Abstract
G-Quadruplexes are noncanonical nucleic acid secondary structures based on guanine association that are readily adopted by G-rich RNA and DNA sequences. Naturally occurring genomic G-quadruplex-forming sequences have functional roles in biology that are mediated through structure. To appreciate how this is achieved, an understanding of the likelihood of G-quadruplex formation and the structural features of the folded species under a defined set of conditions is informative. We previously systematically investigated the thermodynamic stability and folding topology of DNA G-quadruplexes and found a strong dependence of these properties on loop length and loop arrangement [Bugaut, A., and Balasubramanian, S. (2008) Biochemistry 47, 689–697]. Here we report on a complementary analysis of RNA G-quadruplexes using UV melting and circular dichroism spectroscopy that also serves as a comparison to the equivalent DNA G-quadruplex-forming sequences. We found that the thermodynamic stability of G-quadruplex RNA can be modulated by loop length while the overall structure is largely unaffected. The systematic design of our study also revealed subtle loop length dependencies in RNA G-quadruplex structure.
Guanine-rich RNA and DNA sequences are known to fold into quadruple-helical structures called G-quadruplexes.1 The core of a G-quadruplex consists of multiple G-tetrads formed from four guanines that self-associate through hydrogen bonding at both Watson–Crick and Hoogsteen edges. Stacks of planar G-tetrads give rise to characteristic G-quadruplex structures that are stabilized by monovalent cations, especially potassium,2 and may contain connecting loops of different sizes.
The ability of nucleic acids to adopt unusual secondary structures such as G-quadruplexes raises the question of whether these structures serve biological functions. Bioinformatics analyses have estimated that there are as many as 376000 putative quadruplex-forming sequences in the genome.3,4 In addition to repetitive DNA regions that harbor many putative quadruplex-forming sequences, such as telomeres and rDNA, an enrichment of G-rich sequences has been observed in other genomic locations, including gene promoters,5 and in areas of the transcriptome including splice sequences6 and untranslated regions.7 These observations have informed hypothesis-driven investigations into understanding G-quadruplex function. The human telomeres, which are involved in genome stability and cell growth by protecting chromosome ends,8,9 offer one of the most well characterized and validated examples of G-quadruplex involvement in a cellular process.10 The recent discovery that telomeric DNA is transcriptionally active11,12 has further emphasized the importance of G-rich telomeric sequences. The r[(GGGUUA)n] repeat transcripts localize at the telomere,11,13 inhibit telomerase,12 and also have a propensity to form RNA G-quadruplex structures.14–16
A series of recent studies have established that G-quadruplex structures formed in the 5′-UTR of certain mRNAs, including NRAS,17 Bcl-2,18 Zic-1,19 TRF2,20 MT3,21 ESR1,22 and other23 human mRNAs, have the ability to reduce the efficiency of translation, and that the extent of inhibition may have a stability and positional dependence.24 Furthermore, an RNA G-quadruplex “switch” in the 5′-UTR of human VEGF mRNA has been implicated in IRES-mediated translation initiation.25 In bacteria, Wieland et al. demonstrated the ability to modulate in vivo gene expression by artificially coupling RNA G-quadruplex formation with the masking of a ribosome binding site.26
For intramolecular G-quadruplex structures, the largest variability resides in the loops that connect the stacks of G-tetrads. There have been numerous studies addressing the effect of loop lengths and sequences on the formation of DNA G-quadruplexes.27–34 We previously investigated the effect of short loop lengths on DNA G-quadruplex stability and folding topology in a well-defined and sequence-independent manner.35 Herein we report an analogous study using RNA G-quadruplex-forming sequences, with the aim of understanding fundamental differences and similarities between RNA and DNA G-quadruplexes. Specifically, we explored the effect of loop length and loop arrangement on the stability and folding topology of RNA G-quadruplex libraries using UV melting and CD spectroscopy, respectively. We measured the average stability and folding topology properties of RNA oligonucleotide libraries that contain a population of G-quadruplex-forming sequences of defined loop lengths and arrangements. The RNA oligonucleotide libraries represent individual loop lengths of 1–5 nucleotides and total loop lengths of 3–15 nucleotides.
MATERIALS AND METHODS
Oligonucleotides
RNA oligonucleotide libraries were purchased from Biomers and used as supplied. Stock solutions were prepared with nuclease free water (Ambion Biosciences). Concentrations of single-stranded oligonucleotides were determined by UV absorption at 260 nm after they had been heated at 95 °C for 5 min, and molar extinction coefficients were calculated using OligoAnalyzer version 3.1 (http://eu.idtdna.com/analyzer/applications/oligoanalyzer/default.aspx). Molar extinction coefficients are provided in Table S1 of the Supporting Information.
UV Melting
UV melting curves were recorded using a Cary 100 UV–visible spectrophotometer (Agilent) by measuring the absorbance at 295 nm as a function of temperature. RNA oligonucleotide library solutions were prepared with a final concentration of 4 μM in 10 mM lithium cacodylate buffer (pH 7.0) containing 5 mM potassium chloride. The melting experiments were performed in 10 mm path length quartz cuvettes with 1 mL of a buffered RNA oligonucleotide library solution. A steady stream of nitrogen was applied to prevent condensation at low temperatures, and the solutions were covered with a layer of mineral oil to minimize evaporation. Temperature ramps consisted of low → high → low → high cycles with a temperature range of 5–90 or 15–98 °C for libraries with melting temperatures of <60 or >60 °C, respectively. A ramp rate of 0.25 °C/min was maintained for all experiments with data collected every 1 min. van’t Hoff analysis was performed on the melting and annealing curves as previously described.35 This method of analysis relies on the assumption that there is a two-state (folded vs unfolded) equilibrium with no significantly populated intermediates, and the determination of baselines relies on subjective manual selection.
