Abstract
Over the last twenty years there have been great advances in light microscopy with the result that multi-dimensional imaging has driven a revolution in modern biology. The development of new approaches of data acquisition are reportedly frequently, and yet the significant data management and analysis challenges presented by these new complex datasets remains largely unsolved. Like the well-developed field of genome bioinformatics, central repositories are and will be key resources, but there is a critical need for informatics tools in individual laboratories to help manage, share, visualize, and analyze image data. In this article we present the recent efforts by the bioimage informatics community to tackle these challenges and discuss our own vision for future development of bioimage informatics solution.
Keywords: microscopy, file formats, image management, image analysis, image processing
Introduction
Modern imaging systems have enabled a new kind of discovery in cellular and developmental biology. With spatial resolutions running from millimeters to nanometers, analysis of cell and molecular structure and dynamics is now routinely possible across a range of biological systems. The development of fluorescent reporters, most notably in the form of genetically encoded fluorescent proteins (FPs) combined with increasingly sophisticated imaging systems enable direct study of molecular structure and dynamics (6, 54). Cell and tissue imaging assays have scaled to include all three spatial dimensions, a temporal component, and the use of spectral separation to measure multiple molecules such that a single “image” is now a five-dimensional structure-- space, time, and channel. High content screening (HCS) and fluorescence lifetime, polarization, correlation are all examples of new modalities that further increasing the complexity of the modern microscopy dataset. However, this multi-dimensional data acquisition generates a significant data problem: a typical four-year project generates many hundreds of gigabytes of images, perhaps on many different proprietary data acquisition systems, making hypothesis-driven research completely dependent on data management, visualization and analysis.
Bioinformatics is now a mature science, and forms the cornerstone of much of modern biology. Modern biologists routinely use what they refer to as ‘genomic databases’ to inform their experiments. In fact these ‘databases’ are well-crafted multi-layered applications, that include defined data structures, application programming interfaces (APIs), and use standardized user interfaces to enable querying, browsing and visualization of the underlying genome sequences. These facilities serve as a great model of the sophistication that is necessary to deliver complex, heterogeneous datasets to bench biologists. However, most genomic resources work on the basis of defined data structures with defined formats and known identifiers that all applications can access (they also employ expert staff to monitor systems and databases, a resource that is rarely available in individual laboratories). There is no single agreed data format, but a defined number that are used in various applications, depending on the exact application (e.g., FASTA and EMBL files). Access to these files is through a number of defined software libraries that provide data translation into defined data objects that can be used for further analysis and visualization. Because a relatively small number of sequence data generation and collation centers exist, standards have been relatively easy to declare and support. Nonetheless, a key to the successful use of these data was the development of software applications, designed for use by bench biologists as well as by specialist bioinformaticists that enabled querying and discovery based on genomic data held by and served from central data resources.
Given this paradigm, the same facility should in principle be available for all biological imaging data (as well as proteomics and soon, deep sequencing). In contrast to centralized genomics resources, in most cases, these methods are being used for defined experiments in individual laboratories or facilities, and the number of image datasets recorded by a single postdoctoral fellow (hundreds to thousands) can easily rival the number of genomes that have been sequenced to date. For the continued development and application of experimental biology imaging methods, it will be necessary to invest in and develop informatics resources that provide solutions for individual laboratories and departmental facilities. Is it possible to deliver flexible, powerful and usable informatics tools to manage a single laboratory’s data that are comparable to that used to deliver genomic sequence applications and databases to the whole community? Why can’t the tools used in genomics be immediately adapted to imaging? Are image informatics tools from other fields appropriate for biological microscopy?
In this article, we address these questions and discuss the requirements for successful image informatics solutions for biological microscopy and consider the future directions that these applications must take in order to deliver effective solutions for biological microscopy.
Flexible informatics for experimental biology
Experimental imaging data is by its very nature heterogeneous and dynamic. The challenge is to capture the evolving nature of an experiment in data structures that by their very nature are specifically typed and static, for later recall, analysis, and comparison. Achieving this goal in imaging applications means solving a number of problems:
1. Proprietary file formats
There are over 50 different proprietary file formats used in commercial and academic image acquisition software packages for light microscopy (36). This number only increases if electron microscopy, new HCS systems, tissue imaging systems and other new modes of imaging modalities are included. Regardless of the specific application, almost all store data in their own proprietary file format (PFFs). Each of these formats includes the binary data-- the values in the pixels-- and the metadata-- the data that describes the binary data. Metadata includes physical pixel sizes, time stamps, spectral ranges, and any other measurements or values that are required to fully define the binary data. It is critical to note that because of the heterogeneity of microscope imaging experiments, there is no agreed upon community specification for a minimal set of metadata (see below). Regardless, the binary data and metadata combined from the full output of the microscope imaging system, and each software application must contend with the diversity of PFFs and continually update its support for changing formats.
2. Experimental protocols
Sample preparation, data acquisition methods and parameters, and analysis workflow all evolve during the course of a project, and there are invariably differences in approach even between laboratories doing very similar work. This evolution reflects the natural progression of scientific discovery. Recording this evolution (”what exposure time did I use in the experiment last Wednesday?”) and providing flexibility for changing metadata, especially when new metadata must be supported, is a critical requirement for any experimental data management system.
3. Image result management
Many experiments only use a single microscope, but the visualization and analysis of image data associated with a single experiment can generate many additional derived files, of varying formats. Typically these are stored on a hard disk using arbitrary directory structures. Thus an experimental “result” typically reflects the compilation of many different images, recorded across multiple runs of an experiment and associated processed images, analysis outputs, and result spreadsheets. Simply keeping these disparate data linked so that they can be recalled and examined at a later time is a critical requirement and a significant challenge.
4. Remote image access
Image visualization requires significant computational resources. Many commercial image-processing tools use specific graphics CPU hardware (and thus depend on the accompanying driver libraries). Moreover, they often do not work well when analyzing data across a network connection to a data stored on a remote file system. As work patterns move to wireless connections and more types of portable devices, remote access to image visualization tools, coupled with the ability to access and run powerful analysis and processing will be required.
5. Image processing and analysis
Substantial effort has gone into the development of sophisticated image processing and analysis tools. In genome informatics, the linkage of related but distinct resources (e.g., Wormbase (50) and Flybase (14)) is possible due to the availability of defined interfaces that different resources use to provide access to underlying data. This facility is critical to enable discovery and collaboration-- any algorithm developed to ask a specific question should be able to address all available data. This is especially critical as new image methods are developed-- an existing analysis tool should not be made obsolete just because a new file format has been developed that it does not read. When scaled across the large number of analysis tool developers, this is an unacceptable code maintenance burden.
