Summary
Nestin-cre transgenic mice have been widely used to direct recombination to neural stem cells (NSCs) and intermediate neural progenitor cells (NPCs). Here we report that a readily utilized, and the only commercially available, Nestin-cre line is insufficient for directing recombination in early embryonic NSCs and NPCs. Analysis of recombination efficiency in multiple cre-dependent reporters and a genetic mosaic line revealed consistent temporal and spatial patterns of recombination in NSCs and NPCs. For comparison we utilized a knock-in Emx1cre line and found robust recombination in NSCs and NPCs in ventricular and subventricular zones of the cerebral cortices as early as embryonic day 12.5. In addition we found that the rate of Nestin-cre driven recombination only reaches sufficiently high levels in NSCs and NPCs during late embryonic and early postnatal periods. These findings are important when commercially available cre lines are considered for directing recombination to embryonic NSCs and NPCs.
Key words: Neurogenesis, Neural Progenitors, Nestin-cre
Introduction
Cre mediated recombination is a powerful and broadly utilized genetic approach to conditionally delete or express selective genes, and is extensively used to lineage trace distinct progenitor populations during development (reviewed by Branda and Dymecki, 2004; Bouvier and Cheng, 2009). Transgenic Nestin-cre lines of mice have been broadly utilized over the past decade to direct recombination to neural stem cells (NSCs) and intermediate neural progenitor cells (NPCs) (Gavériaux-Ruff and Kieffer, 2007). Several independent Nestin-cre lines have been generated and published (e.g. Isaka et al., 1999; Trumpp et al., 1999; Petersen et al., 2002; Dubois et al., 2006), but only one (Tronche et al., 1999) is available through the Jackson laboratories and has been used in numerous studies. Based on early reports by the originating investigator it has been assumed that this line is highly efficient in directing cre-mediated recombination to neural progenitors. Here we tested whether or not the only commercially available line of Nestin-cre mice is sufficient for recombination in NSCs and NPCs of the forebrain during embryonic and early postnatal development. For comparison we utilized a knock-in Emx1cre line that is also broadly used for cre-mediated recombination in NSCs and NPCs of the dorsal telencephalon.
Materials and Methods
Animals
Animals were used under Institutional Animal Care and Use Committee and North Carolina State University regulations, and were housed in the Laboratory Animal Research facility at the College of Veterinary Medicine. Nestin-cre (B6.Cg-Tg (Nes-cre)1Kln/J; Jackson Lab, #003771; on a mixed C57BL/6/SJL background) (Tronche et al., 1999) and knock-in Emx1cre mice (B6.129S2-Emx1tm1(cre)Krj/J; Jackson Lab, #005628; on a 129S2/SvPas background) (Gorski et al., 2002) were crossed to mice expressing the reporter genes tdTomato (tdTom) (B6.129S6-Gt (ROSA) 26Sortm14 (CAG-tdTomato) Hze/J; Jackson Lab, #007908; on a mixed 129S6/SvEvTac/C57BL/6NCr background) (Madisen et al., 2010) or LacZ (B6.129S4-Gt (ROSA) 26Sortm1Sor/J; Jackson Lab, #003309, on a 129S4/SvJaeSor background) (Soriano, 1999), which are conditionally expressed under a knock-in pCAG promoter into the Rosa locus to track cre-mediated recombination.
For mosaic analyses, MADM11-TG/GT:Nestin-cre or MADM11-TG/GT:Emx1cre mice were generated using breeding schemes previously described (Hippenmeyer et al., 2010). In brief, Nestin-cre or Emx1cre mice were first crossed to the MADM11-GT line (on a 129X1/SvJ background) and the resulting MADM11-GT:Nestin-cre or MADM11-GT: Emx1cre mice were then crossed to the MADM11-TG+ mice (also on a 129X1/SvJ background) to generate the MADM11-TG/GT:Nestin-cre or MADM11-TG/GT:Emx1cre mice for mosaic analyses.
For embryonic analyses, the morning of observed vaginal plug was designated as embryonic day 0.5 (E0.5). Embryos were harvested from time-pregnant females following Avertin overdose (7.5 mg/g body weight), their brain removed and fixed with 4% paraformaldehyde for a minimum of 24 hours prior to processing. New born (P0) mice were anesthetized by hypothermia followed by transcardial perfusion with 4% paraformaldehyde in 0.1 M phosphate buffered saline (PBS). All mice received a pulse of 5-bromo-2′-deoxyuridine (BrdU, Aldrich; 100 mg/kg body weight via intraperitoneal injections to pregnant females or P0 pups) one hour prior to harvestation.