Circular Dichroism
Circular dichroism spectra were recorded on a Chirascan spectropolarimeter (Applied Photophysics, Leatherhead, U.K.) using a 1 mm path length quartz cuvette at an RNA concentration of 10 μM in 10 mM lithium cacodylate buffer (pH 7.0) containing 5 or 100 mM potassium chloride. Sample preparation involved heating at 95 °C for 5 min followed by slow cooling to room temperature. Scans were performed at 25 °C over a range of 320–220 nm, and each final spectrum is the average of four scans taken with a step size of 1 nm, a time per point of 1 s, and a bandwidth of 0.5 nm. A buffer only blank was subtracted from each spectrum, and data were zero-corrected at 320 nm.
Gel Electrophoresis
Nondenaturing gel electrophoresis was performed on a Novex 6% Tris-Glycine Mini Gel (1.0 mm, 10-well, Invitrogen). Folded RNA G-quadruplex was prepared at a concentration of 20 μM in 10 mM lithium cacodylate (pH 7.0) containing 5 mM potassium chloride by being heated at 95 °C for 5 min and slowly cooled to room temperature. Electrophoresis was conducted in 0.5× TBE running buffer with a constant voltage of 100 V at room temperature for 30 min. Gels were stained with Stains-All (Sigma-Aldrich).
RESULTS
Experimental Design
UV and CD spectroscopies, which are well-established biophysical tools for characterizing G-quadruplex formation, were employed to investigate the influence of loop length on the thermodynamic stability and folding topology of RNA oligonucleotide libraries designed to fold into intramolecular G-quadruplexes. Each library contains quadruplex-forming RNA sequences with four tracts of G3 separated by three sets of loop nucleotides (Figure 1). The libraries are designated Ljkl, where j, k, and l denote the number of nucleotides in the first, second, and third loops, respectively, between the G3 tracts. The RNA oligonucleotide libraries presented here encompass G-quadruplexes with mono-, di-, and trinucleotide loops (j, k, or l = 1, 2, or 3). The general sequences of any mononucleotide and dinucleotide loops are H and HH, respectively, where H is A, U, or C with equal probability, and the general sequence of any trinucleotide loop is HNH, where N is A, U, C, or G with equal probability. This prevents bias arising from loop sequence and the possibility of G4 tracts. However, this design does not exclude the possibility that an isolated G in a trinucleotide loop may participate in tetrad formation, which has been observed for DNA G-quadruplexes.36,37 Overall, the design allows the libraries to be categorized according to total loop length (“TLL”, the total number of nucleotides in all three loops) as well as loop length at each loop position. RNA oligonucleotide libraries with the same total loop length and loop nucleotide combinations but different loop arrangements are termed “Ljkl et al.”; for example, L112 et al. denotes RNA oligonucleotide libraries L112, L121, and L211.
Figure 1.
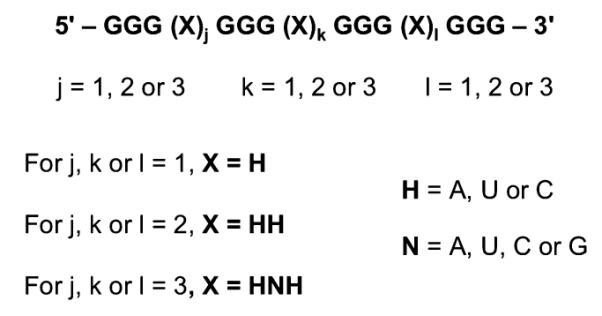
RNA oligonucleotide library design.
UV Melting
The melting of a G-quadruplex structure can be monitored by following a characteristic hypochromic shift at 295 nm.38 All of the RNA oligonucleotide libraries studied here exhibited hypochromic melting transitions at 295 nm in the presence of 5 mM potassium in 10 mM lithium cacodylate (pH 7.0). Unlike the analogous DNA study,35 we could not perform the UV melting experiments at a higher (20 mM) potassium ion concentration because the most stable libraries could not be unfolded (data not shown).
The melting and annealing profiles of each RNA oligonucleotide library were superimposable (Figure S1 of the Supporting Information), supporting a fast and reversible formation of intramolecular G-quadruplex species.39,40 We analyzed the migration behavior of RNA oligonucleotide libraries with the shortest loop lengths through a nondenaturing polyacrylamide gel matrix to confirm molecularity. We found that RNA oligonucleotide libraries L111, L112, and L113 each migrate as a single band at 20 μM RNA in the presence of 5 mM potassium chloride (Figure 2). Additionally, the migration of the selected RNA oligonucleotide libraries is similar to that of the TRF2 (cf. L112),20 MT3 (cf. L113),21 and TERRA (cf. L333)41 RNA G-quadruplexes, each of which has previously been shown to fold into an intramolecular G-quadruplex. Furthermore, the melting temperature of RNA library L111 is invariant over the concentration range of 1– 25 μM (Figure S2 of the Supporting Information). Taken together, these results indicate that all of the RNA oligonucleotide libraries are largely populated by stably folded intramolecular G-quadruplex structures.