6. Distributed processing
As sizes and numbers of images increase, access to larger computing facilities will be routinely required by all investigators. Grid-based data processing is now available for specific analyses of genomic data, but the burden of moving many gigabytes of data even for a single experiment means that distributed computing must also be made locally available, at least in a form that allows laboratories and facilities to access their local clusters or to leverage an investment in multi-cpu, multi-core machines.
7. Image data and interoperability
Strategic collaboration is one of the cornerstones of modern science, and fundamentally consists of one scientist being able to share resources and data with another. Biological imaging is comprised of several specialized sub-disciplines – experimental image acquisition, image processing, and image data mining. Each requires its own domain of expertise and specialization, which is justified because each presents unsolved technical challenges as well as ongoing scientific research. In order for a group specializing in image analysis to make the best use of its expertise, it needs to have access to image data from groups specializing in acquisition. Ideally, this data should comprise current research questions and not historical image repositories that may no longer be scientifically relevant. Similarly, groups specializing in image informatics need to have access to both image data as well as to results produced by image processing groups. Ultimately, this drives the development of useful tools for the community and certainly results in synergistic collaborations that enhance each groups advances.
The delivery of solutions for these problems requires the development of a new emerging field known as “bioimage informatics” (47) that includes the infrastructure and applications that enable discovery of insight using systematic annotation, visualization, and analysis of large sets of images of biological samples. For applications of bioimage informatics in microscopy, we include HCS, where images are collected from arrayed samples, treated with large sets of siRNAs or small molecules (48), as well as large sets of time-lapse images (28), collections of fixed and stained cells or tissues (10, 19) and even sets of generated localization patterns (61) that define specific collections of localization for reference or for analysis. The development and implementation of successful bioimage informatics tools provides enabling technology for biological discovery in several different ways:
management: simply keeping track of data from large numbers of experiments
sharing: with defined collaborators, allowing groups of scientists to compare images and analytic tools with one another
remote access-- ability to query, analyze, and visualize without having to connect to a specific file system or use specific video hardware on the users computer or mobile device;
interoperability: interfacing of visualization and analysis programs with any set of data, without concern for file format
integration of heterogeneous data types; collection of raw data files, analysis results, annotations, and derived figures into a single resource, that is easily searchable and browseable.
Building by and for the community
Given these requirements, how should an image informatics solution be developed and delivered? It certainly will involve the development, distribution and support of software tools that must be acceptable to bench biologists and must also work with all of the existing commercial and academic data acquisition, visualization and analysis tools. Moreover, it must support a broad range of imaging approaches, and if at all possible, include the newest modalities in light and electron microscopy, support extensions into clinical research familiar with microscopy (e.g. histology and pathology), and provide the possibility of extension into modalities that do not use visible light (MRI, CT, ultrasound). Since many commercial image acquisition and analysis software packages are already established as critical research tools, all design, development, and testing must assume and expect integration and interoperability. It therefore seems prudent to avoid a traditional commercial approach and make this type of effort community-led, using open source models that are now well defined. This does not exclude the possibility of successful commercial ventures being formed to provide bioimage informatics solutions to the experimental biological community, but a community-led open-source approach will be best placed to provide interfaces between all existing academic and commercial applications.
Delivering on the promise-- “standardized file formats” vs. “just put it in a database”
In our experience, there are a few commonly suggested solutions for biological imaging. The first is a common, open file format for microscope imaging. A number of specifications for file formats have been presented, including our own (2, 15). Widespread adoption of standardized image data formats has been successful in astronomy (FITS), crystallography (PDB), and in clinical imaging (DICOM), where either most of the acquisition software is developed by scientists or a small number of commercial manufacturers are able to adopt a standard defined by the imaging community. Biological microscopy is a highly fractured market, with at least 40 independent commercial providers. This combined with rapidly developing technical platforms acquiring new kinds of data have stymied efforts at establishing a commonly used data standard.
Against this background, it is worth asking whether defining a standardized format for imaging is at all useful and practical. Standardized file formats and minimum data specifications have the advantage of providing a single or perhaps more realistically, a small number of data structures for the community to contend with. This facilitates interoperability, enabling visualization and analysis-- tools developed by one lab can be used by another. This is an important step for collaboration, and allows data exchange-- moving a large multi-dimensional file from one software application to another, or from one lab or center to another. This only satisfies some of the requirements defined above and provides none of the search, query, remote access, or collaboration facilities discussed above, and thus is not a complete solution. However, the expression of a data model in a file format, and especially the development of software that reads and write that format, is a useful exercise-- it tests the modeling concepts, relationships and requirements (e.g., “if an objective lens is specified, should the numerical aperture be mandatory?”) and provides a relatively easy way for the community to access, use, and comment on the defined data relationships defined by the project. Ultimately, the practical value of such formats may be in the final publishing and release of data to the community. Unlike gene sequence and protein structure data, there is no requirement for release of images associated with published results, but the availability of standardized formats may facilitate this.
Alternatively, why not use any number of commercial database products (Microsoft Access, Filemaker Pro, etc) to build local databases in indvidual laboratories that, as well-developed commercial applications, provide tools for building customized local databases? This is certainly a solution, but to date, these local database efforts have not simultaneously dedicated themselves to addressing interoperability, allowing broad support for alternative analysis and visualization tools that were not specifically supported when the database was built. However, perhaps most importantly, single lab efforts often emphasize specific aspects of their own research (e.g., the data model supports individual cell lines, but not yeast or worm strains), and the adaptability necessary to support a range of disciplines across biological research, or even their own evolving repertoire of methods and experimental systems is not included.
In no way does this preclude the development of local or targeted bioimage informatics solutions. In genomics, there are several community-initiated informatics projects focused on specific resources that support the various biological model systems (14, 52, 59). It seems likely that similar projects will grow up around specific bioimage informatics projects, following the models of the Allen Brain Atlas , , E-MAGE, and the Cell Centered Database (11, 20, 23). In genomics, there is underlying interoperability between specialized sources - ultimately all of the sequence data as well as the specialized annotation exists in common repositories and formats (GenBank, etc.). Common repositories may not be practical with images, but there will be value in linking through the gene identifiers themselves, or ontological annotations, or perhaps, localization maps or sets of phenotypic features, once these are standardized (61). Once these links are made to images stored in common formats, then distributed storage will effectively accomplish the same thing as centralized storage.
