Abstract
A marked escalation in the rate of discovery of new types of human polyomavirus has occurred over the last five years largely owing to recent technological advances in their detection. Among the newly discovered viruses, Merkel Cell Polyomavirus (MCPyV or MCV) has gained the most attention due to its link with a rare human cancer. Infection with MCPyV is common in the human population, and the virus is detected in several anatomical locations, but most frequently in skin. Study of MCPyV molecular virology has been complicated by the lack of straightforward cell culture models, but recent in vitro studies are making strides towards understanding the virus life cycle, its cellular tropism, and mode of transmission. While MCPyV shares several traditional traits with other human polyomaviruses, the burst of research since its discovery reveals insight into a virus with many unique genetic and mechanistic features. The evidence for a causal link between MCPyV and the rare neuroendocrine cancer, Merkel Cell Carcinoma (MCC), is compelling. A majority of MCCs contain clonally integrated viral DNA, express viral T antigen transcripts and protein, and exhibit an addiction to the viral large T and small t antigen oncoproteins. The MCPyV large T antigen contains MCC tumor-specific mutations that ablate its replication capacity but preserve its oncogenic functions, and the small t antigen promotes an environment favorable for cap-dependent translation. The mechanisms of MCPyV-induced transformation have not been fully elucidated, but the likely etiological role of this new polyomavirus in human cancer provides a strong opportunity to expand knowledge of virus-host interactions and viral oncology.
Introduction
For over fifty years, study of the Polyomaviridae family of viruses has yielded significant contributions to the understanding of basic cell biology, cancer biology, and viral oncogenesis. There are currently three genera and twenty-two different species of polyomaviruses, all of which are non-enveloped, icosahedral viruses that contain a circular, double-stranded DNA genome of approximately 5,000 base pairs (Johne et al., 2011). The transforming potential of these viruses has long been appreciated since isolation of the first polyomavirus by Ludwik Gross in 1953. Gross demonstrated that a transmissible agent, the now-archetypal murine polyomavirus (MPyV), induced tumors in mice (Gross, 1953, 1954). The field of polyomavirus virology expanded in 1960 with the discovery of simian vacuolating virus 40 (SV40) in monkey kidney cell cultures used for production of human poliovirus vaccines (Sweet and Hilleman, 1960). Subsequent studies reporting the oncogenic potential of SV40 (Eddy et al., 1961; Eddy et al., 1962; Rabson and Kirschstein, 1962; Shein and Enders, 1962) thrust this virus into the sphere of human health and sparked concern over the administration of a vaccine with an oncogenic contaminant to a large percentage of the human population. Over the next several decades, intense research efforts attempted to establish a causal relationship between SV40 and human cancers, but to date no definitive link has been made (Reviewed in Poulin and DeCaprio, 2006). Regardless, the seminal contributions to science that arose from these efforts and from the study of polyomaviruses in general cannot be understated, and include discoveries of p53 (Lane and Crawford, 1979; Linzer and Levine, 1979), the first nuclear localization signal (Lanford and Butel, 1984), tyrosine phosphorylation (Eckhart et al., 1979) and PI3K signaling (Whitman et al., 1985), and basic mechanisms in the process of DNA replication (Li and Kelly, 1984; Waga and Stillman, 1994).
The detection of polyomaviruses that infect humans has rapidly escalated in the last decade. There are currently eleven identified human polyomaviruses, nine of which have been discovered in the last five years (Reviewed in Buck et al., 2012; Chang and Moore, 2012; Siebrasse et al., 2012). Nearly half of these human polyomaviruses are linked to human diseases, although many of these associations remain tenuous. To date, Merkel Cell Polyomavirus (MCPyV; also referred to as MCV) is the only human polyomavirus with a robust collection of scientific evidence supporting its classification as a causative agent of a human malignancy, Merkel Cell Carcinoma (MCC).
Merkel Cells and Merkel Cell Carcinoma
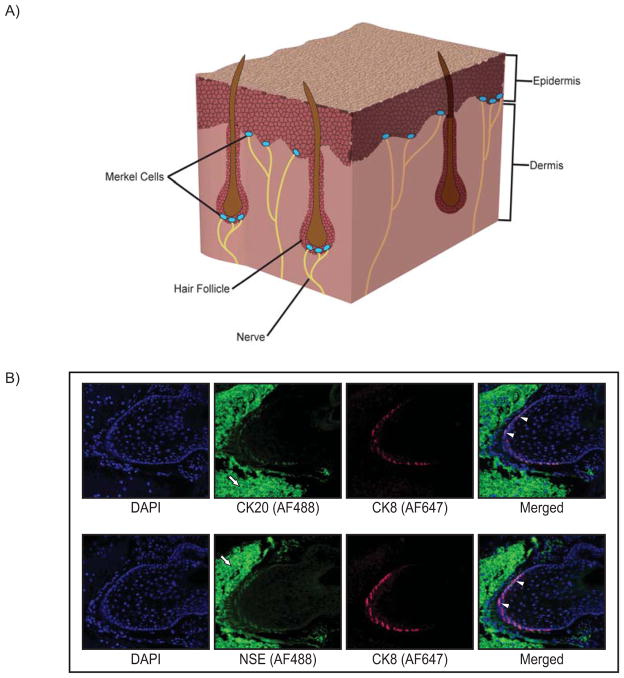
First described as ‘touch cells’ by Friedrich Sigmund Merkel in 1875, Merkel cells are found in hair follicles, certain mucosal tissues, and are most abundant in areas of the skin involved in the sensation of touch (Fig. 1A). In vertebrate skin, Merkel cells are located in the basal layer of the epidermis, and it is here where the structure and function of these cells are best characterized. At their position adjacent to the dermis, Merkel cells are bound to the terminal ends of sensory nerve fibers. These nerve-associated Merkel cells serve as mechanoreceptors that transduce mechanical stimuli from the skin. Morphological and ultrastructural characteristics of Merkel cells include a round or oval shape, electron-dense core secretory granules surrounded by a clear ‘halo’, and spinous projections on their surface. A panel of immunohistochemical markers is used to detect and distinguish Merkel cells, and typically includes cytokeratins 8, 18, 19, and 20 and the neuroendocrine markers chromogranin A, synaptophisin, and neuron-specific enolase (Fig. 1B). Of these markers, cytokeratin 20 is considered the most sensitive and specific diagnostic marker of Merkel cells. (Reviewed in Gould et al., 1985; Halata et al., 2003; Lucarz and Brand, 2007; Moll et al., 2005; Sidhu et al., 2005).
Figure 1. Merkel cell location and immunoreactivity.
A) Cutaneous Merkel cells located in the basal layer of the epidermis associated with the terminal ends of nerves, and also enriched in the bulge region of hair follicles, as depicted in a section of skin tissue. B) Whisker pad tissue was harvested from mice, paraffin-embedded, and representative sections analyzed by indirect immunofluorescence using Merkel cell-specific markers. Top panel: Images of a murine vibrissae follicle co-stained with antibodies specific to the Merkel cell markers cytokeratin 8 (CK8; red Alexa-Fluor 647) and cytokeratin 20 (CK20; green Alexa-Fluor 488). Bottom panel: A vibrissae follicle co-stained with the Merkel cell-specific markers CK8 (red; Alexa-Fluor 647) and neuron-specific enolase (NSE; green Alexa-Fluor 488). In each panel, representative cells staining positive for both Merkel cell markers are indicated with white arrowheads. High background staining with the Alexa-Fluor 488 secondary antibody is evident in cells surrounding the follicle (white arrows). This background staining was also seen in no primary antibody controls (data not shown). Both immunostaining analyses highlight the hair/whisker follicle as a prominent location of Merkel cells.
Merkel cell carcinoma (MCC) is a malignancy of cutaneous neuroendocrine cells, and is therefore generally believed to originate from the transformation of Merkel cells (Gould et al., 1985). It was first described by Toker in 1972 (Toker, 1972), yet the pathogenesis and etiology of MCC remains complex and poorly understood. Approximately 1,500 new cases of MCC are diagnosed per year in the US, representing a relatively rare cancer; however, the incidence of MCC has tripled in the last two decades and annual diagnoses continue to climb by 5–10% (Agelli and Clegg, 2003; Becker et al., 2009b; Calder and Smoller, 2010; Hodgson, 2005; Lemos and Nghiem, 2007). Moreover, despite the rarity of MCC, it is one of the most lethal and aggressive skin cancers with a mortality rate greater than 30% (Hodgson, 2005).
The most common clinical features of MCC are summarized by the acronym “AEIOU” (Heath et al., 2008): Asymptomatic, Expanding rapidly, Immune suppression, Older than age 50, and UV-exposed site on a fair-skinned individual. There is a striking increase in the incidence of MCC in HIV-infected individuals (Engels et al., 2002), chronic lymphocytic leukemia patients (Vlad and Woodlock, 2003), and following organ transplantation (Buell et al., 2002; Heath et al., 2008; Penn and First, 1999). MCC is primarily a disease of the elderly, with the mean age of onset for men and women being 74 and 76 years of age, respectively (Agelli and Clegg, 2003). In addition, MCC occurs almost exclusively in Caucasians at sites of the body that are frequently sun-exposed, and disease rates are correlated with equatorial proximity (Agelli and Clegg, 2003; Albores-Saavedra et al., 2010; Heath et al., 2008; Miller and Rabkin, 1999). Interestingly, Merkel cells are more concentrated in sun-exposed compared to non-exposed skin (Moll et al., 1990) and UV-specific mutations have been observed in the p53 gene of MCCs (Popp et al., 2002). Taken together, these statistics suggest that a combination of immunosuppression and the potential oncogenic pathways elicited by UV exposure are involved in MCC pathogenesis, yet these factors alone do not fully account for the etiology of MCC.