Figure 2.
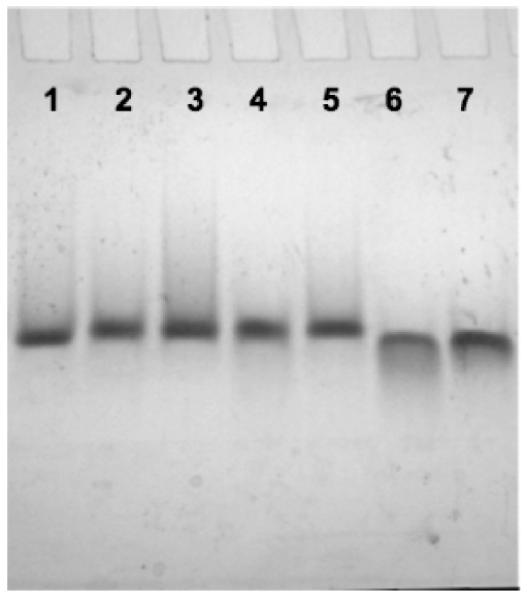
Nondenaturing gel electrophoresis of RNA oligonucleotide libraries (20 μM) and naturally occurring intramolecular RNA G-quadruplexes. Lanes 1–7: L111, L112, TRF2, L113, MT3, L333, and TERRA, respectively.
The melting and annealing curves for each RNA oligonucleotide library were subjected to thermodynamic analysis using the van’t Hoff model.42 Melting temperatures (Tm) of the RNA oligonucleotide libraries are listed in Table 1, along with the apparent van’t Hoff free energy of G-quadruplex formation at 37 °C. An overlay of the folded fraction curves as a function of temperature for the RNA oligonucleotide libraries is provided in Figure 3.
Table 1.
Melting Temperatures and van’1t Hoff Thermodynamic Parameters for RNA and DNA G-Quadruplex-Forming Librariesa and Naturally Occurring RNA G-Quadruplex-Forming Sequencesb
| Tm(VH)/Tmc (°C) | ΔHvh (kJ mol−1) | ΔSvh (kJ mol−1 K−1) | ΔGvh(37 °C) (kJ mol−1) | |
|---|---|---|---|---|
| L111 | 86/86 | −249 ± 16 | −0.692 ± 0.045 | −33.8 ± 1.8 |
| L112 | 78/78 | −237 ± 10 | −0.674 ± 0.030 | −27.6 ± 1.2 |
| L121 | 76/76 | −239 ± 15 | −0.685 ± 0.042 | −26.7 ± 1.4 |
| L211 | 77/77 | −247 ± 4 | −0.703 ± 0.013 | −28.3 ± 0.1 |
| L113 | 74/74 | −241 ± 17 | −0.701 ± 0.067 | −25.7 ± 1.4 |
| L131 | 73/73 | −279 ± 16 | −0.798 ± 0.064 | −25.7 ± 1.6 |
| L311 | 75/75 | −238 ± 2 | −0.684 ± 0.007 | −26.0 ± 0.1 |
| L122 | 72/72 | −266 ± 17 | −0.768 ± 0.075 | −26.5 ± 1.0 |
| L212 | 72/72 | −245 ± 1 | −0.711 ± 0.002 | −24.6 ± 0.8 |
| L221 | 70/71 | −251 ± 1 | −0.730 ± 0.006 | −24.4 ± 0.3 |
| L222 | 67/67 | −236 ± 19 | −0.693 ± 0.056 | −20.8 ± 1.6 |
| L133 | 63/63 | −242 ± 5 | −0.721 ± 0.014 | −18.4 ± 0.2 |
| L313 | 65/65 | −233 ± 4 | −0.687 ± 0.012 | −19.2 ± 0.6 |
| L331 | 63/64 | −243 ± 0 | −0.722 ± 0.000 | −19.1 ± 0.1 |
| L223 | 63/63 | −241 ± 5 | −0.717 ± 0.017 | −18.6 ± 0.4 |
| L232 | 61/62 | −238 ± 14 | −0.712 ± 0.041 | −17.4 ± 1.2 |
| L322 | 59/59 | −224 ± 4 | −0.673 ± 0.012 | −14.9 ± 0.2 |
| L233 | 59/59 | −231 ± 3 | −0.693 ± 0.008 | −15.4 ± 0.5 |
| L323 | 59/59 | −218 ± 9 | −0.657 ± 0.027 | −14.2 ± 0.3 |
| L332 | 57/58 | −220 ± 6 | −0.665 ± 0.016 | −13.5 ± 0.6 |
| L333 | 55/55 | −232 ± 7 | −0.707 ± 0.021 | −12.9 ± 0.6 |
| L444 | 49/50 | −159 ± 0 | −0.493 ± 0.001 | −6.1 ± 0.1 |
| L555 | 46/47 | −165 ± 4 | −0.518 ± 0.013 | −4.6 ± 0.1 |
| dL111 | 77/78 | −190 ± 9 | −0.543 ± 0.027 | −21.9 ± 0.6 |
| dL222 | 41/41 | −116 ± 3 | −0.368 ± 0.008 | −1.4 ± 0.1 |
| dL333 | 38/38 | −132 ± 1 | −0.425 ± 0.004 | −0.3 ± 0.0 |
| TRF2 | 77/78 | −255 ± 6 | −0.728 ± 0.016 | −29.4 ± 0.9 |
| MT3 | 71/71 | −247 ± 6 | −0.719 ± 0.016 | −24.1 ± 1.0 |
| NRAS | 69/68 | −244 ± 4 | −0.713 ± 0.012 | −22.7 ± 0.5 |
| TERRA | 57/58 | −265 ± 5 | −0.799 ± 0.016 | −16.3 ± 0.3 |
For the libraries, these measurements represent apparent values derived from melting curves of the mixtures.