Several large-scale bioinformatics projects related to interoperability between large biological information datasets have emerged, including caBIG focusing on cancer research (7), BIRN focusing on neurobiology with a substantial imaging component (4), BioSig (46) providing tools for large-scale data analysis, and myGrid focusing on simulation, workflows, and “in silico” experiments (27). Projects specifically involved in large-scale imaging infrastructure include the Protein Subcellular Location Image Database (PSLID; (17, 26)), Bisque (5), the Cell-Centered Database (CCDB (9, 23)), and our own, the Open Microscopy Environment (OME (33, 57)). All of these projects were initiated to support the specific needs of the biological systems and experiments in each of the labs driving the development of each project. For example, studies in neuroscience absolutely depend on a proper specification for neuroanatomy so that any image and resulting analysis can be properly oriented with respect to the physiological source. In this case, an ontological framework for neuroanatomy is then needed to support and compare the results from many different laboratories (23). A natural progression is a resource that enables sharing of specific images, across many different resolution scales, that are as well-defined as possible (9). PSLID is an alternative repository that provides a well-annotated resource for subcellular localization by fluorescence microscopy. It is important to point out that in all cases, these projects are the result of dedicated, long-term collaboration between computer scientists and biologists, indicating that the challenges presented by this infrastructure development represent the state of the art not only in biology but in computing as well. Many if not most of these projects make use of at least some common software and data models, and although full interoperability is not something that can be claimed today, key members of these projects regularly participate in the same meetings and working groups. In the future, it should be possible for these projects to interoperate to enable, for example, OME software to upload to PSLID or CCDB.
OME-- a community-based effort to develop image informatics tools
Since 2000, the Open Microscopy Consortium has been working to deliver tools for image informatics for biological microscopy. Our original vision (57), to provide software tools to enable interoperability between as many image data storage, analysis, and visualization applications as possible remains unchanged. However, the project has evolved and grown since its founding to encompass a much broader effort, and now includes sub-projects dedicated to data modeling (39), file format specification and conversion (36, 37), data management (29) and image-based machine learning (31). The Consortium (30) also maintains links with many academic and commercial partners (34). While the challenges of running and maintaining a larger Consortium are real, the major benefits are synergies and feedback that develop when our own project has to use its own updates to data models and file formats. Within the Consortium, there is substantial expertise in data modeling and software development and we have adopted a series of project management tools and practices to make the project as professional as possible, within the limits of working within academic laboratories. Moreover, our efforts occur within the context of our own image-based research activities. We make no pretense that this samples the full range of potential applications for our specifications and software, just that our ideas and work are actively tested and refined before release to the community. Most importantly, the large Consortium means that we can interact with a larger community, gathering requirements and assessing acceptance and new directions as widely as possible.
The Foundation—The OME Data Model
Since its inception in 2000, the OME Consortium has dedicated itself to developing a specification for the metadata associated with the acquisition of a microscope image. Initially, our goal was to specify a single data structure that would contain spatial, temporal, and spectral components (often referred to as “Z”, “C”, “T”, which together form a “5D” image (1)), This has evolved into specifications for the other elements of the digital microscope system including objective lenses, fluorescence filter sets, illumination systems, detectors, etc. This effort has been greatly aided by many discussions about configurations and specifications with commercial imaging device manufacturers (34). This work is ongoing, with our current focus being the delivery of specifications for regions-of-interest (based on existing specifications from the geospatial community (44) and a clear understanding of what data elements are required to properly define a digital microscope image. This process is most efficient when users or developers request updates to OME data model—the project’s web-site (39) accepts request for new or modified features and fixes.
OME File Formats
The specification of an open, flexible file format for microscope imaging provides a tool for data exchange between distinct software applications. It is certainly the lowest level of interoperability, but for many situations, it suffices in its provision of readable, defined structured image metadata. OME’s first specification cast a full 5D image—binary and metadata—in an XML file (15). While conceptually sound, a more pragmatic approach is to store binary data as TIFF and then link image metadata represented as OME-XML by including it within the TIFF image header or as a separate file (37). To ensure that these formats are in fact defined, we have delivered an OME-XML and OME-TIFF file validator (38) that can be used by developers to ensure files follow the OME-XML specification. As of this writing there are now five commercial companies supporting these file formats in their software with a “Save as…”, thus enabling export of image data and metadata to a vendor neutral format.
Support for Data Translation-- Bio-Formats
PFFs are perhaps the most common informatics challenge faced by bench biologists. Despite the OME-XML and OME-TIFF specifications, PFFs will continue to be the dominant source of raw image for visualization and analysis applications for some time. Since all software must contend with PFFs, the OME Consortium has dedicated its resources to developing a software library that can convert PFFs to a vendor neutral data structure—OME-XML. This led to development, release, and continued maintenance of Bio-Formats, a standalone Java library for reading and writing life sciences image formats. The library is general, modular, flexible, extensible and accessible. The project originally grew out of efforts to add support for file formats to the LOCI VisBio software (42, 51) for visualization and analysis of multidimensional image data, when we realized that the community was in acute need of a broader solution to the problems created by myriad incompatible microscopy formats.
Utility
Over the years we have repeatedly observed software packages reimplement support for the same microscopy formats (i.e., ImageJ (18), MIPAV (24), BioImageXD (3), and many commercial packages). The vast majority of these efforts focus exclusively on adaptation of formats into each program’s specific internal data model; Bio-Formats (36), in contrast, unites popular life sciences file formats under a broad, evolving data specification provided by the OME Data Model. This distinction is a critical: Bio-Formats does not adapt data into structures designed for any specific visualization or analysis agenda, but rather expresses each format’s metadata in an accessible data model built from the ground up to encapsulate a wide range of scientifically relevant information. We know of no other effort within the life sciences with as broad a scope as Bio-Formats and dedicated toward delivering the following features.
Modularity
The architecture of the Bio-Formats library is split into discrete, reusable components that work together, but are fundamentally separable. Each file format reader is implemented as a separate module extending a common IFormatReader interface; similarly, each file format writer module extends a common IFormatWriter interface. Both reader and writer modules utilize the Bio-Formats MetadataStore API to work with metadata fields in the OME Data Model. Shared logic for encoding and decoding schemes (e.g., JPEG and LZW) are structured as part of the Bio-Formats codecs package, so that future readers and writers that need those same algorithms can leverage them without reimplementing similar logic or duplicating any code.