Discovery of Merkel Cell Polyomavirus
Based mainly on the aforementioned link between MCC and immune suppression, the research team led by Patrick Moore and Yuan Chang at the University of Pittsburgh sought to determine whether MCC has an infectious origin. This group, in particular, was well-suited for the search given their previous discovery of Kaposi’s sarcoma-associated herpesvirus (KSHV) in AIDS-associated Kaposi’s sarcoma tissue using a technique known as representational difference analysis (RDA) (Chang et al., 1994). RDA uses PCR amplification of restriction fragments to create ‘representations’ of the DNA genome of normal or diseased tissue. After ligating the diseased tissue representation with priming sequences, it is mixed with unligated representation DNA from normal tissue. Following multiple rounds of PCR amplification in a subtractive hybridization process, those sequences that fail to hybridize with normal tissue counterparts become doubly primed and thus enriched during PCR. While the RDA technique was used to successfully identify KSHV, it relies heavily on the presence of non-diseased tissue controls and is rather non-quantitative.
To begin the search for viral sequences in MCC, the Chang-Moore research group built on the principles of RDA and modernized the technique by combining it with the availability of sequences from the entire human genome. The product was a related but distinct method known as digital transcriptome substraction (DTS) (Feng et al., 2007). DTS uses a combination of high-throughput cDNA sequencing and in silico methods to sequentially exclude human sequences from a tissue sample transcriptome. For each normal or diseased sample, cDNA libraries are created and then subjected to sequencing, and the results compared to human sequence databases. In 2008, the DTS technique was applied to four MCC tumors and revealed previously unidentified viral transcripts with high sequence-identity to archival polyomavirus tumor antigen (T antigen) sequences. Ultimately, a full-length, ~5.4 kilobase circular viral genome (prototype MCV350) was sequenced from a MCC tumor and was aptly named Merkel cell polyomavirus (Feng et al., 2008).
The Merkel cell polyomavirus (MCPyV) genome is similar to other human polyomaviruses in that it includes a conserved replication origin, as well as opposing early and late gene regions that express small and large T antigens (sT and LT) and VP1, VP2, and VP3, respectively (Feng et al., 2008; Shuda et al., 2008). While many domains of MCPyV share homology with their well-studied human polyomavirus counterparts, the MCPyV genome is most similar to the African Green Monkey lymphotropic polyomavirus and is phylogenetically classified in the murine polyomavirus clade (Dalianis et al., 2009; Feng et al., 2008). However, based on the most recent and revised taxonomical classification of the Polyomaviridae family, MCPyV is now classified in the Orthopolyomavirus genus designated for mammalian virus species sharing high nucleotide sequence similarity (Johne et al., 2011).
MCPyV Molecular Virology: Genome Organization
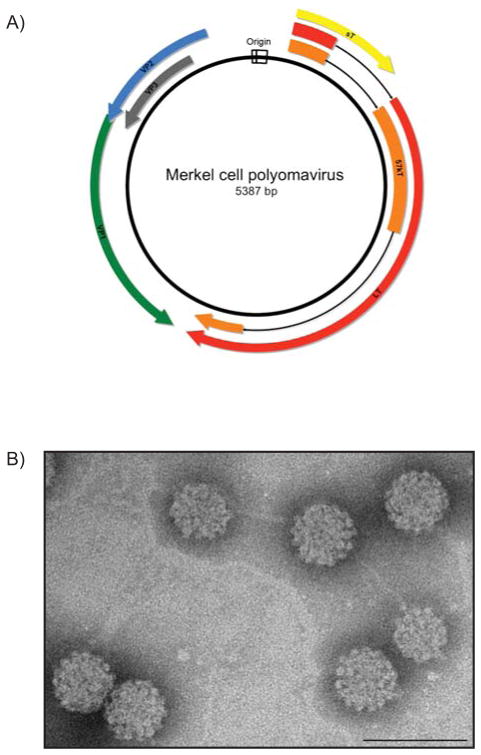
As mentioned above, the genetic components and organization of the MCPyV genome are largely similar to other polyomaviruses (Reviewed in Chang and Moore, 2012; Feng et al., 2008). The prototype virus MCV350 has a circular, double-stranded DNA genome of 5,387 base-pairs, and the genomes of all subsequent MCPyV strains sequenced to date average ~5.4 kilobases (Fig. 2). As is customary for polyomaviruses, the MCPyV genome contains bidirectional early and late coding regions, separated by a non-coding regulatory region that houses the viral replication origin. The segregation of the early and late transcriptional units promotes a temporally-regulated cascade of gene expression distinguished by the onset of viral DNA replication, a replication scheme consistent with other DNA viruses (Feng et al., 2011). The early genes, namely the products of the T antigen locus, are expressed immediately upon infection and are generally involved in replication of the viral DNA genome. Following DNA replication, the late region is transcriptionally activated to express gene products that are structural components of the viral capsid and contribute to progeny virion production during the late phase of infection (Cole and Conzen, 2001). Despite these similarities, the MCPyV viral genome has several unique genetic characteristics compared to other human polyomaviruses. As one example, gene products of both the early (large and small T antigens) and late regions (VP1 and VP2) of Merkel cell polyomavirus are more closely related to murine polyomaviruses than to the other human polyomaviruses (Feng et al., 2008).
Figure 2. The Merkel Cell Polyomavirus (MCPyV) genome and capsids.
A) Schematic of the MCPyV prototype MCV350. The double-stranded DNA genome is approximately 5.4 kilobases in length. The bi-directional origin of replication (white) promotes temporal expression of discrete regions of the genome. The early region encodes three viral T antigens: large T antigen (LT; red), 57 kT antigen (57 kT; orange) and small t antigen (sT; yellow). The late region is transcribed in the opposite direction from the origin and encodes the viral structural proteins: the major capsid protein Virus Protein 1 (VP1; green), and the minor capsid proteins Virus Protein 2 and Virus Protein 3 (VP2; blue, VP3; gray). B) Transmission electron micrograph of MCPyV virus-like particles composed of VP1 and VP2 proteins. The VP1 and VP2 proteins self-assemble into ~50 nm icosahedral particles. (Scale bar=100 nm)
Origin of Replication
The non-coding regulatory region (NCRR) of MCPyV contains a 71-base pair minimum origin of replication that, like other polyomaviruses, has an AT-rich tract, LT-binding domain, and early enhancer region. However, the MCPyV origin of replication has several unique characteristics. Polyomavirus origins of replication traditionally contain pentanucleotide consensus sequences G(A/G)GGC that help direct assembly of LT proteins on the DNA by specifically binding the origin-binding domain (OBD) within the central portion of LT, an event that in turn allows LT to function as a helicase and direct the initiation of viral DNA replication (Reviewed in Cole and Conzen, 2001; Fanning and Zhao, 2009; Pipas, 1992). In the MCPyV origin, however, there are more pentanucleotide sequences than any other polyomavirus, and these pentameric elements exist in closer proximity to one another than in other polyomaviral origins (Johnson, 2010; Kwun et al., 2009). Kwun and colleagues suggested that this close proximity likely allows OBD-OBD interaction between LT proteins bound at the origin. More recently, a crystal structure of the MCPyV LT OBD in complex with a segment of the MCPyV origin of replication was reported and verifies that OBD-OBD intermolecular interactions occur on LT proteins bound to the origin of replication (Harrison et al., 2011). As determined by mutational analysis, these OBD-OBD interactions are not required for LT binding to the origin, nor do they appear to affect the structure of the viral DNA. Such intermolecular interactions do not occur on the SV40 origin, as the OBD-binding pentanucleotide sequences are more spatially separated (Bochkareva et al., 2006; Meinke et al., 2007). Secondary-structure matching analysis reveals a striking similarity between MCPyV LT and the E1 origin-binding protein of bovine papillomavirus (Harrison et al., 2011). The crystal structure also confirmed the previous report by Kwun et. al. of the unique combination of pentanucleotide sequences that are required for efficient MCPyV replication. Taken together, these features indicate that replication at the MCPyV viral origin and the process of LT seeding may be somewhat distinct and more complex in comparison to other polyomaviruses.
Early Region: The T Antigen Locus
The MCPyV early region spans nearly 3 kilobases of the 5.4 kilobase genome, and its genes are the first to be expressed upon infection (Feng et al., 2011; Feng et al., 2008; Neumann et al., 2011). Polyomavirus early region gene products historically target cellular proteins that function in cell cycle regulation and tumor suppression (DeCaprio et al., 1988; Lane and Crawford, 1979; Linzer and Levine, 1979; Pallas et al., 1990). Because these gene products are designated as tumor antigens, or T antigens, the early region is frequently referred to as the “T antigen locus”. The polyomavirus T antigens also perform key functions in the initiation of viral DNA synthesis. The MCPyV early region expresses three T antigens: large T antigen (LT), small T antigen (sT), and 57kT antigen (57 kT) (Feng et al., 2008; Shuda et al., 2009; Shuda et al., 2008). There is no evidence that MCPyV expresses proteins analogous to middle T antigen or agnoprotein expressed by other polyomaviruses. The basic genetic features and functional domains of the three MCPyV T antigens will be reviewed in the following section. Discussion of their roles in viral replication and their potential roles in transformation and MCPyV-induced carcinogenesis are also described in subsequent sections.