All errors are given as the standard deviation. Tm values are accurate to ±1 °C.
Tm(VH) values are melting temperatures determined from van’t Hoff analysis, whereas Tm values are determined as the point at which the folded fraction equals one-half.
Figure 3.
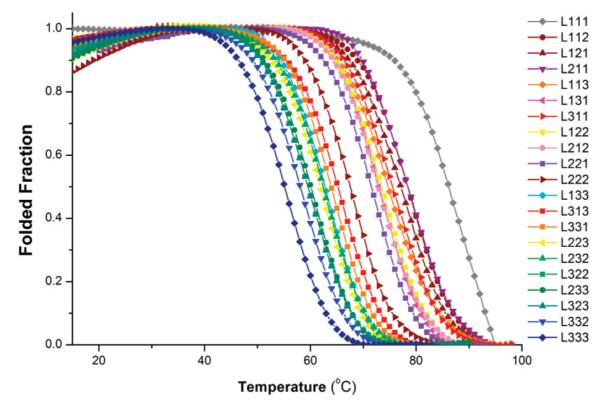
Fraction folded of RNA oligonucleotide libraries as a function of temperature.
A general trend emerged indicating that the thermodynamic stability of RNA G-quadruplexes decreases as the total loop length increases (Figure 4). RNA library L111 formed the most stable G-quaduplexes under the conditions of the study [Tm = 86 °C; ΔGVH(37 °C) = –33.8 kJ mol−1]. Replacing one mononucleotide loop with a dinucleotide loop resulted in a decrease in free energy of ~6.5 kJ mol−1. Each subsequent increase of one nucleotide in total loop length led to an average reduction in free energy of G-quadruplex formation of ~2–4 kJ mol−1. The trend in thermal stability was essentially identical (Figure S3 of the Supporting Information), with an incremental decrease of ~3–6 °C per one-nucleotide increase in total loop length.
Figure 4.
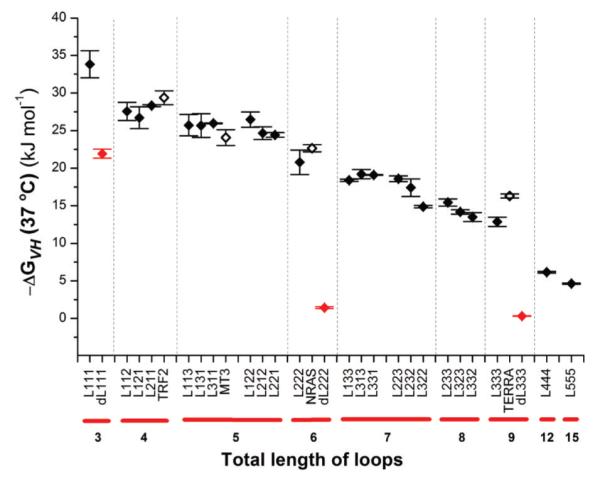
ΔGVH as a function of total loop length: RNA libraries (◆), naturally occurring RNA G-quadruplex sequences (◇), and DNA libraries (red diamonds). Errors are given as the standard deviation of triplicate experiments.
To determine whether a plateau in stability is reached when loop lengths are further increased, UV melting experiments were performed on RNA libraries L444 and L555 with total loop lengths of 12 and 15 nucleotides, respectively. The free energies of G-quadruplex formation for RNA libraries L444 and L555 decreased by 6.8 and 8.3 kJ mol−1, respectively, compared to that of L333. However, the magnitude of the difference between L444 and L555 (1.5 kJ mol−1) is markedly smaller than that between L333 and L444, suggesting that thermodynamic stability may become less dependent on total loop length near this point.
We then tested the utility and relevance of our framework of averaged thermodynamic stabilities by performing UV melting experiments with four individual RNA G-quadruplex-forming sequences (Figure 4 and Tables 1 and 2). The TRF2,20 MT3,21 and NRAS17 RNA G-quadruplexes, which naturally occur in the 5′-UTR of the corresponding human mRNA, were selected for their distinct loop length arrangements of L112 (TLL 4), L113 (TLL 5), and L213 (TLL 6), respectively. Additionally, the human telomeric RNA sequence TERRA43 was also selected to represent a loop length arrangement of L333 with a total loop length of nine nucleotides. The stability of 5′-UTR RNA G-quadruplexes TRF2, MT3, and NRAS was similar to the stability (within experimental error) of RNA oligonucleo- tide libraries L112, L113, and L222, respectively. For TERRA, the stability of the sequence was ~3 kJ mol−1 greater than the average thermodynamic stability of L333.