When reading data from a dataset, Bio-Formats provides a tiered collection of reader modules for extracting or restructuring various types of information from the dataset. For example, a client application can instruct Bio-Formats to compute minimum and maximum pixel values using a MinMaxCalculator, combine channels with a ChannelMerger, split them with a ChannelSeparator, or reorder dimensional axes with a DimensionSwapper. Performing several such operations can be accomplished merely by stacking the relevant reader modules one atop the other.
Several auxiliary components are also provided, the most significant being a caching package for intelligent management of image planes in memory when storage requirements for the entire dataset would be too great; and a suite of graphical components for common tasks such as presenting the user with a file chooser dialog box, or visualizing hierarchical metadata in a tree structure.
Flexiblity
Bio-Formats has a flexible metadata API, built in layers over the OME Data Model itself. At the lowest level, the OME data model is expressed as an XML schema called OME-XML, which is continually revised and expanded to support additional metadata fields. An intermediate layer known as the OME-XML Java library is produced using code generation techniques, which provides direct access to individual metadata fields in the OME-XML hierarchy. The Bio-Formats metadata API, which provides a simplified, flattened version of the OME Data Model for flexible implementation by the developer, leverages the OME-XML Java library layer, and is also generated automatically from underlying documents to reduce errors in the implementation.
Extensibility
Adding a new metadata field to the data model is done at the lowest level, to the data model itself via the OME-XML schema. The supporting code layers -- both the OME-XML Java library and the Bio-Formats metadata API -- are programmatically regenerated to include the addition. The only remaining task is to add a small amount of code to each file format reader mapping the original data field into the appropriate location within the standardized OME data model.
Though the OME data model specifically targets microscopy data, in general, the Bio-Formats model of metadata extensibility is ideal for adaptation to alternative data models unrelated to microscopy. By adopting a similar pattern for the new data model, and introducing code generation layers corresponding to the new model, the Bio-Formats infrastructure could easily support additional branches of multidimensional scientific imaging data and in the future will provide significant interoperability between the multiple established data models at points where they overlap, by establishing a common base layer between them.
Bio-Formats is written in Java so that the code can execute on a wide variety of target platforms, and code and documentation for interfacing Bio-Formats with a number of different tools including ImageJ, MATLAB and IDL are available (36). We provide documentation on how to use Bio-Formats as both an end user and as a software developer, including hints on leveraging Bio-Formats from other programming environments such as C++, Python, or a command shell. We have successfully integrated Bio-Formats with native acquisition software written in C++, using ICE middleware (see below, (60)).
Data Management Applications: OME & OMERO
Data management is a critical application for modern biological discovery, and in particular necessary for biological imaging because of the large heterogeneous data sets generated during data acquisition and analysis. We define data management as the collation, integration, annotation, and presentation of heterogeneous experimental and analytic data in ways that enable the physical temporal, and conceptual relationships in experimental data to be captured and represented to users. The OME Consortium has built two data management tools— the original OME Server (31) and the recently released OMERO application platform (32). Both applications are now heavily used worldwide, but our development focus has shifted from the OME Server towards OMERO and that is where most future advances will occur.
The OME data management applications are specifically designed to meet the requirements and challenged described above, enabling the storage, management, visualization, and analysis of digital microscope image data and metadata. The major focus of this work is not on creating novel analysis algorithms, but instead on development of a structure that ultimately allows any application to read and use any data associated with or generated from digital imaging microscopes.
A fundamental design concept in the OME data management applications is the separation of image storage, management, analysis, and visualization functions between a lab’s or imaging facility’s server and a client application (e.g., web browser or Java user interface (UI)). This concept mandates the development of two facilities: a server, that provides all data management, access control, and storage, and a client, that runs on a user’s desktop workstation or laptop, that provides access to the server and the data via a standard internet connection. The key to making this strategy work is judicious choice of the functionality placed on client and server to ensure maximal performance.
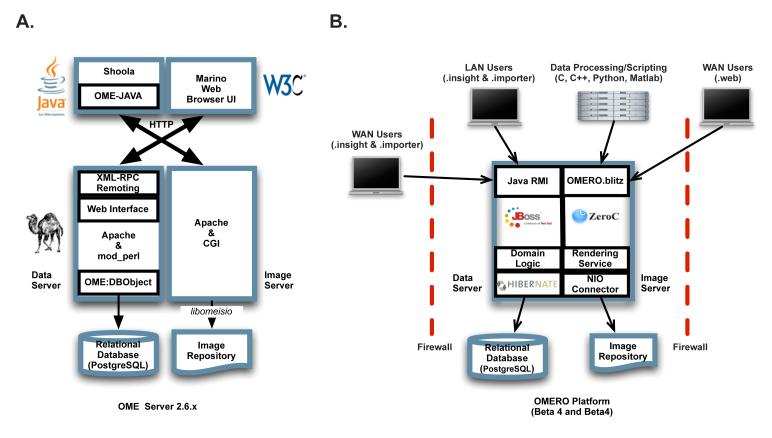
The technical design details and principles of both systems have recently been described (25) and are available on-line (41). In brief, both the OME Server and OMERO platform (see Figure 1) use a relational database management system (RDMS) (PostgreSQL, (49)) to provide all aspects of metadata management and an image repository to house all the binary pixel data. Both systems then use a middleware application to interact with the RDMS and read and write data from the image repository. The middleware applications include a rendering engine that reads binary data from the image repository and renders it for display by the client, and if necessary, compresses the image to reduce the bandwidth requirements for transferring across a network connection to a client. The result is access to high performance data visualization, management, and analysis in a remote setting. Both OME Server (Figure 1A) and OMERO (Figure 1B) also provide well-developed data querying facilities to access metadata, annotations and analytics from the RDMS. For user interfaces, the OME Server includes a web browser-based interface that provides access to image, annotation, analytics, and visualization and also a Java interface (“OME-JAVA”) and remote client (“Shoola”) to support access from remote client applications. OMERO includes separate Java-based applications for uploading data to and OMERO server (“OMERO.importer”), visualizing and managing data (“OMERO.insight”) and web-browser-based server administration tool (“OMERO.webadmin”).
Figure 1.
Architecture of OME and OMERO server and client applications. (A) Architecture of the OME Server, built using Perl for most of the software code and an Apache web server. The main client application for the server is a web browser-based interface. (B) The architecture of the OMERO platform, including OMERO.server and the OMERO clients. OMERO is based on the JBOSS JavaEE framework, but also includes an alternative remoting architecture called ICE (60). For more detail, see (41).