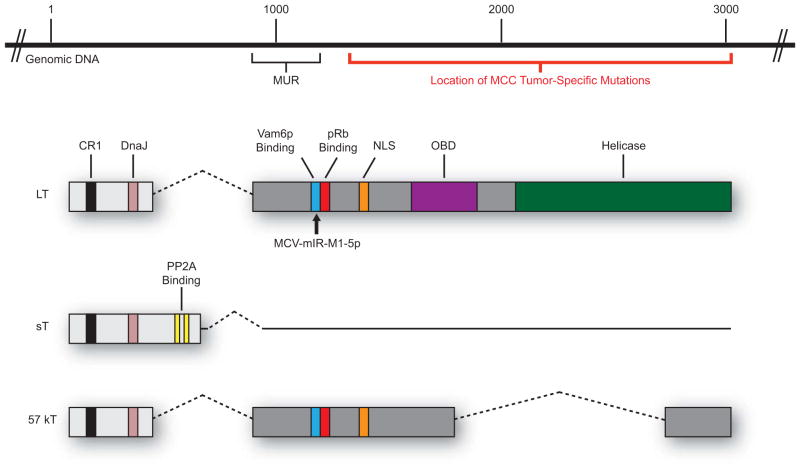
All three MCPyV T antigens share a short, common amino-terminus, but alternative splicing downstream of the first exon ultimately gives rise to three unique proteins (Fig. 3). The MCPyV LT protein is the largest of the three T antigens and is composed of two exons that together encode for a protein of 816 amino acids. As shown in Figure 3, MCPyV LT has conserved features important for well-characterized LT functions, including CR1 (LXXLL) and DnaJ (HPDKGG) domains, a pRb-binding domain (LXCXE), an origin-binding domain (OBD), and a helicase domain (Reviewed in Pipas, 1992). For this reason, it is inferred that MCPyV LT generally shares many functions with other polyomavirus LTs. A nuclear localization signal (NLS) is located between the pRb-binding LXCXE motif and the OBD and is conserved in other MCPyV isolates (Nakamura et al., 2010). The carboxyl terminal half of LT contains most of the domains involved in viral DNA replication, including the OBD, ATPase domain, and helicase domains. This region of LT is also the site of tumor-specific mutations, the significance of which will be discussed in further detail in later sections. It should be noted that MCPyV LT is clearly distinct from other human polyomaviruses and shares only 30% amino acid identity with their human polyomavirus counterparts (Feng et al., 2008; Johnson, 2010). One unique aspect of MCPyV LT is the presence of approximately 200 additional amino acids at the beginning of exon 2 just proximal to the pRb-binding LXCXE domain called the “MCV-Unique Region”, or MUR. The MUR region of LT binds a cellular factor, Vam6p, and redistributes it to the nucleus. This function appears to be unique to MCPyV LT (Liu et al., 2011). The MCPyV LT pRb-binding domain is unique in that it contains a ‘spacer region’ that effectively separates the ‘Psycho’ domain from the LXCXE motif, and also contains complementary sequence for a MCPyV viral microRNA (Johnson, 2010; Seo et al., 2009). This region is unique to MCPyV LT and is contained within the MUR described by Liu and colleagues, but the effects of this additional sequence on LT function have not yet been determined.
Figure 3. The MCPyV T antigen locus.
The MCPyV T antigen locus is located in the early region of the genome and encodes three T antigen isoforms that arise by alternative splicing. The large T antigen (LT) shares many traditional domains found in other polyomaviruses: CR1 (LXXLL; black), DnaJ (HPDKGG; pink), pRb-binding (LXCXE; red), a nuclear localization signal (NLS; orange), origin-binding domain (OBD; purple), and a helicase domain (green). LT also contains unique features, such as a binding site for Vam6p (blue) and a 200-amino acid “MCV-Unique Region” (MUR). The location of reported tumor-specific mutations in the 3′ region of LT is depicted with a red bar. The site of MCPyV microRNA (MCV-mIR-M1-5p) complementarity in LT is denoted (black arrow). The small t antigen (sT) shares the first exon with LT, but due to alternative splicing also contains a protein phosphatase 2A-binding site (PP2A; yellow). The 57 kT antigen is identical to LT up to the NLS motif, at which point differential splicing precludes the inclusion of complete OBD and helicase domains.
In addition to LT, the T antigen locus of MCPyV encodes for a small T (sT) protein. As mentioned previously, the MCPyV sT protein shares the same amino terminus with LT and 57 kT (Fig. 3) and therefore has conserved region 1 (CR1) and heat shock protein-binding (DnaJ) motifs. However, transcriptional read-through of the first exon splice site used by LT and 57 kT gives rise to a small protein of 186 amino acids with a unique carboxyl terminus that harbors a protein phosphatase 2A (PP2A) binding site. The ability to bind PP2A is a conserved function of polyomavirus sT proteins and this association has been shown to play an important role in virus-induced cellular transformation by other polyomaviruses (Pallas et al., 1990). A recent study by Shuda and colleagues provided initial insight into the functional characteristics of MCPyV sT protein, which include physical association between sT and the PP2A A3 subunit in co-immunoprecipitation assays and promoting sustained phosphorylation of the cellular translation factor eIF4E binding protein 1 (4E-BP1) (Shuda et al., 2011). So far, the MCPyV sT protein has been implicated in the process of viral DNA replication (Feng et al., 2011; Kwun et al., 2009) and cellular transformation (Shuda et al., 2011), both functions that will be explored in further detail in upcoming sections.
The early region also expresses a third T antigen species, 57 kT. The 57 kT protein is the product of a transcript that undergoes alternative splicing to join three exons encoding for 432 amino acids, and this protein shares many of the same motifs present in LT and sT. The 57 kT protein has the same amino terminus as both LT and sT, including the CR1 and DnaJ domains, and also contains the MUR, pRb-binding LXCXE motif, and the most C-terminal 100 amino acids that are present in LT. Information on the role of 57 kT in the viral life cycle or MCPyV-induced tumorigenesis is sparse. The 57 kT antigen is frequently described as an analogue of the SV40 17 kT antigen (Zerrahn et al., 1993), which appears to function both independently of and in cooperation with the other T antigens in the process of cellular proliferation in vivo (Comerford et al., 2012).
The Late Region: Structural Proteins
The late region of the MCPyV genome contains open reading frames for the major capsid protein viral protein 1 (VP1) and the minor capsid proteins 2 and 3 (VP2 and VP3). Interestingly, MCPyV lacks the conserved genetic architecture that normally gives rise to the VP3 protein. This sequence aberration, coupled with failed attempts to create viable VP3 expression vectors (Pastrana et al., 2009) and abnormal VP1 localization reminiscent of polyomaviruses lacking a VP3 protein (Neumann et al., 2011), suggests that MCPyV either expresses a non-functional VP3 or does not express this protein at all. Regardless, MCPyV particles composed of VP1 and VP2, either as a result of their ectopic expression and self-assembly into virus-like particles (VLPs) (Tolstov et al., 2009) or from replication of genetically engineered consensus genomes (Feng et al., 2011; Neumann et al., 2011; Schowalter et al., 2011), have an average particle size (~40–55nm) consistent with other polyomavirus particles (Fig. 2B).
MCPyV MicroRNA
The MCPyV genome expresses a 22-nucleotide viral miRNA (MCV-miR-M1-5p) from the late strand that most likely autoregulates early viral gene expression during the late phase of infection, as it was shown to reduce the level of reporter transcripts containing MCPyV early region sequences (Lee et al., 2011; Seo et al., 2009). This miRNA has complete reverse complementarity to a sequence in LT adjacent to the LXCXE motif, implying that it targets this early protein for autoregulation, and is also predicted to affect a variety of cellular targets with potentially relevant functions in transformation (Johnson, 2010; Lee et al., 2011; Seo et al., 2009). One study reported that miRNA expression is preserved in approximately 50% of MCPyV-positive MCC tumors, and the level of miRNA expression correlates with viral genome copy number in the tumor (Lee et al., 2011). The presence of MCPyV miRNA in MCC tumors certainly warrants further investigation into its role, if any, in the pathogenesis of MCC.
Overall, while the MCPyV genome sequence and organization is reminiscent of other members of the Polyomaviridae family, there are clearly some unique characteristics that may suggest this new human polyomavirus has evolved to have divergent mechanisms and functions for reasons not yet fully appreciated.
MCPyV Molecular Virology: Virus Life Cycle and Replication
MCPyV was originally discovered in MCC, and it is within this malignant milieu where most of its molecular virology is currently being characterized. Details regarding the naturally occurring life cycle and replication of MCPyV are limited as there is no cell culture system yet identified in which virus can be propagated, except for one in which viral LT and sT genes are expressed in trans. Moreover, the precise cellular tropism and cell type that supports productive MCPyV infection remain unclear. These limitations have resulted in a reliance on cell culture-based systems, combined with genetically-engineered consensus genomes or pseudoviruses, for the study of the MCPyV life cycle and replication.
Attachment and Entry
A report by Erickson et. al. first identified a potential cellular receptor for MCPyV (Erickson et al., 2009). Through the use of hemagluttination and sucrose gradient flotation assays, the authors found that MCPyV capsomeres composed of recombinant VP1 proteins bound sialic acid components of the ganglioside Gt1b. The results of this initial study suggested that MCPyV abides by the typical attachment and uptake processes used by other polyomaviruses, which primarily utilize carbohydrates with sialic acid as primary receptors (Reviewed in Neu et al., 2009). However, subsequent studies suggest a more nuanced scenario. Schowalter, Pastrana, and Buck tested the ability of MCPyV pseudovirions (composed of VP1, VP2, and a luciferase reporter plasmid) to bind and transduce encapsidated reporter DNA to the nucleus of cultured A549 cells (Schowalter et al., 2011). They presented compelling evidence that MCPyV requires sulfated glycosaminoglycans, particularly heparan sulfate, for infectious entry. This is not unlike what is thought to be the case for papillomaviruses (Reviewed in Horvath et al., 2010). Interestingly, while MCPyV pseudovirions efficiently bound to Lec2 cells that lack sialylated glycans like Gt1b, the particles were deficient in gene transduction. These results led the authors to speculate that MCPyV utilizes a glycosaminoglycan as a primary receptor, but then requires a sialylated glycan as a post-attachment co-receptor for gene transduction. Determination of the MCPyV VP1 crystal structure ultimately supports this model (Neu et al., 2012). Neu and colleagues identified sialic acid binding sites on VP1 that are required for a post-attachment step in MCPyV infectious entry; however, mutation of these sites did not preclude initial cellular attachment of MCPyV pseudovirions. Taken together, these initial reports suggest a novel infectious entry process that requires glycosaminoglycans for initial attachment and subsequent association with sialic acid for gene transduction. The use of sialic acid as a secondary receptor by MCPyV differs from the current consensus for other polyomaviruses (Neu et al., 2011), and provides further evidence that this virus is unique among Polyomaviridae family members.
