] [Google Scholar]