OME Server has been installed in hundreds of facilities worldwide, however after significant development effort it became clear that the application, which we worked on for five years (2000-2005) had three major flaws:
installation was too complex, and too prone to failure;
our object-relational mapping (ORM) library (“DBObject”) was all custom code, developed by OME, and required significant code maintenance effort to maintain compatibility with new versions of Linux and Perl (see Figure 1A). Support for alternative RDMSs (e.g., Oracle) were possible in principle, but required significant work;
The data transport mechanisms available to us in a Perl-based architecture amounted to XML-RPC and SOAP. While totally standardized and promoting interoperability, this mechanism, with its requirement for serialization/deserialization of large data objects, was too slow for working with remote client applications—simple queries with well-populated databases could take minutes to transfer from server to client.
With work (a) became less of a problem, but (b) and (c) remained significant fundamental barriers to delivery of a great image informatics application to end-users. For these reasons, we initiated work on OME Remote Objects (“OMERO”), a port of the basic image data management functionality to a Java enterprise application. In taking on this project, it was clear that the code maintenance burden needed to be substantially reduced, the system must be simple to install, and the performance of the remoting system must be significantly improved. A major design goal was the reduction of self-written code through the re-use of existing middleware and tools where possible. In addition, OMERO must support as broad a range of client applications as possible, enabling the development of new user interfaces, but also a wide range of data analysis applications.
We based the initial implementation of OMERO’s architecture (Figure 1B) on the JavaEE5 specification as it appeared to have wide uptake, clear specifications and high performance libraries in active development from a number of projects. A full specification and description of the OMERO.server is available (25). The architecture follows accepted standards and consists of services implemented as EJB3 session beans (55) that make use of Hibernate (16), a high-performance object-relational mapping solution, for metadata retrieval from the RDMS. Connection to clients is via Java Remote Method Invocation (Java RMI; (56)). All released OMERO remote applications are written in Java and cross-platform. OMERO.importer uses the Bio-Formats library to read a range of file formats and load the data into an OMERO.server, along with simple annotations and assignment to the OME Project-Dataset-Image experimental model (for demonstrations, see (35)). OMERO.insight includes facilities for managing, annotating, searching, and visualizing data in an instance of OMERO.server. OMERO.insight also includes simple line and region-of-interest measurements, and thus supports the simplest forms of image analysis.
OMERO Enhancements— Beta3 and Beta4
Through 2007, the focus of the OMERO project has been on data visualization and management, all the while laying the infrastructure for data analysis. With the release of OMERO3-Beta2, we began adding functionality that has the foundation for delivering a fully developed image informatics framework. In this section, we summarize the major functional enhancements that are being delivered in OMERO-Beta3 (released June 2008) and OMERO-Beta4 (released December 2008). Further information on all the items described below is available at the OMERO documentation portal (41).
OMERO.blitz
Starting with OMERO-Beta3, we provided interoperability with many different programming environments. We chose an ICE-based framework (60), rather than the more popular web services-based GRID approaches because of the absolute performance requirements we had for the passage of large binary objects (image data) and large data graphs (metadata trees) between server and client. Our experience using web services and XML-based protocols with the Shoola remote client and the OME Server showed that web services, while standardized in most genomic applications, was inappropriate for client-server transfer of the much larger data graphs we required. Most importantly, the ICE framework provided immediate support for multiple programming environments (C, C++, and Python are critical for our purposes) and a built-in distribution mechanism (IceGRID; (60)) that we have adapted to deliver OMERO.grid (41), a process distribution system. OMERO.blitz is 3-4x faster than JavaRMI and we are currently examining migrating our Java API and the OMERO clients from JBOSS to OMERO.blitz. This framework provides substantial flexibility-- interacting with data in an OMERO Server can be as simple as starting the Python interpreter and interacting with OMERO via the console. Most importantly, this strategy forms the foundation for our future work as we can now leverage the advantages and existing functionality in cross-platform Java, native C and C++ and scripted Python for rapidly expanding the functionality in OMERO.
Structured Annotations
As of OMERO-Beta3, users can attach any type of data to an image or other OMERO data container—text, URL, or other data files (.doc, .pdf, .xls, .xml, etc.) providing essentially the same flexibility as email attachments. The installation of this facility followed feedback from users and developers concerning the strategy for analysis management built into the OME Server. The underlying data model supported ‘hard semantic typing’ where each analysis result was stored in relational tables with names that could be defined by the user (25, 57). This approach, while conceptually desirable, proved too complex and burdensome. As an alternative in OMERO, Structured Annotations can be used to store any kind of analysis result as untyped data, defined only by a unique name to ensure that multiple annotations are easily distinguished. The data is not queryable by standard SQL, but any text-based file can be indexed and therefore found by users. Interestingly, BISQUE has implemented a similar approach (5), enabling ‘tags’ with defined structures that are otherwise completely customized by the user. In both cases, whether this very flexible strategy provides enough structure to manage large sets of analysis results will have to be assessed.
OMERO.search
As of OMERO-Beta3, OMERO includes a text indexing engine based on Lucene (21), which can be used to provide indexed based search for all text-based metadata in an OMERO database. This includes metadata and annotations stored within the OMERO database and also any text based documents or results stored as Structured Annotations.
OMERO.java
As of OMERO-Beta3, we have released OMERO.java which provides access for all external Java applications via the OMERO.blitz interface. As a first test of this facility, we are using analysis applications written in Matlab as client applications to read from and write to OMERO. server. As a demonstration of the utility of this library, we have adapted the popular open source Matab-based image analysis tool Cellprofiler (8) to work as a client of OMERO, using the Matlab Java interface.
OMERO.editor
In OMERO-Beta3, we also released OMERO.editor, a tool to help experimental biologists define their own experimental data models and if desired, use other specified data models in their work. It allows users to create a protocol template and to populate this with experimental parameters. This creates a complete experimental record in one XML file, which can be used to annotate a microscope image or exchanged with other scientists. OMERO.editor supports the definition of completely customized experimental protocols, but also includes facilities to easily import defined data model (e.g., MAGE-ML (58), OME-XML (15)) and also includes support for all ontologies included in the Ontology Lookup Service (12).