In Vitro Replication Studies
Other recent efforts have focused on characterization of MCPyV tissue tropism and replication. The ability of a cell type or tissue to support efficient infectious entry of a virus particle and ultimately support the production of infectious progeny virions can imply tropism of a virus. So far, no clear tropism has been defined for MCPyV. In one report, the infectivity of MCPyV pseudovirions was determined on a panel of sixty human cancer cell lines (NCI-60) by measuring infectious entry of an encapsidated reporter gene (Schowalter et al., 2012). While MCPyV transduced a wide spectrum of cell types and no clear tropism emerged, human skin-derived primary keratinocytes (HEKa) and transformed melanocyte (melanoma) cell lines supported infectious entry of MCPyV pseudovirions. On the other hand, a transformed keratinocyte cell line (HaCat) and primary melanocytes were resistant to MCPyV pseudovirus entry. These results suggest the need for cautious interpretation of research using cultured cell lines to study MCPyV tropism, but may also imply that the skin is a preferential site for MCPyV infection.
Others have used genetically-engineered consensus viral genomes to study the MCPyV life cycle and the roles of cellular and host proteins in virus replication. Multiple reports were published around the same time attempting to recapitulate the virus life cycle in cultured cells (Feng et al., 2011; Neumann et al., 2011; Schowalter et al., 2011). In the reports by Feng and Neumann, consensus MCPyV genomes were synthesized that each share high homology to DNA found within encapsidated virions shed from healthy human skin (Schowalter et al., 2010). In the report by Feng et. al., weak viral DNA replication and expression of all three T antigen species (LT, sT, 57 kT) and VP1 were detected by Southern blot and immunoblot, respectively, 2–5 days following transfection of circularized MCV-HF DNA genome into 293 cells. Likewise, Neumann et. al. observed replicated genomic DNA and expression of both LT and VP1 transcripts and protein following transfection of circularized MCV-Syn DNA genome. Acknowledging widely variable transfection efficiencies between cell lines, only 3 out of 22 lines tested supported early and late gene expression: human embryonic kidney cells (293), human non-small cell lung carcinoma cells (H1299), and a human neuroectodermal cell line (PFSK-1) (Neumann et al., 2011). Both groups observed gene expression kinetics consistent with a systematic early to late cascade of gene expression that culminated in the detection of virus particles by approximately 4 days post-transfection, although Neumann et. al. observed smaller and more irregularly shaped MCPyV particles than the SV40 counterparts synthesized in a parallel control experiment. In the study by Schowalter and colleagues (Schowalter et al., 2011), MCPyV virions produced in 293TT cells cotransfected with a viral genome isolated from healthy skin and VP1/2 expression vectors were used to infect a cell line expressing the MCPyV LT and sT genes in trans (293-4T). Over a several week period, modest replication of the input viral DNA genome and synthesis of VP1 protein was observed, as well as production of new virus particles (as judged by quantifying yield of virions from cell culture following lysis of the cells). This experimental system, while artifical in that LT and sT are being expressed in trans, comes the closest, to date, to providing the means to study the full life cycle of MCPyV in a laboratory setting, and an efficient means for producing the virus.
Viral and Cellular Factors Involved in Replication
The cellular and viral genes required for efficient MCPyV replication are also coming into focus. Based on other polyomavirus replication schemes, the MCPyV T antigens would be expected to play critical roles in the virus life cycle. Results of in vitro studies on the MCPyV origin of replication confirm these predictions, revealing that LT activity is required for efficient DNA replication and is greatly enhanced by sT (Kwun et al., 2009). The 57 kT antigen does not appear to play a role in replication, and its function in the viral life cycle remains unknown. Mutational analysis of sT indicates that association with PP2A is required for its ability to enhance LT’s viral DNA replication function. Interestingly, the shared DnaJ motif in the amino terminal portion of LT/sT (Fig. 3) is required for LT function in replication, but is dispensable in the context of sT’s ability to augment LT’s DNA replication functions. The role of LT and sT in viral DNA replication is supported by in vitro replication studies. Replication of consensus genomes is greatly enhanced by co-expressing sT, and to a lesser extent LT (Feng et al., 2011), or by supplying the early region in trans (Neumann et al., 2011). And as described above, 293-4T cells, which constitutively express both MCPyV LT and sT proteins, could support not only replication but also produce virions when infected with MCPyV virus (Schowalter et al., 2011). So far, Vam6p is the only cellular factor shown to affect MCPyV replication, which greatly antagonizes MCPyV replication through its interaction with LT (Feng et al., 2011).
Transmission Studies
Despite the advances made in understanding MCPyV replication through in vitro replication assays, all experiments have so far failed to detect cytopathic effect (CPE) or robust serial transmission of virions produced in these culture systems. Co-culture of 293 cells transfected with the MCV-HF DNA genome, in which early and late genes are expressed and virus particles were detected, with naïve cell populations failed to promote secondary virus infection or CPE (Feng et al., 2011). Moreover, repeated freeze/thaw cycles to release cell-associated virions also failed to promote horizontal spread of MCPyV virus (Neumann et al., 2011). These findings indicate that a bona fide experimental system for MCPyV virus propagation in the laboratory has not yet been fully developed. It has now been established that MCPyV DNA (Feng et al., 2008; Foulongne et al., 2010a; Foulongne et al., 2010b; Loyo et al., 2010; Wieland et al., 2009) and encapsidated virions (Schowalter et al., 2010) can be detected on normal human skin. The location of MCPyV detection supports the notion that MCPyV is part of the normal microflora of human skin. In fact, the primary events of MCPyV replication have been shown to occur in cultured monolayers of primary keratinocytes, which are a prominent cell type of the skin and other stratified epithelia (Schowalter et al., 2011). Taken together with the difficulty in reproducing the MCPyV virus life cycle in monolayer cell culture, especially the production of infectious progeny virions and subsequent virus transmission, these findings may imply a dependence on the process of differentiation in the stratified epithelium of the skin. Through this consideration, the potential to formulate a better model of MCPyV tissue tropism and virus propagation can be developed. The understanding of the context of a natural MCPyV infection, including its life cycle, host range, and the isolation of intact virus, remains limited but these discoveries will certainly provide insight into the relation of MCPyV to other polyomaviruses.
MCPyV Epidemiology
Since its discovery in 2008, epidemiological studies have established MCPyV as a common virus infecting the human population. Enzyme immunoassays specific for the immunogenic determinant of MCPyV, the major capsid protein VP1, have been used to determine that up to 80% of the adult population contains serum antibodies to MCPyV (Carter et al., 2009; Kean et al., 2009; Pastrana et al., 2009; Tolstov et al., 2011; Tolstov et al., 2009; Touze et al., 2010). Seropositivity to MCPyV is essentially nonexistent in prenatal blood spots (Gustafsson et al., 2012), yet becomes widely prevalent in children in their first decade of life and continues to increase with age, suggesting exposure to the virus occurs during early childhood (Chen et al., 2011; Kean et al., 2009; Tolstov et al., 2009; Viscidi et al., 2011). By monitoring overt symptoms alongside seroconversion of MCPyV-seronegative individuals, it appears that MCPyV infection is mostly asymptomatic (Tolstov et al., 2011). Viral DNA has been detected in a wide variety of anatomical locations, including the respiratory tract (Babakir-Mina et al., 2010; Bialasiewicz et al., 2009; Goh et al., 2009; Kantola et al., 2009), saliva (Foulongne et al., 2010b; Loyo et al., 2010), lymphoid tissues (Sharp et al., 2009; Shuda et al., 2009; Toracchio et al., 2010), urine (Bofill-Mas et al., 2010; Husseiny et al., 2010; Laude et al., 2010; Mertz et al., 2010), and the gastrointestinal tract (Campello et al., 2011; Feng et al., 2008). Viral DNA detection in these locations is relatively low compared to the skin, where both viral DNA and encapsidated virions can be detected (Feng et al., 2008; Foulongne et al., 2010a; Foulongne et al., 2010b; Loyo et al., 2010; Schowalter et al., 2010; Wieland et al., 2009). No route of transmission for MCPyV has been validated, but available data supports several possibilities. For instance, the presence of viral DNA in the gut, gastrointestinal tract, and urban sewage suggests a fecal-oral mode of transmission (Bofill-Mas et al., 2010; Campello et al., 2011; Loyo et al., 2010), while presence in the aerodigestive tract supports a respiratory route (Babakir-Mina et al., 2010; Kantola et al., 2009; Tolstov et al., 2009). The discovery that nuclease-resistant MCPyV DNA (presumably encapsidated virus) is chronically shed from the skin (Schowalter et al., 2010) and is present on a variety of environmental surfaces (Foulongne et al., 2011), along with the emerging hypothesis that the MCPyV life cycle may be linked to cellular differentiation, could imply a mode of transmission that stems from cutaneous shedding of MCPyV virus.
MCPyV and MCC: Association and Evidence for Causality
Merkel cell polyomavirus was originally isolated from MCC tissue samples. In the hallmark report announcing their discovery of MCPyV, Feng and colleagues established the initial framework for a causal relationship between the virus and MCC (Feng et al., 2008). There are now several lines of evidence that suggest MCPyV is an etiological agent of MCC. One of the most unequivocal findings is the presence of the viral genome in a majority of MCCs. Using PCR to detect MCPyV-specific sequences, Feng et. al. reported the presence of MCPyV in 8 out of 10 (80%) MCC tumors tested. Since this original report, there has been widespread confirmation of these results that together indicate around 75% of MCCs contain MCPyV DNA (Andres et al., 2009; Becker et al., 2009a; Busam et al., 2009; Carter et al., 2009; Duncavage et al., 2009; Fischer et al., 2010; Garneski et al., 2009; Kassem et al., 2008; Katano et al., 2009; Loyo et al., 2010; Sastre-Garau et al., 2009; Varga et al., 2009). In the 8 MCPyV-positive MCCs originally analyzed, the viral genome copy number was subsequently found to average at least 1 copy per tumor cell (Shuda et al., 2009), although a vast range of viral copy numbers have since been reported (Bhatia et al., 2010b; Houben et al., 2010a; Katano et al., 2009; Laude et al., 2010). Along these lines, MCC patients were found to have much higher MCPyV antibody titers than the general public, even if the tumor tested negative for the virus, indicating a strong association between MCPyV and MCC and a possible viral role in tumor initiation (Carter et al., 2009; Pastrana et al., 2009; Tolstov et al., 2009; Viscidi et al., 2011).