Table 2.
Naturally Occurring RNA G-Quadruplex-Forming Sequences Used in This Study
To allow a direct comparison between the RNA oligonucleotide libraries analyzed here and their equivalent DNA sequences, we performed UV melting experiments with DNA oligonucleotide libraries dL111, dL222, and dL333 under experimental conditions identical to those used for the RNA oligonucleotide libraries (Figure 4 and Table 1). DNA library dL111 [ΔGVH (37 °C) = −21.9 kJ mol−1] exhibited a 12 kJ mol−1 lower free energy of formation than RNA library L111. This represents an ~1.6-fold average increase in ΔGVH for RNA library members with three mononucleotide loops as compared to their DNA counterpart. For libraries L222 and L333, the values of ΔGVH increase by ~15- and ~43-fold when compared to those of DNA libraries dL222 and dL333, respectively.
CD Spectroscopy
G-Quadruplex structures possess an overall chirality and therefore exhibit circular dichroism (CD). Interpretations of CD spectra of G-quadruplexes are typically achieved empirically. When the polarity of all the strands is oriented in one direction, the G-quadruplex is said to be parallel and will generally give a characteristic maximum near 265 nm and a minimum near 240 nm.44,45 Antiparallel G-quadruplexes with alternating strand polarity give a distinctly different spectrum with a maximum near 295 nm accompanied by a minimum near 260 nm.44,46,47 Unstructured single-stranded nucleic acids or minimally structured states do not exhibit either of these characteristic signals. The composite case is also possible where maxima near both 295 and 265 nm are present, and this is indicative of the presence of both parallel and antiparallel folds in solution, or a mixed-hybrid-type G-quadruplex of three parallel strands and one antiparallel strand.48–50 This latter case is exemplified well by the quaduplex-forming human telomeric DNA sequence, which has shown a high propensity for polymorphism under different conditions.51–55
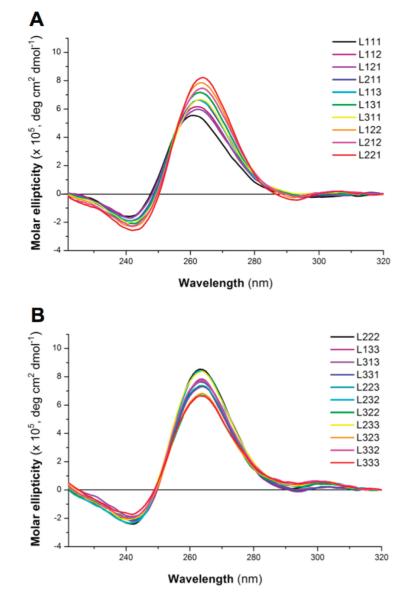
Overlays of the CD spectra recorded in 100 mM potassium showed that all of the 21 RNA oligonucleotide libraries generated CD positive peaks at ~265 nm and negative peaks at ~240 nm (Figure 5). Similar qualitative data were also obtained in 5 mM KCl [the same monovalent ion concentration used for UV melting experiments (Figure S4 of the Supporting Information)]. These data are consistent with the formation of G-quadruplexes with a parallel topology, which is in agreement with all biophysical analyses of individual RNA G-quadruplexes to date. However, we cannot exclude the possibility that some of the sequences may adopt alternative folds that cannot be detected in the CD spectra of the mix- tures. Our systematic study also revealed a subtle difference in CD maxima and minima as a function of total loop length. While the majority of libraries exhibit identical peak wavelengths [242 nm (maximum) and 263 nm (minimum)], we observed a small but observable shift of the maxima and minima toward shorter wavelengths for libraries with three mononucleotide loops [L111; 240 nm (minimum) and 261 nm (maximum)] or two mononucleotide loops and one dinucleotide loop [L112 et al.; 241 nm (maximum) and 262 nm (minimum)].
Figure 5.
Overlays of CD spectra generated from RNA oligonucleotide libraries with total loop lengths of (A) 3–5 and (B) 6–9.
DISCUSSION
This study explores the thermodynamic stability and folding topology of RNA G-quadruplexes as a function of systematically varied loop lengths. Twenty-one G-quadruplex-forming RNA oligonucleotide libraries were examined by UV melting and CD spectroscopy. The libraries were designed to contain four tracts of G3 separated by different combinations of mono-, di-, or trinucleotide loops.