OMERO.web
Staring with OMERO-Beta4, we will release a web browser-based client for OMERO.server. This new client is targeted specifically to truly remote access (different country, limited bandwidth connections), especially where collaboration with other users is concerned. OMERO.web includes all the standard functions for importing, managing, viewing, and annotating image data. However, a new function is the ability to “share” specific sets of data with another user on the system—this allows password-protected access to a specific set of data that can initiate or continue data sharing between two lab members or two collaborating scientists. OMERO.web also supports a “publish” function, where a defined set of data is publish to the world, via a public URL. OMERO.web uses the Python API in OMERO.blitz for access to OMERO.server using the Django framework (13).
OMERO.scripts
In OMERO-Beta4, we will extend the analysis facility provided by OMERO.java to provide a scripting engine, based on Python Scripts and the OMERO.blitz interface. OMERO.scripts is a scripting engine that reads and executes functions cast in Python scripts. Scripts are passed to processors specified by OMERO.grid which can be on the local server or on networked compute facilities. This is the facility that will provide support for analysis of large image sets or of calculations that require simple linear or branched workflows.
OMERO.fs
Finally, a fundamental design principle of OMERO.server is the presence of a single image repository for storing binary image data that is tightly integrated with the server application. This is the basis of the “import” model that is the only way to get image data into an OMERO.server installation—data is uploaded to the server, and binary data stored in the single image repository. In many cases, as the storage space required expands, multiple repositories must be supported. Moreover, data import takes times, and especially with large data sets, can be prohibitive. A solution to this involves using the OMERO.blitz Python API to access the filesystem search and notification facilities that are now provided as part of the Windows, Linux, and OS X operating systems. In this scenario, an OMERO client application, OMERO.fs, sits between the filesystem and OMERO.blitz and provides a metadata service that scans user-specified image folders or filesystems and reads image metadata into an OMERO relational database using proprietary file format translation provided by Bio-Formats. As the coverage of Bio-Formats expands, this approach means that essentially any data can be loaded into an OMERO.server instance.
Workflow-based Data Analysis: WND-CHARM
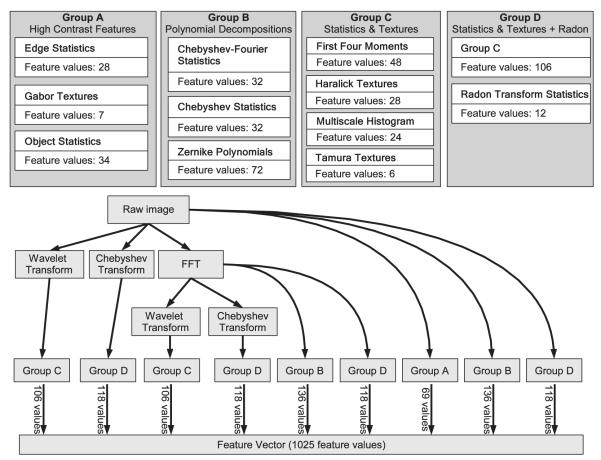
WND-CHARM is an image analysis algorithm based on pattern recognition (45). It relies on supervised machine learning to solve image analysis problems by example rather than by using a pre-conceived perceptual model of what is being imaged. An advantage of this approach is its generality. Because the algorithms used to process images are not task-specific, they can be used to process any image regardless of the imaging modality or the image’s subject. Similarly to other pattern recognition algorithms, WND-CHARM first decomposes each image to a set of pre-defined numeric image descriptors. Image descriptors include measures of texture, factors in polynomial decompositions, various statistics of the image as a whole as well as measurements and distribution of “high-contrast” objects in the image. The algorithms that extract these descriptors (”features”) operate on both the original image pixels as well as transforms of the original pixels (Fourier, wavelet, etc). Together, there are 17 independent algorithms comprising 53 computational nodes (algorithms used along specific upstream data flows), with 189 links (data flows) producing 1025 numeric values modeled as 48 Semantic Types in OME (see Figure 2). Although the entire set of features can be modeled as a single algorithm, this set is by no means complete and will grow to include other algorithms that extract both more specific and more general image content. The advantage of being able to model this complex workflow as independently functional units is that new units can be easily added to the existing ones. This workflow model is therefore more useful to groups specializing in pattern recognition. Conversely, a monolithic representation of this workflow is probably more practical when implemented in a biology lab that would use a standard set of image descriptors applied to various imaging experiments. In neither case however, should anyone be particularly concerned with what format was used to capture these images, or how they are represented in a practical imaging system. WND-CHARM is an example of a highly complex image-processing work-flow and as such represents an important application for any system capable of managing work-flows and distributed processing for image analysis. Currently the fully modularized version of WND-CHARM runs only on OME Server. In the near future, the monolithic version of WND-CHARM (53) will be implemented using OMERO.blitz.
Figure 2.
Workflows in WND-CHARM. (A) List of feature types calculated by WND-CHARM. (B) Workflow of feature calculations in WND-CHARM. Note that different feature groups use different sets of processing tools.
The raw results from a pattern recognition application are annotations assigned to whole images or image regions. These annotations are probabilities (or simply scores) that the image or ROI belongs to a previously defined training set. In a dose-response experiment, for example, the training set may consist of control doses defining a standard curve, and the experimental images would be assigned an equivalent dose by the pattern-recognition algorithm. Interestingly, while the original experiment may be concerned with characterizing a collection of chemical compounds, the same image data could be analyzed in the context of a different set of training images - one defined by RNA-interference, for example. In practice when using these algorithms, our group has found that performing these “in silico” experiments to re-process existing image data in different contexts can be extremely fruitful.
Usability
All of the functionality discussed above must be built into OMERO.server and then delivered in a functional, usable fashion within the OMERO client applications, OMERO.importer, OMERO.insight, and OMERO.web. This development effort is achieved by the OMERO development team. This is invariably an iterative process, that requires testing by our local community, but also sampling feedback from the broader community as well. Therefore, the OMERO project has made software usability a priority throughout the project. A key challenge for the OME Consortium has been to improve the quality of the ‘end user’ (i.e. the life scientist at their bench) experience. The first versions of OME software, the OME Server, provided substantial functionality, but never received wide acceptance, despite dedicated work, mostly because its user interfaces were too complicated and the developed code, while open and available, was too complex for other developers to adopt and extend. In response to this failure, we initiated the Usable Image project (43) to apply established methods from the wider software design community, such as user-centered design and design ethnography (22), to the OME development process. Our goals were to initially improve the usability and accessibility of the OMERO client software, and to provide a paradigm useful for the broader e-science and bioinformatics communities. The result of this combined usability and development effort has been an high level of success and acceptance of OMERO software. A wholly unanticipated outcome has been the commitment to the user-centered design process by both users and developers. The investment in iterative, agile development practice has produced rapid, substantial improvements that the users appreciate, which in turn makes them more enthusiastic about the software. On the other hand, the developers have reliable, well-articulated requirements, that when implemented in software, are rewarded with more frequent use. This positive feedback loop has transformed our development process, and made usability analysis a core part of our development cycles. It has also forced a commitment to the development of usable code—readable, well-documented, tested, and continuously integrated—and the provision of up-to-date resources defining architecture and code documentation (40, 41).