Perhaps one of the more persuasive pieces of evidence supporting the theory that MCPyV is an etiological factor of MCC is clonal integration of the viral genome into the cellular genomes of MCC tumors (Feng et al., 2008; Laude et al., 2010; Martel-Jantin et al., 2012; Sastre-Garau et al., 2009) and some MCC cell lines (Fischer et al., 2010). MCC tumor metastases are also MCPyV-positive and share the same integration pattern as the original tumor (Feng et al., 2008; Laude et al., 2010). Collectively, these findings strongly suggest MCPyV virus infection occurs in a cell that ultimately gives rise to neoplastic growth and clonal expansion. The virus appears to integrate as head-to-tail concatemers at distinct locations randomly distributed throughout the cellular genome, and any evidence for cellular ‘hot-spots’ of viral integration does not currently exist (Feng et al., 2008; Laude et al., 2010; Martel-Jantin et al., 2012; Sastre-Garau et al., 2009). The precise mechanisms that direct integration of the MCPyV genome and the contribution of exogenous influences in this process are not yet understood.
Another line of evidence supporting causality is the retention of MCPyV gene expression in MCCs. Transcripts for MCPyV T antigens are consistently detected in MCC tumors (Feng et al., 2008; Sastre-Garau et al., 2009; Shuda et al., 2008). Moreover, MCPyV T antigen protein expression has been observed in MCC tumor sections by immunohistochemistry (Bhatia et al., 2010a; Busam et al., 2009; Shuda et al., 2009; Shuda et al., 2011). Most, but not all, MCC tumors are positive for MCPyV LT, and only cells within the tumor are reactive to LT antibodies while neighboring tissue is negative (Shuda et al., 2009). Interestingly, many tumors negative for LT exhibit positive staining with an antibody specific to sT, and additional analysis suggests that sT is actually more frequently expressed in MCC tumors than LT (Shuda et al., 2011). The significance of T antigen expression to MCPyV-induced carcinogenesis is strongly reinforced by the finding that their expression is required for growth of MCPyV-infected MCC cell lines. In a study by Houben et. al., shRNAs targeting the first exon shared by all T antigens were used to knockdown T antigen expression in MCC cell lines, resulting in impeded cell proliferation and survival (Houben et al., 2010b). The individual requirements for LT and sT in MCPyV-positive MCC cell growth were further dissected using shRNA specifically targeting sT antigen. Whereas sT knockdown inhibited cell growth to a level comparable to knockdown of all T antigens, it did not promote the robust cell cycle arrest or cell death seen in cells with pan-T antigen knockdown (Shuda et al., 2011). These data suggest that LT and sT most likely contribute to MCPyV-induced carcinogenesis via distinct mechanisms, but are both required for optimal MCC tumor cell growth.
A common characteristic of viral oncogenesis is a combination of events that renders the virus incapable of replication, yet still proficient in the induction of cellular proliferation (Reviewed in Moore and Chang, 2010). Clearly, integration of the MCPyV genome creates a barrier to virus replication, as explained below, but there are also features of tumor-associated MCPyV genomes that further suggest a strong selective pressure within MCCs to abrogate virus replication while preserving proliferative functions. Several groups have reported mutations in the origin of replication and VP1 gene in MCC-derived MCPyV genomic sequences that prevent efficient replication and progeny virion production, respectively (Feng et al., 2011; Kassem et al., 2008; Kwun et al., 2009; Neumann et al., 2011). However, the most common tumor-specific MCPyV mutations occur in the LT antigen. The LT antigen sequences in almost all tumor-derived MCPyV isolates and MCC cell lines analyzed to date contain mutations within exon 2 that prematurely truncate the LT protein (Fischer et al., 2010; Katano et al., 2009; Laude et al., 2010; Shuda et al., 2008) (Fig. 3). Strikingly, these mutations all occur within the carboxy terminus of LT at sites downstream of the LXCXE Rb-binding motif. This consistent, tumor-specific mutational pattern gives rise to truncated LT antigens in which the CR1, DnaJ, MUR, NLS, and pRb-binding motifs are preserved, yet domains associated with replication are lost. Indeed, truncated LT proteins can still co-immunoprecipitate pRb but are defective for origin-binding and viral helicase activities, and are thus no longer functional in replication assays (Shuda et al., 2008). These mutations in LT lie within a region that does not affect the coding capacity for sT. The presence of tumor-specific mutations within the MCPyV genome is compelling evidence that the virus has lost the capacity to replicate in MCC tumors, but retains motifs with potential to contribute to uncontrolled proliferation.
Given the preponderance of evidence associating MCPyV with MCC, the knowledge that MCPyV genes are required for the growth of MCC-derived cell lines, and the transforming potential of MCPyV genes, it is not surprising that the World Health Organization’s International Agency for Research on Cancer has recently classified MCPyV as a group 2A carcinogen, a designation meaning it is “probably carcinogenic to humans” (Bouvard et al., 2012).
Potential Mechanisms of MCPyV Transformation
Considering the status of the MCPyV genome and T antigens found in MCC tissue, several mechanisms of virus-induced transformation are plausible. The most obvious pathways involve the constitutive expression of LT and sT proteins. Using SV40 as a reference (Reviewed in Ahuja et al., 2005 and Khalili, 2008 #279; Hahn et al., 2002), one might hypothesize that MCPyV LT is the main viral oncoprotein and sT plays an accessory role in transformation by MCPyV. However, emerging data is starting to support the already accumulating evidence that MCPyV is unique with respect to other polyomaviruses. Multiple groups have observed that pan-T antigen knockdown in MCC cell lines contributes to robust cell death and cell cycle arrest (Houben et al., 2010b; Shuda et al., 2011). This data suggests that MCC tumor cells are ‘addicted’ to the MCPyV T antigen proteins. But, is one T antigen more oncogenic than the other? Is either T antigen sufficient for transformation? Many of the studies directed at discerning a role for T antigens in MCPyV-induced transformation have been complicated by the fact that all T antigens were initially targeted for knockdown simultaneously through the use of shRNAs specific to the common first exon. While one report did pinpoint a requirement for the pRb-binding motif in the ability of LT to promote growth of MCPyV-positive MCC cell lines and xenografts (Houben et al., 2012a), most of these studies could not distinguish between the independent roles of LT and sT in transformation. In 2011, Shuda and colleagues specifically targeted sT with shRNA, and the individual oncogenic potential of MCPyV LT and sT began to come into focus (Shuda et al., 2011). In several different traditional in vitro transformation assays performed in both rodent and human cells, sT exhibited an increased transformation capacity compared to LT, which had very little phenotype in these assays. Any transformation capacity of LT was minimal compared to sT, although truncated LT showed more proliferative capacity than full-length LT. Moreover, the sT transformation activity was not enhanced by the presence of LT, although this was only tested in one of many assays. These results make an extremely convincing argument that sT is the dominant MCPyV oncoprotein; however, the authors caution that sT alone did not fully transform human cells and pose a reminder that specific sT knockdown does not phenocopy knockdown of all T antigens in MCC cell lines. So while this data suggests that LT expression alone is not sufficient for MCPyV-induced transformation, it appears necessary for optimal transformation by the MCPyV T antigen locus. How these in vitro transformation phenotypes relate to in vivo tumorigenicity properties of MCPyV remains unclear.
The most widely characterized function of polyomavirus sT antigens is the ability to bind and reprogram the cellular protein phosphatase PP2A, a function that is intimately tied to its ability to promote cell cycle progression and transformation (Reviewed in Khalili et al., 2008; Pallas et al., 1990). Interestingly, mutational disruption of the PP2A-binding site of MCPyV sT did not affect its ability to promote transformation, which represents a major departure from dogma on polyomavirus biology (Shuda et al., 2011). Rather, sT was found to preserve hyperphosphorylation of the cellular translation factor eIF4E-binding protein 1 (4E-BP1) in an mTOR-dependent manner. Hyperphosphorylation of 4E-BP1 remained high in the presence of sT with a mutated PP2A-binding site, suggesting this function of sT does not require sT:PP2A association. This post-translational modification was confirmed in four MCPyV-positive MCC cell lines and was abolished following shRNA-mediated sT knockdown. The 4E-BP1 protein binds to the cap-binding protein eIF4E, but upon phosphorylation liberates eIF4E to participate in a rate-limiting step of cap-dependent translation (Pause et al., 1994). That sT increases association of eIF4E and another accessory factor, eIF4G, with the 5′-methyl-7-guanosine cap in cap-binding assays is consistent with its ability to preserve 4E-BP1 hyperphosphorylation. It therefore seems likely that sT promotes a favorable cellular environment for active cap-dependent translation. Most importantly, the ability of sT to preserve 4E-BP1 phosphorylation, which occurs independent of its association with PP2A, is necessary for its role in driving cellular transformation. Given that sT is more frequently expressed in MCC than LT, together with its transformation capacity in vitro, it seems likely that MCPyV sT may play a more dominant role than LT in MCPyV-mediated tumorigenicity.
Additional studies have added insight into other potential mechanisms of MCPyV-induced carcinogenesis. For instance, although Akt is phosphorylated in almost 90% of MCCs, this modification is not associated with MCPyV status and is not affected by LT/sT antigen knockdown in MCPyV-positive MCCs (Hafner et al., 2012). These results mirror those reported by Shuda et. al. showing that MCPyV sT interaction with PP2A did not activate Akt (Shuda et al., 2011). Together, these data suggest that MCPyV oncoproteins are not involved in promoting the high Akt activation observed in MCC.