The Thermodynamic Stability of RNA G-Quadruplex Libraries Decreases with Increasing Loop Length and Is Generally Greater Than That of Their DNA Counterpart
The melting temperatures and thermodynamic values obtained for the 21 RNA oligonucleotide libraries show a clear trend toward less stable folded structures as the length of the loops increases. The apparent van’t Hoff free energies of G-quadruplex formation ranged from −33.8 kJ mol−1 for the most stable RNA library (L111) to −4.6 kJ mol−1 for the least stable RNA library (L555). When melting analysis was performed with DNA libraries dL111, dL222, and dL333 under the same experimental conditions, each of these DNA libraries had a markedly lower thermodynamic stability compared to those of their RNA equivalents. The decrease in thermodynamic stability was ~1.6-fold (L111 → dL111), ~15-fold (L222 → dL222), and ~43-fold (L333 → dL333). This disparity between RNA and DNA G-quadruplex stability has already been reported in several cases40,41,56,57 and has in part been attributed to the participation of the ribose 2′-OH groups in an extended network of intramolecular hydrogen bonds, including interactions with phosphate and sugar oxygen atoms (ribose 2′- OH…O3′, …O4′, and …O5′) and H-bond acceptors in bases (ribose 2′-OH…N2 of guanine).16,58 These interactions are believed to enhance the favorable enthalpic contribution to the free energy of RNA G-quadruplex formation. In good agreement, our study reveals consistently larger ΔHVH values for RNA libraries L111 (−249 kJ mol−1), L222 (−236 kJ mol−1), and L333 (−232 kJ mol−1) versus those of their corresponding DNA counterparts dL111 (−191 kJ mol−1), dL222 (−116 kJ mol−1), and dL333 (−132 kJ mol−1), respectively.
In contrast to our complementary study of DNA G-quadruplexes,35 we did not observe any difference in thermodynamic stability among RNA oligonucleotide libraries that differed in their loop length arrangements but possessed the same total lengths of loops. For example, DNA libraries with two mononucleotide loops and one trinucleotide loop were all more stable than the libraries with one mononucleotide loop and two dinucleotide loops. Here, the stabilities of all RNA oligonucleotide libraries with a total loop length of five nucleotides overlap with each other regardless of whether the loop arrangement belongs to the one-trinucleotide and two-mononucleotide loop family (L113 et al.) or the two-dinucleotide and one-mononucleotide loop family (L122 et al.). The same is also true for libraries with a total loop length of seven nucleotides (L133 et al. and L223 et al.). The differential trends between RNA and DNA G-quadruplex stability, when considered as a function of loop length arrangement, may reflect a stronger propensity of mononucleotide loops to affect the thermal stability of DNA G-quadruplexes as compared to more stable and less polymorphic RNA G-quadruplexes. The overall shape of the trends generated from ΔGVH versus total loop length also differs between RNA and DNA G-quadruplex libraries. While the equivalent DNA libraries reached a plateau in stability near a total loop length of six nucleotides,35 the stability of the RNA libraries was still decreasing at this total loop length (Figure S5 of the Supporting Information).
The Averaged Thermodynamic Stabilities of the RNA G-Quadruplex Libraries Fairly Reflect the Stabilities of Naturally Occurring Individual Quadruplex-Forming Sequences
The Tm and van’t Hoff Gibbs free energies of formation of three naturally occurring RNA G-quadruplexes, TRF2 (TLL 4), MT3 (TLL 5), and NRAS (TLL 6), slotted well into the overall trend in stability of the RNA G-quadruplex libraries according to the total loop length (Figure 4 and Tables 1 and 2). This indicates that the thermodynamic trends and boundaries derived from this study may serve as indicators of the thermodynamic stability of individual RNA G-quadruplex-forming sequences. However, we also found that the ΔGVH determined for TERRA (TLL 9, −16.3 ± 0.6 kJ mol−1) was higher than that of RNA library L333 (−12.9 ± 0.3 kJ mol−1), suggesting that in some cases the stability of an individual RNA G-quadruplex-forming sequence can significantly differ from the stability determined from the RNA G-quadruplex mixtures. In the case of mixtures of two G-quadruplex sequences with similar melting temperatures, it has been demonstrated that the slope of the apparent single transition in the melting curve is typically lower than that of the component G-quadruplexes, thereby leading to an overall lower ΔH value.59 The heterogeneity of sequences in the RNA G-quadruplex libraries studied here may therefore lead to thermodynamic values lower than those of the component sequences, where the effects of heterogeneity may become more relevant with increasing loop length.
RNA G-Quadruplex Topology Is Predominantly Parallel and Independent of Loop Length
Under our experimental conditions, we consistently observed CD signals that can be attributed to G-quadruplexes with a parallel conformation for all of the RNA oligonucleotide libraries. However, we cannot rule out the possibility that a minor population of sequences within a library may adopt another G-quadruplex conformation or an alternative structure, and this may be more applicable in the case of libraries with longer loop lengths, which are more complex. The general adherence of all of the RNA oligonucleotide libraries to parallel G-quadruplex formation is in agreement with all published circular dichroism analyses of individual RNA G-quadruplexes to date.17–22,26,41,56,57,60 This study is in contrast to our previous study of DNA35 in which a total loop length of more than five nucleotides served as a minimum threshold for the formation of an antiparallel or mixed-type hybrid population. This topological difference between DNA and RNA G-quadruplex structures is also exemplified by the G-quadruplexes formed from human telomeric repeats of d(GGGTTA)n and r(GGGUUA)n. While the structural polymorphism of the human telomeric DNA G-quadruplex has been exhaustively demonstrated to be influenced by the monovalent ion identity (K+ or Na+), the flanking sequences, and whether the G-quadruplex is in crystal or in solution,41,51–53,61 the NMR solution and crystal structures of TERRA RNA G-quadruplexes have consistently shown a parallel folding topology whether in a K+ solution,15 a Na+ solution,14 or a K+ crystal in the absence16 and presence58 of ligand.