Summary and Future Impact
In this article we have focused on the OME Consortium’s efforts (namely OME-XML, Bio-Formats, OMERO and WND-CHARM) as we feel they are representative of the attempts community-wide to address many of the most pressing challenges in bioimaging informatics. While OME is committed to developing and releasing a complete image informatics infrastructure focused on the needs of the end user bench biologist, we are at least equally committed to the concept that beyond our software, our approach is part of a critical shift in how the challenges of data analysis, management, sharing and visualization have been traditionally addressed in biology.. In particular the OME Consortium has put an emphasis on flexibility, modularity, and inclusiveness that targets not only the bench biologist but importantly the informatics developer as well to help ensure maximum implementation and penetration in the bioimaging community. Key to this has been a dedication to allowing the biologist to retain and capture all available metadata and binary data from disparate sources including proprietary ones, map this to a flexible data model, and analyze this data in whatever environment he or she chooses. This ongoing effort requires an interdisciplinary approach combining concepts from traditional bioinformatics, ethnography, computer science and data visualization. It is our intent and hope that the bioimage informatics infrastructure that is being developed by the OME Consortium will continue to have utility not only for its principal target community of the experimental bench biologist, but also serve as a collaborative framework for developers and researchers from other closely related fields who might want to adopt the methodologies and code-based approaches for informatics challenges that exist in other communities. Interdisciplinary collaboration between biologists, physicists, engineers, computer scientists, ethnographers, and software developers is absolutely necessary for the successful maturation of the bioimage informatics community and it will play an even larger role as this field evolves to fully support the continued evolution of imaging in modern experimental biology.
Summary Points.
Advances in digital microscopy have driven the development of a new field, bioimage informatics. This field encompasses the storage, querying, management, analysis and visualization of complex image data from digital imaging systems used in biology.
While standardized file formats have often been proposed to be sufficient to provide the foundation for bioimage informatics, the prevalence of PFFs and the rapidly evolving data structures needed to support new developments in imaging make this impractical.
Standardized APIs and software libraries enable the interoperability that is a critical unmet need in cell and developmental biology.
A community-driven development project is best-placed to define, develop, release and support these tools.
A number of bioimage informatics initiatives are underway, and collaboration and interaction is developing
The OME Consortium has released specifications and software tools to support bioimage informatics in the cell and developmental biology community.
The next steps in software development will deliver increasingly sophisticated infrastructure applications and should deliver powerful data management and analysis tools to experimental biologists.
Future Issues.
8. Further development of the OME Data Model to keep pace with and include advances in biological imaging with a particular emphasis on improving support for image analysis metadata and enabling local extension of the OME Data Model to satisfy experimental requirements with good documentation and examples.
9. Development of Bio-formats to include as many biological image file formats as possible and extension to include data from non-image based biological data.
10. Continue OMERO development as an image management system with a particular emphasis on ensuring client application usability and the provision of sophisticated image visualization and analysis tools.
11. Support both simple and complex analysis workflow as a foundation for common use of data analysis and regression in biological imaging.
12. Drive links between the different bioimage informatics enabling transfer of data between instances of the systems so that users can make use of the best advantages of each.
Acknowledgements
JRS is a Wellcome Trust Senior Research Fellow and work in his lab on OME is supported by the Wellcome Trust (Ref 080087 and 085982), BBSRC (BB/D00151X/1), and EPSRC (EP/D050014/1). Work in IGG’s lab is supported by the National Institutes of Health.
Terms/Definitions list
- Bioimage informatics
infrastructre including specifications, software, and interfaces to support experimental biological imaging
- Proprietary file formats
image file data formats defined and used by individual entities
- Application programming interface
an interface provide one software program or library to provide easy access to its functionality with full knowledge of the underlying code or data structures
Acronyms
- OME
Open Microscopy Environment
- API
application programming interface
- XML
Extensible Markup Language
- HCS
high content screen
- PFF
proprietary file format
- RDMS
relational database management system
- OMERO
OME Remote Objects
- CCDB
Cell-Centred Database
- PSLID
Protein Subcellular Location Image Database
Literature Cited
- 1.Andrews PD, Harper IS, Swedlow JR. To 5D and beyond: Quantitative fluorescence microscopy in the postgenomic era. Traffic. 2002;3:29–36. doi: 10.1034/j.1600-0854.2002.30105.x. [DOI] [PubMed] [Google Scholar]
- 2.Berman J, Edgerton M, Friedman B. The tissue microarray data exchange specification: A community-based, open source tool for sharing tissue microarray data. BMC Medical Informatics and Decision Making. 2003;3:5. doi: 10.1186/1472-6947-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BioImageXD BioImageXD - open source software for analysis and visualization of multidimensional biomedical data. 2008 http://www.bioimagexd.net/
- 4.BIRN BIRN-Biomedical Informatics Research Network. 2008 http://www.nbirn.net/
- 5.Centre for Bio-Image Informatics Bisque Database. 2008 http://www.bioimage.ucsb.edu/downloads/Bisque%20Database.