The general lack of p53 mutations in MCPyV-positive MCCs is also noteworthy. As mentioned previously, p53 mutations that bear signatures consistent with UV-B exposure have been detected in MCC cell lines (Popp et al., 2002). However, this study was conducted prior to discovery of MCPyV and thus the viral status of these MCC cell lines is unclear. More recently, the status of p53 was analyzed in both MCPyV-positive and MCPyV-negative MCC tumors (Sihto et al., 2011). Interestingly, an inverse correlation between p53 expression and viral DNA copy number was observed, and only 7% of MCPyV-positive MCC tumors expressed detectable p53. Moreover, any p53 mutations observed in MCCs were almost exclusively detected in MCPyV-negative MCCs. These findings suggest that aberrant expression of mutated p53 is most likely not a major driver of MCPyV-associated tumorigenesis. It remains unclear whether MCPyV is functioning to inactivate the p53 pathway, thereby eliminating any selective advantage that mutations in p53 might provide to MCPyV-positive MCCs.
Finally, Houben et. al. recently described a MCPyV-positive MCC cell line in which proliferation is completely unaffected by T antigen knockdown (Houben et al., 2012b), which may suggest a hit-and-run mechanism of transformation. The link between MCC and ultraviolet exposure may also provide clues to viral pathogenesis. For instance, the MCPyV non-coding regulatory region can be activated in vitro by UV exposure, and human skin samples exposed to solar simulated radiation exhibited a dose-dependent increase in sT transcript levels (Mogha et al., 2010). Moreover, a MCPyV-infected MCC cell line showed impaired DNA damage repair and cell cycle arrest following exposure to UV radiation that was subsequently linked to LT, suggesting the presence of the virus affects normal repair mechanisms in cells exposed to UV (Demetriou et al., 2012). Clearly, the ongoing study of this new human tumor virus will generate valuable knowledge about the pathogenesis of MCPyV-induced MCC.
Conclusions and Future Perspectives
The biological and epidemiological characteristics of MCPyV culminate in the following hypothetical model for MCPyV-associated tumorigenesis (For more thorough reviews, see Bhatia et al., 2011; Chang and Moore, 2012; Moore and Chang, 2010). By all accounts, the virus is obtained during early childhood via transmission by either fecal-oral, respiratory, or cutaneous routes and results in a mostly asymptomatic primary infection. The virus is maintained as part of the normal microflora of mainly the skin, where it is chronically shed in the form of encapsidated virions. Localized or systemic compromises to the immune system, brought on by UV radiation, natural aging, AIDS, or drug-induced suppression of the immune system such as occurs in organ transplant recipients, are likely to lead to less effective host immune surveillance and subsequent virus reactivation. At this point, at least two mutational events occur that are critical steps in the proposed model of MCPyV-induced carcinogenesis. One mutational event is the integration of the viral genome into the host chromosome. The exogenous forces responsible for this event have not yet been determined, but hypotheses range from UV and ionizing radiation exposure to a defect in the virus itself. Moreover, the question of whether Merkel cells are in fact the cell type where this integration occurs, or if another prominent cell type in the skin is targeted, has not been answered. An additional event important in MCPyV-induced carcinogenesis is mutation of the viral genome in a manner that renders the virus unable to replicate. The most common mutations occur in the carboxy terminus of LT, which generate a truncated LT that lacks the helicase domain required for replication yet preserve its oncogenic functions. Because essentially all MCPyV-positive MCC tumors express truncated LT, it seems likely a strong selective pressure is implemented for replication-defective viruses. The exact order of these two events has not been directly tested or elucidated. One could envision viral genome integration as the first event, which would lead to unlicensed DNA synthesis and provocation of a cellular response leading to cell death, and LT truncation as the second event that prevents this replication and thereby protects the nascent tumor cell from its deleterious effects. An alternative scenario where LT truncations occur first followed by viral genome integration also seems possible. It has been shown that expression of truncated LT leads to defective DNA repair and cell cycle arrest in response to UV radiation (Demetriou et al., 2012). Consequentially, this increase in genomic instability may promote integration of the viral genome harboring mutated LT sequences. Regardless of the sequence of events, persistent expression of truncated LT and sT from the integrated MCPyV genome can inactivate pRb tumor suppressor function and promote and/or deregulate cap-dependent translation initiation, respectively. Ultimately, the cells in which integration of a replication-defective MCPyV is successful become addicted to viral oncogene expression and undergo clonal expansion and neoplastic progression to cause development of MCC.
It should be noted that this hypothetical model might not be exclusive to the development of MCC. The presence of MCPyV DNA in a variety of other cancers has been evaluated since its link to MCC was reported. Many such analyses have discounted a role of MCPyV in some cancers, including mesotheliomas (Bhatia et al., 2010c), hematolymphoid cancers (Shuda et al., 2009), melanomas (Varga et al., 2009) and colon cancer (Campello et al., 2011). However, for other cancers there are contradictory findings. A great amount of conflicting information exists regarding the role of MCPyV in the pathogenesis of chronic lymphocytic leukemia (Reviewed in Chang and Moore, 2012). Additionally, contradictory data also exists for the relationship between squamous cell carcinoma (SCC) and MCPyV. Some groups report no significant association (Andres et al., 2010; Reisinger et al., 2010), whereas others point to the presence of MCPyV DNA in 40% of cutaneous SCCs, which significantly correlates with MCPyV seropositivity (Rollison et al., 2012), and to the presence of MCPyV DNA in SCCs prior to the development of MCC (Kaibuchi-Noda et al., 2011). Strikingly, Dworkin et. al. detected mutations within exon 2 of nearly all MCPyV-positive SCCs, and these mutations theoretically produce truncated LT (Dworkin et al., 2009). Another report observed MCPyV at a high frequency in both basal cell and squamous cell carcinomas (Kassem et al., 2009). Finally, a full-length MCPyV genome (MCV-TKS) was isolated from a Kaposi’s sarcoma that expresses a truncated LT reminiscent of isolates detected in MCCs (Katano et al., 2009). In conclusion, MCC may not be the end of the story for MCPyV and human cancers.
The discovery of MCPyV and its apparent contribution to MCC serves to highlight the continued significance of the polyomavirus and DNA tumor virus fields. The current renaissance of collective interest in polyomavirus research and tumor virology, coupled with continuous technical advances, has already stimulated the discovery of new human viruses. Through the use of rolling-circle amplification, consensus PCR, and deep sequencing, six new human polyomaviruses have been discovered since the isolation of MCPyV in 2008 (Buck et al., 2012; Sauvage et al., 2011; Schowalter et al., 2010; Scuda et al., 2011; Siebrasse et al., 2012; van der Meijden et al., 2010). While most details of its molecular virology are still being elucidated, it is clear that MCPyV is unique within the Polyomaviridae family in many respects. Isolation of a naturally-occurring, intact MCPyV will certainly excite and expedite the process of learning more about its life cycle, lytic infection, and host range. These molecular details will help reconcile the known mechanisms of MCC pathogenesis with the etiological role of MCPyV in this human cancer. More thrilling, however, is the inherent potential this new human polyomavirus has to expose pre-existing, yet uncovered, molecular mechanisms involved in cell biology, virology, and carcinogenesis.
Highlights.