Our systematic study also revealed other features that appear to be dependent on loop length. We observed a shift in the maximal and minimal peaks toward shorter wavelengths for RNA oligonucleotide libraries L111 and L112 et al. in comparison to the RNA libraries with longer loops. This is suggestive of subtle differences in the overall G-quadruplex structure that could be due to differences in tetrad stacking, for example, the degree of twist of tetrads about the helical axis or the tilt of tetrads relative to each other. It is conceivable that the restricted dynamics of shorter loops exert a greater influence in dictating the nature of these structural features compared to longer loops.
Biological Significance
With the recent emergence of RNA G-quadruplex structures as potential active players in the regulation of gene expression, the physical properties of RNA G-quadruplexes stimulate questions about how their high thermodynamic stability and undeviating overall structures are compatible with functional roles in the transcriptome. We and others previously noticed that most of the intramolecular DNA quadruplex-forming sequences with assigned putative biological functions to date contained at least one mononucleotide loop, for example, c-myc,62 VEGF,63 Bcl-2,48 and HIF-1α.64 These sequences, however, represent the upper end of stability among DNA G-quadruplexes, and these studies altogether suggest that stability could be an indicator of biological function(s) for DNA G-quadruplexes. In the case of RNA, published reports of naturally occurring RNA G-quadruplex sequences cover a larger diversity of loop lengths and arrangements; however, all possess remarkably thermodynamically stable structures irrespective of loop length, and this appears to facilitate their role as robust, but mechanically challenging, regulatory elements for translation. There are some examples of enzymes with a preference for G-quadruplex RNA substrates such as the RHAU helicase65 and the mXRN1p exoribonuclease,66 which could represent mechanisms that evolved so that the cell could cope with the thermodynamic properties of RNA G-quadruplexes.
The combined evidence so far indicates that the parallel folding topology of RNA G-quadruplexes is preserved irrespective of the exact loop sequence, loop length, molecularity, stabilizing cation, number of tetrads, or physical state (dilute solution, molecularly crowded, or crystal). The topological control exerted by G-quadruplex-forming RNA sequences may confer a potential biological advantage. Indeed, some G-quadruplex-binding proteins have been shown to select between folding topologies, including the nucleolin67 and insulin68 proteins that preferentially bind parallel folded DNA G-quadruplexes. Small molecule binding discrimination between G-quadruplexes formed from human telomeric DNA and RNA sequences has also been demonstrated for naphthalene diimides,69 while the ability of G-quadruplex-binding small molecules to modulate gene expression in in vitro reporter assays has been shown for the NRAS70 and TRF220 RNA G-quadruplexes. Although it is generally thought that loop regions are not stabilized, it is significant that hydrogen bonding involving the 2′-OH group has been shown to actively participate in stabilizing ligand-induced loop conformations in RNA G-quadruplexes.58
By employing libraries designed to report on systematic, averaged, and sequence-independent variations in loop lengths of RNA G-quadruplex-forming sequences, we have established trends in stability and structural topology that can be exploited for their predictive value. Our framework of thermodynamic stabilities could potentially be used to aid the development of RNA folding algorithms that integrate RNA G-quadruplex formation.
Supplementary Material
Supporting Information UV melting and annealing curves for RNA–DNA oligonucleotide libraries and naturally occurring individual RNA G-quadruplex sequences, Tm values of L111 melting as a function of RNA concentration, Tm versus loop length, CD overlays of representative RNA oligonucleotide libraries folded in the presence in 5 mM potassium, comparison of ΔGVH trends between RNA and DNA G-quadruplex libraries, and molar extinction coefficients. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
We thank Dr. Neil Bell and Dr. Pierre Murat for proofreading the manuscript.
Funding This work was supported by the BBSRC and Cancer Research UK. A.Y.Q.Z. is funded by Trinity College.
ABBREVIATIONS
- CD
circular dichroism
- G
guanine
- UTR
untranslated region
- Ljkl et al.