- 6.Bullen A. Microscopic imaging techniques for drug discovery. Nat Rev Drug Discov. 2008;7:54–67. doi: 10.1038/nrd2446. [DOI] [PubMed] [Google Scholar]
- 7.caBIG Community Website http://cabig.nci.nih.gov/
- 8.Carpenter A, Jones T, Lamprecht M, Clarke C, Kang I, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cell Centered Database Cell Centered Database. 2008 http://ccdb.ucsd.edu/CCDBWebSite/
- 10.Christiansen JH, Yang Y, Venkataraman S, Richardson L, Stevenson P, et al. EMAGE: a spatial database of gene expression patterns during mouse embryo development. Nucl. Acids Res. 2006;34:D637–41. doi: 10.1093/nar/gkj006. %R 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christiansen JH, Yang Y, Venkataraman S, Richardson L, Stevenson P, et al. EMAGE: a spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Res. 2006;34:D637–41. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cote RG, Jones P, Martens L, Apweiler R, Hermjakob H. The Ontology Lookup Service: more data and better tools for controlled vocabulary queries. Nucl. Acids Res. 2008;36:W372–76. doi: 10.1093/nar/gkn252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Django Project Django: the web framework for perfectionists with deadlines. 2008 http://www.djangoproject.com/
- 14.Drysdale R, the FlyBase C FlyBase. Drosophila. 2008. pp. 45–59. [DOI] [PubMed]
- 15.Goldberg IG, Allan C, Burel J-M, Creager D, Falconi A, et al. The Open Microscopy Environment (OME) Data Model and XML File: Open Tools for Informatics and Quantitative Analysis in Biological Imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.hibernate.org hibernate.org - Hibernate. 2008 http://www.hibernate.org/
- 17.Huang K, Lin J, Gajnak JA, Murphy RF. Image Content-based Retrieval and Automated Interpretation of Fluorescence Microscope Images via the Protein Subcellular Location Image Database. Proc IEEE Symp Biomed Imaging. 2002:325–28. [Google Scholar]
- 18.ImageJ ImageJ. 2008 http://rsbweb.nih.gov/ij/
- 19.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 20.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 21.Apache Lucene Apache Lucene - Overview. 2008 http://lucene.apache.org/java/docs/
- 22.Macaulay C, Benyon D, Crerar A. Ethnography, theory and systems design: from intuition to insight. International Journal of Human-Computer Studies. 2000;53:35–60. [Google Scholar]
- 23.Martone ME, Tran J, Wong WW, Sargis J, Fong L, et al. The Cell Centered Database project: An update on building community resources for managing and sharing 3D imaging data. Journal of Structural Biology. 2008;161:220–31. doi: 10.1016/j.jsb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MIPAV Medical Image Processing, Analysis, and Visualization. 2008 http://mipav.cit.nih.gov/
- 25.Moore J, Allan C, Burel J-M, Loranger B, MacDonald D, et al. Open Tools for Storage and Management of Quantitative Image Data. Methods Cell Biol. 2008;85 doi: 10.1016/S0091-679X(08)85024-8. [DOI] [PubMed] [Google Scholar]
- 26.Murphy RF. Murphy Lab | Imaging services | PSLID. 2008 http://murphylab.web.cmu.edu/services/PSLID/
- 27.myGRID myGRID >> Home. 2008 http://www.mygrid.org.uk/
- 28.Neumann B, Held M, Liebel U, Erfle H, Rogers P, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–90. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 29.OME Consortium OME - Data Management. 2008 http://www.openmicroscopy.org/site/documents/data-management.
- 30.OME Consortium OME - Development Teams. 2008 http://openmicroscopy.org/site/about/development-teams.
- 31.OME Consortium OME - OME Server. 2008 http://openmicroscopy.org/site/documents/data-management/ome-server.
- 32.OME Consortium OME - OMERO Platform. 2008 http://openmicroscopy.org/site/documents/data-management/omero.
- 33.OME Consortium OME - Open Microscopy Environment. 2008 http://openmicroscopy.org.
- 34.OME Consortium OME - Partners. 2008 http://openmicroscopy.org/site/about/partners.
- 35.OME Consortium OME - Videos for Quicktime. 2008 http://openmicroscopy.org/site/videos.
- 36.OME Consortium OME at LOCI - Bio-Formats. 2008 http://www.loci.wisc.edu/ome/formats.html.
- 37.OME Consortium OME at LOCI - OME-TIFF - Overview and Rationale. 2008 http://www.loci.wisc.edu/ome/ome-tiff.html. [Google Scholar]
- 38.OME Consortium OME Validator. 2008 http://validator.openmicroscopy.org.uk/
- 39.OME Consortium OME-XML: The data model for the Open Microscopy Environemnt-- Trac. 2008 http://ome-xml.org/
- 40.OME Consortium OMERO Continuous Integration System. 2008 http://hudson.openmicroscopy.org.uk.
- 41.OME Consortium ServerDesign - OMERO.server - Trac. 2008 http://trac.openmicroscopy.org.uk/omero/wiki/ServerDesign.
- 42.OME Consortium VisBio - Introduction. 2008 http://validator.openmicroscopy.org.uk/
- 43.OME Consortium Welcome to Usable Image. 2008 http://usableimage.org.
- 44.OpenGIS® OpenGIS® Standards and Specifications. 2008 http://www.opengeospatial.org/standards.
- 45.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognition Letters. 2008;29:1684–93. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parvin B, Yang Q, Fontenay G, Barcellos-Hoff MH. BioSig: An Imaging and Informatic System for Phenotypic Studies. IEEE Transaction on Systems, Man and Cybernetics, Part B. 2003 doi: 10.1109/TSMCB.2003.816929. [DOI] [PubMed] [Google Scholar]
- 47.Peng H. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008;24:1827–36. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–96. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 49.PostgreSQL PostgreSQL: The world’s most advanced open source database. 2008 http://www.postgresql.org/
- 50.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, et al. WormBase 2007. Nucl. Acids Res. 2008;36:D612–17. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rueden C, Eliceiri KW, White JG. VisBio: a computational tool for visualization of multidimensional biological image data. Traffic. 2004;5:411–7. doi: 10.1111/j.1600-0854.2004.00189.x. [DOI] [PubMed] [Google Scholar]
- 52.Saccharomyces Genome Database Saccharomyces Genome Database. 2008 http://www.yeastgenome.org/
- 53.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG. WND-CHARM - an open source utility for biological image analysis. Source Code for Biology and Medicine. 2008;3 doi: 10.1186/1751-0473-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 55.Sun Microsystems Enterprise Javabeans Technology. 2008 http://java.sun.com/products/ejb.
- 56.Sun Microsystems, Inc. Remote method Invocation Home. 2008 http://java.sun.com/javase/technologies/core/basic/rmi/index.jsp.
- 57.Swedlow JR, Goldberg I, Brauner E, Sorger PK. Informatics and quantitative analysis in biological imaging. Science. 2003;300:100–2. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whetzel PL, Parkinson H, Causton HC, Fan L, Fostel J, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22:866–73. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 59.WormBase WormBase - Home Page. 2008 http://www.wormbase.org/
- 60.Zeroc, Inc. Welcome to ZeroC, the Home of Ice. 2008 http://www.zeroc.com/
- 61.Zhao T, Murphy RF. Automated learning of generative models for subcellular location: Building blocks for systems biology. Cytometry Part A. 2007;71A:978–90. doi: 10.1002/cyto.a.20487. [DOI] [PubMed] [Google Scholar]