Merkel Cell Polyomavirus (MCPyV) is first PyV clearly associated with human cancer
MCPyV is found integrated in Merkel Cell Carcinoma (MCC)
MCPyV small and large tumor antigens (sT and LT) are expressed in MCC
In MCC, LT is mutated, looses its DNA helicase activity but retains binding to pRb
Amongst human polyomaviruses, McPyV has some unique characteristics
Acknowledgments
We would like to thank Chris Buck (NIH, Bethesda) for the electron micrograph of the MCPyV virus-like particles (Fig. 2B) and for critically reading the manuscript. We also thank Genesis Perez for use of immunofluorescence images (Fig. 1B) generated during her rotation with M.E.S. in the P.F.L. laboratory. M.E.S. is supported by a T32 training grant from NIAID (AI078985). P.F.L is supported by grants from the NCI (CA022443, CA141583, CA098428).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Agelli M, Clegg LX. Epidemiology of primary Merkel cell carcinoma in the United States. J Am Acad Dermatol. 2003;49:832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: a population based study. Journal of cutaneous pathology. 2010;37:20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- Andres C, Belloni B, Puchta U, Sander CA, Flaig MJ. Prevalence of MCPyV in Merkel cell carcinoma and non-MCC tumors. Journal of cutaneous pathology. 2010;37:28–34. doi: 10.1111/j.1600-0560.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- Andres C, Ihrler S, Puchta U, Flaig MJ. Merkel cell polyomavirus is prevalent in a subset of small cell lung cancer: a study of 31 patients. Thorax. 2009;64:1007–1008. doi: 10.1136/thx.2009.117911. [DOI] [PubMed] [Google Scholar]
- Babakir-Mina M, Ciccozzi M, Lo Presti A, Greco F, Perno CF, Ciotti M. Identification of Merkel cell polyomavirus in the lower respiratory tract of Italian patients. Journal of medical virology. 2010;82:505–509. doi: 10.1002/jmv.21711. [DOI] [PubMed] [Google Scholar]
- Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009a;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- Becker JC, Schrama D, Houben R. Merkel cell carcinoma. Cell Mol Life Sci. 2009b;66:1–8. doi: 10.1007/s00018-008-8483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Immunological detection of viral large T antigen identifies a subset of Merkel cell carcinoma tumors with higher viral abundance and better clinical outcome. International journal of cancer Journal international du cancer. 2010a;127:1493–1496. doi: 10.1002/ijc.25136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. International journal of cancer Journal international du cancer. 2010b;126:2240–2246. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia K, Modali R, Goedert JJ. Merkel cell polyomavirus is not detected in mesotheliomas. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2010c;47:196–198. doi: 10.1016/j.jcv.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Current oncology reports. 2011;13:488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialasiewicz S, Lambert SB, Whiley DM, Nissen MD, Sloots TP. Merkel cell polyomavirus DNA in respiratory specimens from children and adults. Emerg Infect Dis. 2009;15:492–494. doi: 10.3201/eid1503.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Martynowski D, Seitova A, Bochkarev A. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. Embo J. 2006;25:5961–5969. doi: 10.1038/sj.emboj.7601452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S, Rodriguez-Manzano J, Calgua B, Carratala A, Girones R. Newly described human polyomaviruses Merkel cell, KI and WU are present in urban sewage and may represent potential environmental contaminants. Virol J. 2010;7:141. doi: 10.1186/1743-422X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K. Carcinogenicity of malaria and of some polyomaviruses. The lancet oncology. 2012;13:339–340. doi: 10.1016/s1470-2045(12)70125-0. [DOI] [PubMed] [Google Scholar]
- Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. Complete genome sequence of a tenth human polyomavirus. J Virol. 2012;86:10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell JF, Trofe J, Hanaway MJ, Beebe TM, Gross TG, Alloway RR, First MR, Woodle ES. Immunosuppression and Merkel cell cancer. Transplant Proc. 2002;34:1780–1781. doi: 10.1016/s0041-1345(02)03065-8. [DOI] [PubMed] [Google Scholar]
- Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, Arora R, Hanson NC, Tassello JA, Frosina D, Moore P, Chang Y. Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. The American journal of surgical pathology. 2009;33:1378–1385. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder KB, Smoller BR. New insights into merkel cell carcinoma. Advances in anatomic pathology. 2010;17:155–161. doi: 10.1097/PAP.0b013e3181d97836. [DOI] [PubMed] [Google Scholar]
- Campello C, Comar M, D’Agaro P, Minicozzi A, Rodella L, Poli A. A molecular case-control study of the Merkel cell polyomavirus in colon cancer. Journal of medical virology. 2011;83:721–724. doi: 10.1002/jmv.22004. [DOI] [PubMed] [Google Scholar]
- Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. Journal of the National Cancer Institute. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chang Y, Moore PS. Merkel cell carcinoma: a virus-induced human cancer. Annual review of pathology. 2012;7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. Serological evidence of Merkel cell polyomavirus primary infections in childhood. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2011;50:125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Cole CN, Conzen SD. Polyomaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Field’s Virology. 4. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 2141–2174. [Google Scholar]
- Comerford SA, Schultz N, Hinnant EA, Klapproth S, Hammer RE. Comparative analysis of SV40 17kT and LT function in vivo demostrates that LT’s C-terminus re-programs hepatic gene expression and is necessary for tumorigenesis in the liver. Oncogenesis. 2012 doi: 10.1038/oncsis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalianis T, Ramqvist T, Andreasson K, Kean JM, Garcea RL. KI, WU and Merkel cell polyomaviruses: a new era for human polyomavirus research. Seminars in cancer biology. 2009;19:270–275. doi: 10.1016/j.semcancer.2009.04.001. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Demetriou SK, Ona-Vu K, Sullivan EM, Dong TK, Hsu SW, Oh DH. Defective DNA repair and cell cycle arrest in cells expressing Merkel cell polyomavirus T antigen. International journal of cancer Journal international du cancer. 2012;131:1818–1827. doi: 10.1002/ijc.27440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncavage EJ, Zehnbauer BA, Pfeifer JD. Prevalence of Merkel cell polyomavirus in Merkel cell carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:516–521. doi: 10.1038/modpathol.2009.3. [DOI] [PubMed] [Google Scholar]
- Dworkin AM, Tseng SY, Allain DC, Iwenofu OH, Peters SB, Toland AE. Merkel cell polyomavirus in cutaneous squamous cell carcinoma of immunocompetent individuals. J Invest Dermatol. 2009;129:2868–2874. doi: 10.1038/jid.2009.183. [DOI] [PubMed] [Google Scholar]
- Eckhart W, Hutchinson MA, Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Eddy BE, Borman GS, Berkeley WH, Young RD. Tumors induced in hamsters by injection of rhesus monkey kidney cell extracts. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1961;107:191–197. doi: 10.3181/00379727-107-26576. [DOI] [PubMed] [Google Scholar]
- Eddy BE, Borman GS, Grubbs GE, Young RD. Identification of the oncogenic substance in rhesus monkey kidney cell culture as simian virus 40. Virology. 1962;17:65–75. doi: 10.1016/0042-6822(62)90082-x. [DOI] [PubMed] [Google Scholar]
- Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Merkel cell carcinoma and HIV infection. Lancet. 2002;359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- Erickson KD, Garcea RL, Tsai B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J Virol. 2009;83:10275–10279. doi: 10.1128/JVI.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, Chang Y, Moore PS. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Brandner J, Fuchs F, Moll I, Grundhoff A. Detection of Merkel cell polyomavirus (MCPyV) in Merkel cell carcinoma cell lines: cell morphology and growth phenotype do not reflect presence of the virus. International journal of cancer Journal international du cancer. 2010;126:2133–2142. doi: 10.1002/ijc.24877. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Courgnaud V, Champeau W, Segondy M. Detection of Merkel cell polyomavirus on environmental surfaces. Journal of medical virology. 2011;83:1435–1439. doi: 10.1002/jmv.22110. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Dereure O, Kluger N, Moles JP, Guillot B, Segondy M. Merkel cell polyomavirus DNA detection in lesional and nonlesional skin from patients with Merkel cell carcinoma or other skin diseases. The British journal of dermatology. 2010a;162:59–63. doi: 10.1111/j.1365-2133.2009.09381.x. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Kluger N, Dereure O, Mercier G, Moles JP, Guillot B, Segondy M. Merkel cell polyomavirus in cutaneous swabs. Emerg Infect Dis. 2010b;16:685–687. doi: 10.3201/eid1604.091278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246–248. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Lindau C, Tiveljung-Lindell A, Allander T. Merkel cell polyomavirus in respiratory tract secretions. Emerg Infect Dis. 2009;15:489–491. doi: 10.3201/eid1503.081206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould VE, Moll R, Moll I, Lee I, Franke WW. Neuroendocrine (Merkel) cells of the skin: hyperplasias, dysplasias, and neoplasms. Laboratory investigation; a journal of technical methods and pathology. 1985;52:334–353. [PubMed] [Google Scholar]
- Gross L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1953;83:414–421. doi: 10.3181/00379727-83-20376. [DOI] [PubMed] [Google Scholar]
- Gross L. Transmission of Ak leukemic agent into newborn mice of the C57 brown/cd inbred line. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1954;86:734–739. doi: 10.3181/00379727-86-21217. [DOI] [PubMed] [Google Scholar]
- Gustafsson B, Honkaniemi E, Goh S, Giraud G, Forestier E, von Dobeln U, Allander T, Dalianis T, Bogdanovic G. KI, WU, and Merkel cell polyomavirus DNA was not detected in guthrie cards of children who later developed acute lymphoblastic leukemia. Journal of pediatric hematology/oncology. 2012;34:364–367. doi: 10.1097/MPH.0b013e318241fb52. [DOI] [PubMed] [Google Scholar]
- Hafner C, Houben R, Baeurle A, Ritter C, Schrama D, Landthaler M, Becker JC. Activation of the PI3K/AKT pathway in Merkel cell carcinoma. PLoS One. 2012;7:e31255. doi: 10.1371/journal.pone.0031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halata Z, Grim M, Bauman KI. Friedrich Sigmund Merkel and his “Merkel cell”, morphology, development, and physiology: review and new results. The anatomical record Part A, Discoveries in molecular, cellular, and evolutionary biology. 2003;271:225–239. doi: 10.1002/ar.a.10029. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. Journal of molecular biology. 2011;409:529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson NC. Merkel cell carcinoma: changing incidence trends. Journal of surgical oncology. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- Horvath CA, Boulet GA, Renoux VM, Delvenne PO, Bogers JP. Mechanisms of cell entry by human papillomaviruses: an overview. Virol J. 2010;7:11. doi: 10.1186/1743-422X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. International journal of cancer Journal international du cancer. 2012a;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- Houben R, Grimm J, Willmes C, Weinkam R, Becker JC, Schrama D. Merkel cell carcinoma and Merkel cell polyomavirus: evidence for hit-and-run oncogenesis. J Invest Dermatol. 2012b;132:254–256. doi: 10.1038/jid.2011.260. [DOI] [PubMed] [Google Scholar]