RNA oligonucleotide libraries Ljlk, Lljk, and Lkjl
- TLL
total loop length
- Tm
melting temperature
- UV
ultraviolet
- ΔHVH
van’t Hoff enthalpy
- ΔSVH
van’t Hoff entropy
- ΔGVH
van’t Hoff free energy change
REFERENCES
- (1).Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- (2).Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- (3).Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: A database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- (9).Lansdorp PM. Major cutbacks at chromosome ends. Trends Biochem. Sci. 2005;30:388–395. doi: 10.1016/j.tibs.2005.05.004. [DOI] [PubMed] [Google Scholar]
- (10).Mergny J-L, Riou J-F, Maillieet P, Teulade-Richou M-P, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat-Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- (12).Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- (13).Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xu Y, Kaminaga K, Komiyama M. G-Quadruplex Formation by Human Telomeric Repeats-Containing RNA in Na+ Solution. J. Am. Chem. Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- (15).Martadinata H, Phan AT. Structure of Propeller-Type Parallel-Stranded RNA G-Quadruplexes, Formed by Human Telomeric RNA Sequences in K+ Solution. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- (16).Collie GW, Haider SM, Neidle S, Parkinson GN. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5′ Untranslated Region Contains an RNA G-Quadruplex-Forming Motif That Modulates Protein Expression. Biochemistry. 2010;49:8300–8306. doi: 10.1021/bi100957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gomez D, Guedin A, Mergny J-L, Salles B, Riou J-F, Teulade-Fichou M-P, Calsou P. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Morris MJ, Basu S. An Unusually Stable G-Quadruplex within the 5′-UTR of the MT3Matrix Metalloproteinase mRNA Represses Translation in Eukaryotic Cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- (22).Balkwill GD, Derecka K, Garner TP, Hodgman C, Flint APF, Searle MS. Repression of Translation of Human Estrogen Receptor α by G-Quadruplex Formation. Biochemistry. 2009;48:11487–11495. doi: 10.1021/bi901420k. [DOI] [PubMed] [Google Scholar]
- (23).Beaudoin J-D, Perreault J-P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-Quadruplex Is Essential for Cap-Independent Translation Initiation in Human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- (26).Wieland M, Hartig JS. RNA Quadruplex-Based Modulation of Gene Expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- (27).Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- (28).Rachwal PA, Brown T, Fox KR. Effect of G-Tract Length on the Topology and Stability of Intramolecular DNA Quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- (29).Rachwal PA, Findlow IS, Werner JM, Brown T, Fox KR. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Guedin A, Gros J, Alberti P, Mergny J-L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Guedin A, Alberti P, Mergny J-L. Stability of intramolecular quadruplexes: Sequence effects in the central loop. Nucleic Acids Res. 2009;37:5559–5567. doi: 10.1093/nar/gkp563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Guedin A, De CA, Gros J, Lacroix L, Mergny J-L. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- (33).Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-Length-Dependent Folding of G-Quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- (34).Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bugaut A, Balasubramanian S. A Sequence-Independent Study of the Influence of Short Loop Lengths on the Stability and Topology of Intramolecular DNA G-Quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kuryavyi V, Patel DJ. Solution Structure of a Unique G-Quadruplex Scaffold Adopted by a Guanosine-Rich Human Intronic Sequence. Structure. 2010;18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- (39).Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- (40).Sacca B, Lacroix L, Mergny J-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Arora A, Maiti S. Differential Biophysical Behavior of Human Telomeric RNA and DNA Quadruplex. J. Phys. Chem. B. 2009;113:10515–10520. doi: 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- (42).Mergny J-L, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- (43).Luke B, Lingner J. TERRA: Telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- (45).Jin R, Gaffney BL, Wang C, Jones RA, Breslauer KJ. Thermodynamics and structure of a DNA tetraplex: A spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl. Acad. Sci. U.S.A. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel G-Strands Formed in the Human BCL-2 Promoter Region in Solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Rezler EM, Seenisamy J, Bashyam S, Kim M-Y, White E, Wilson WD, Hurley LH. Telomestatin and Diseleno Sapphyrin Bind Selectively to Two Different Forms of the Human Telomeric G-Quadruplex Structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- (50).Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution Structure of the Biologically Relevant G-Quadruplex Element in the Human c-MYC Promoter. Implications for G-Quadruplex Stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- (51).Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- (53).Phan AT. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2009;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- (54).Heddi B, Phan AT. Structure of Human Telomeric DNA in Crowded Solution. J. Am. Chem. Soc. 2011;133(25):9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- (55).Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- (56).Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- (57).Zhang D-H, Fujimoto T, Saxena S, Yu H-Q, Miyoshi D, Sugimoto N. Monomorphic RNA G-Quadruplex and Polymorphic DNA G-Quadruplex Structures Responding to Cellular Environmental Factors. Biochemistry. 2010;49:4554–4563. doi: 10.1021/bi1002822. [DOI] [PubMed] [Google Scholar]
- (58).Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural Basis of Telomeric RNA Quadruplex-Acridine Ligand Recognition. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- (59).Rachwal PA, Fox KR. Quadruplex melting. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- (60).Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5′-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Phan AT, Modi YS, Patel DJ. Propeller-Type Parallel-Stranded G-Quadruplexes in the Human c-myc Promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the Presence of a Guanine Quadruplex Forming Region within a Polypurine Tract of the Hypoxia Inducible Factor 1α Promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- (65).Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 Resolvase 1 Binds Both DNA and RNA Tetramolecular Quadruplex with High Affinity and Is the Major Source of Tetramolecular Quadruplex G4-DNA and G4-RNA Resolving Activity in HeLa Cell Lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Bashkirov VI, Scherthan H, Solinger JA, Buerstedde J-M, Heyer W-D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Gonzalez V, Guo K, Hurley L, Sun D. Identification and Characterization of Nucleolin as a c-myc G-quadruplex-binding Protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Schonhoft JD, Das A, Achamyeleh F, Samdani S, Sewell A, Mao H, Basu S. ILPR repeats adopt diverse G-quadruplex conformations that determine insulin binding. Biopolymers. 2009;93:21–31. doi: 10.1002/bip.21289. [DOI] [PubMed] [Google Scholar]
- (69).Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- (70).Bugaut A, Rodriguez R, Kumari S, Hsu S-TD, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;8:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information UV melting and annealing curves for RNA–DNA oligonucleotide libraries and naturally occurring individual RNA G-quadruplex sequences, Tm values of L111 melting as a function of RNA concentration, Tm versus loop length, CD overlays of representative RNA oligonucleotide libraries folded in the presence in 5 mM potassium, comparison of ΔGVH trends between RNA and DNA G-quadruplex libraries, and molar extinction coefficients. This material is available free of charge via the Internet at http://pubs.acs.org.