- Houben R, Schrama D, Alb M, Pfohler C, Trefzer U, Ugurel S, Becker JC. Comparable expression and phosphorylation of the retinoblastoma protein in Merkel cell polyoma virus-positive and negative Merkel cell carcinoma. International journal of cancer Journal international du cancer. 2010a;126:796–798. doi: 10.1002/ijc.24790. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010b;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husseiny MI, Anastasi B, Singer J, Lacey SF. A comparative study of Merkel cell, BK and JC polyomavirus infections in renal transplant recipients and healthy subjects. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2010;49:137–140. doi: 10.1016/j.jcv.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. Taxonomical developments in the family Polyomaviridae. Archives of virology. 2011;156:1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM. Structural evaluation of new human polyomaviruses provides clues to pathobiology. Trends in microbiology. 2010;18:215–223. doi: 10.1016/j.tim.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibuchi-Noda K, Yokota K, Matsumoto T, Sawada M, Sakakibara A, Kono M, Tomita Y, Watanabe D, Fukumoto H, Katano H, Akiyama M. Detection of Merkel cell polyomavirus in cutaneous squamous cell carcinoma before occurrence of Merkel cell carcinoma. J Am Acad Dermatol. 2011;65:e152–154. doi: 10.1016/j.jaad.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, Rahiala J, Saarinen-Pihkala U, Riikonen P, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: implications for respiratory transmission and latency. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2009;45:292–295. doi: 10.1016/j.jcv.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer research. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- Kassem A, Technau K, Kurz AK, Pantulu D, Loning M, Kayser G, Stickeler E, Weyers W, Diaz C, Werner M, Nashan D, Zur Hausen A. Merkel cell polyomavirus sequences are frequently detected in nonmelanoma skin cancer of immunosuppressed patients. International journal of cancer Journal international du cancer. 2009;125:356–361. doi: 10.1002/ijc.24323. [DOI] [PubMed] [Google Scholar]
- Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, Matsuo K, Nakagawa H, Sata T. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi’s sarcoma. Journal of medical virology. 2009;81:1951–1958. doi: 10.1002/jmv.21608. [DOI] [PubMed] [Google Scholar]
- Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K, Sariyer IK, Safak M. Small tumor antigen of polyomaviruses: role in viral life cycle and cell transformation. J Cell Physiol. 2008;215:309–319. doi: 10.1002/jcp.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Butel JS. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Laude HC, Jonchere B, Maubec E, Carlotti A, Marinho E, Couturaud B, Peter M, Sastre-Garau X, Avril MF, Dupin N, Rozenberg F. Distinct merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with merkel cell carcinoma. PLoS Pathog. 2010;6:e1001076. doi: 10.1371/journal.ppat.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J Invest Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- Liu X, Hein J, Richardson SC, Basse PH, Toptan T, Moore PS, Gjoerup OV, Chang Y. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. The Journal of biological chemistry. 2011;286:17079–17090. doi: 10.1074/jbc.M110.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, Sharma R, Liegeois NJ, Koch WM, Califano JA, Westra WH, Sidransky D. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. International journal of cancer Journal international du cancer. 2010;126:2991–2996. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarz A, Brand G. Current considerations about Merkel cells. European journal of cell biology. 2007;86:243–251. doi: 10.1016/j.ejcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Martel-Jantin C, Filippone C, Cassar O, Peter M, Tomasic G, Vielh P, Briere J, Petrella T, Aubriot-Lorton MH, Mortier L, Jouvion G, Sastre-Garau X, Robert C, Gessain A. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. Virology. 2012;426:134–142. doi: 10.1016/j.virol.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Meinke G, Phelan P, Moine S, Bochkareva E, Bochkarev A, Bullock PA, Bohm A. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS Biol. 2007;5:e23. doi: 10.1371/journal.pbio.0050023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz KD, Junt T, Schmid M, Pfaltz M, Kempf W. Inflammatory monocytes are a reservoir for Merkel cell polyomavirus. J Invest Dermatol. 2010;130:1146–1151. doi: 10.1038/jid.2009.392. [DOI] [PubMed] [Google Scholar]
- Miller RW, Rabkin CS. Merkel cell carcinoma and melanoma: etiological similarities and differences. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8:153–158. [PubMed] [Google Scholar]
- Mogha A, Fautrel A, Mouchet N, Guo N, Corre S, Adamski H, Watier E, Misery L, Galibert MD. Merkel cell polyomavirus small T antigen mRNA level is increased following in vivo UV-radiation. PLoS One. 2010;5:e11423. doi: 10.1371/journal.pone.0011423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Bladt U, Jung EG. Presence of Merkel cells in sun-exposed and not sun-exposed skin: a quantitative study. Archives of dermatological research. 1990;282:213–216. doi: 10.1007/BF00371638. [DOI] [PubMed] [Google Scholar]
- Moll I, Roessler M, Brandner JM, Eispert AC, Houdek P, Moll R. Human Merkel cells--aspects of cell biology, distribution and functions. European journal of cell biology. 2005;84:259–271. doi: 10.1016/j.ejcb.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sato Y, Watanabe D, Ito H, Shimonohara N, Tsuji T, Nakajima N, Suzuki Y, Matsuo K, Nakagawa H, Sata T, Katano H. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology. 2010;398:273–279. doi: 10.1016/j.virol.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Neu U, Bauer J, Stehle T. Viruses and sialic acids: rules of engagement. Curr Opin Struct Biol. 2011;21:610–618. doi: 10.1016/j.sbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu U, Hengel H, Blaum BS, Schowalter RM, Macejak D, Gilbert M, Wakarchuk WW, Imamura A, Ando H, Kiso M, Arnberg N, Garcea RL, Peters T, Buck CB, Stehle T. Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection. PLoS Pathog. 2012;8:e1002738. doi: 10.1371/journal.ppat.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu U, Stehle T, Atwood WJ. The Polyomaviridae: Contributions of virus structure to our understanding of virus receptors and infectious entry. Virology. 2009;384:389–399. doi: 10.1016/j.virol.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F, Borchert S, Schmidt C, Reimer R, Hohenberg H, Fischer N, Grundhoff A. Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome. PLoS One. 2011;6:e29112. doi: 10.1371/journal.pone.0029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5:e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Penn I, First MR. Merkel’s cell carcinoma in organ recipients: report of 41 cases. Transplantation. 1999;68:1717–1721. doi: 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- Pipas JM. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp S, Waltering S, Herbst C, Moll I, Boukamp P. UV-B-type mutations and chromosomal imbalances indicate common pathways for the development of Merkel and skin squamous cell carcinomas. International journal of cancer Journal international du cancer. 2002;99:352–360. doi: 10.1002/ijc.10321. [DOI] [PubMed] [Google Scholar]
- Poulin DL, DeCaprio JA. Is there a role for SV40 in human cancer? Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:4356–4365. doi: 10.1200/JCO.2005.03.7101. [DOI] [PubMed] [Google Scholar]
- Rabson AS, Kirschstein RL. Induction of malignancy in vitro in newborn hamster kidney tissue infected with simian vacuolating virus (SV40) Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1962;111:323–328. doi: 10.3181/00379727-111-27780. [DOI] [PubMed] [Google Scholar]
- Reisinger DM, Shiffer JD, Cognetta AB, Jr, Chang Y, Moore PS. Lack of evidence for basal or squamous cell carcinoma infection with Merkel cell polyomavirus in immunocompetent patients with Merkel cell carcinoma. J Am Acad Dermatol. 2010;63:400–403. doi: 10.1016/j.jaad.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollison DE, Giuliano AR, Messina JL, Fenske NA, Cherpelis BS, Sondak VK, Roetzheim RG, Iannacone MR, Michael KM, Gheit T, Waterboer T, Tommasino M, Pawlita M. Case-control study of Merkel cell polyomavirus infection and cutaneous squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:74–81. doi: 10.1158/1055-9965.EPI-11-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre-Garau X, Peter M, Avril MF, Laude H, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Couturaud B, Dupin N. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. The Journal of pathology. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, Caro V, Eloit M. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis. 2011;17:1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Buck CB. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7:e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell host & microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Reinhold WC, Buck CB. Entry Tropism of BK and Merkel Cell Polyomaviruses in Cell Culture. PLoS One. 2012;7:e42181. doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. The Journal of infectious diseases. 2009;199:398–404. doi: 10.1086/596062. [DOI] [PubMed] [Google Scholar]
- Shein HM, Enders JF. Transformation induced by simian virus 40 in human renal cell cultures. I. Morphology and growth characteristics. Proc Natl Acad Sci U S A. 1962;48:1164–1172. doi: 10.1073/pnas.48.7.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. International journal of cancer. Journal international du cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. The Journal of clinical investigation. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu GS, Chandra P, Cassai ND. Merkel cells, normal and neoplastic: an update. Ultrastructural pathology. 2005;29:287–294. doi: 10.1080/01913120590951284. [DOI] [PubMed] [Google Scholar]
- Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. Identification of MW Polyomavirus, a Novel Polyomavirus in Human Stool. J Virol. 2012;86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihto H, Kukko H, Koljonen V, Sankila R, Bohling T, Joensuu H. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:4806–4813. doi: 10.1158/1078-0432.CCR-10-3363. [DOI] [PubMed] [Google Scholar]
- Sweet BH, Hilleman MR. The vacuolating virus, S.V. 40. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- Toker C. Trabecular carcinoma of the skin. Archives of dermatology. 1972;105:107–110. [PubMed] [Google Scholar]
- Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg Infect Dis. 2011;17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. International journal of cancer. Journal international du cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toracchio S, Foyle A, Sroller V, Reed JA, Wu J, Kozinetz CA, Butel JS. Lymphotropism of Merkel cell polyomavirus infection, Nova Scotia, Canada. Emerg Infect Dis. 2010;16:1702–1709. doi: 10.3201/eid1611.100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touze A, Gaitan J, Arnold F, Cazal R, Fleury MJ, Combelas N, Sizaret PY, Guyetant S, Maruani A, Baay M, Tognon M, Coursaget P. Generation of Merkel cell polyomavirus (MCV)-like particles and their application to detection of MCV antibodies. Journal of clinical microbiology. 2010;48:1767–1770. doi: 10.1128/JCM.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga E, Kiss M, Szabo K, Kemeny L. Detection of Merkel cell polyomavirus DNA in Merkel cell carcinomas. The British journal of dermatology. 2009;161:930–932. doi: 10.1111/j.1365-2133.2009.09221.x. [DOI] [PubMed] [Google Scholar]
- Viscidi RP, Rollison DE, Sondak VK, Silver B, Messina JL, Giuliano AR, Fulp W, Ajidahun A, Rivanera D. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clinical and vaccine immunology: CVI. 2011;18:1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad R, Woodlock TJ. Merkel cell carcinoma after chronic lymphocytic leukemia: case report and literature review. Am J Clin Oncol. 2003;26:531–534. doi: 10.1097/01.coc.0000037108.86294.5E. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- Wieland U, Mauch C, Kreuter A, Krieg T, Pfister H. Merkel cell polyomavirus DNA in persons without merkel cell carcinoma. Emerg Infect Dis. 2009;15:1496–1498. doi: 10.3201/eid1509.081575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Knippschild U, Winkler T, Deppert W. Independent expression of the transforming amino-terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. Embo J. 1993;12:4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]