Tissue processing and imaging
Following perfusion, brains were removed and sectioned in the sagittal plane at 50 µm on a vibratome (Leica VT 1000 S). Floating sections were immunohistochemically processed using standard procedures with rabbit anti-RFP (Abcam; 1:1000), chicken anti-GFP (Abcam; 1:1000), mouse anti-BrdU (BD Bioscience; 13:1000), rabbit anti-cre (Covance; 1:500), and rabbit anti-PH3 (Millipore; 1:500) antibodies. Immunoreactivity was visualized using Alexafluor conjugatged secondary antibodies (Invitrogen). When needed, sections were counterstained with TO-PRO-3 Iodide (Invitrogen; 1:2000) or DAPI (VECTASHIELD mounting medium, Vector labs) for cytoarchitectonic characterization of sections.
Staining of whole embryos for LacZ was as described previously (Nagy et al., 2007). For tissue preparation, embryos were cryopreserved within 30% sucrose in 0.1 M PBS overnight at 4°C followed by overnight freezing in tissue freezing medium (Triangle Biomedical Sciences) at −80°C. Embryos were then sectioned at 20 µm on a cryostat (Leica).
Data Analyses
Percentages of tdTom+ recombined cells in cycling progenitors in various regions/tissues were calculated by dividing numbers of either BrdU+/tdTom+ pulse labeled progenitors, or PH3+/tdTom+ mitotic progenitors, by total number of counted BrdU+ or PH3+ nuclei, respectively (n = 3 animals; 5 sections per animal; a total of 225 and 217 PH3+ cells at E12.5 and E14.5 respectively; a total of 789 BrdU+ cells at P0). Significance was determined using Student's t-test and all values were expressed as mean ± standard error of the mean (SEM).
Results and Discussion
The efficiency of recombination in the central nervous system (CNS) was assessed in multiple reporter lines using the only Nestin-cre line of mice that was commercially available at the time of this report (Fig. 1A). Similar to past reports on the same line, we found that Nestin-cre mediated recombination commences in the CNS during embryogenesis (Fig. 1B). However, much to our surprise, in mice on a tdTomato (tdTom) reporter background the recombination rate was extremely insufficient in the ventricular (VZ) and subventricular (SVZ) zones of the neocortex during early (E12.5) and mid stages of forebrain development (E14.5) (Fig. 1B). Additional expression analyses using cre-specific antibodies confirmed the observed patterns at the various stages of development (Fig. 1C). High-magnification imaging revealed an extremely low recombination rate in PH3 or BrdU immunoreactive cycling NSCs and NPCs in the E14.5 lateral ganglionic eminence (LGE), olfactory ventricle (OV) and the cerebral cortex (Ctx) (Fig. 1D) (1.3±0.7% at E12.5 and 7.4±4% at E14.5 of PH3+ cells in the VZ and SVZ, respectively; n = 3 mice/group). Consistent with these findings, Nestin-cre on a different reporter line (Rosa-lacZ) also yielded low levels of reporter activity in the VZ of E12.5 cerebral cortices (Fig. 1D).
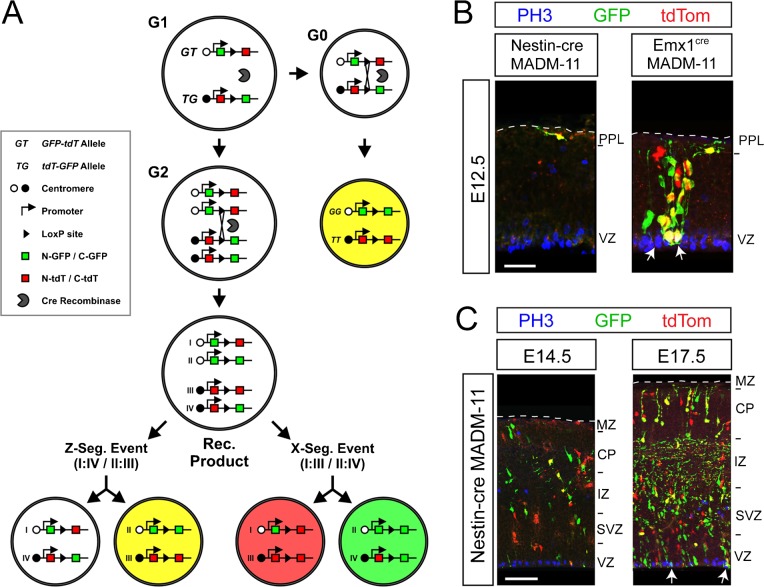
Fig. 1. Insufficient rate of recombination in NSCs and NPCs of Nestin-cre transgenic mice.
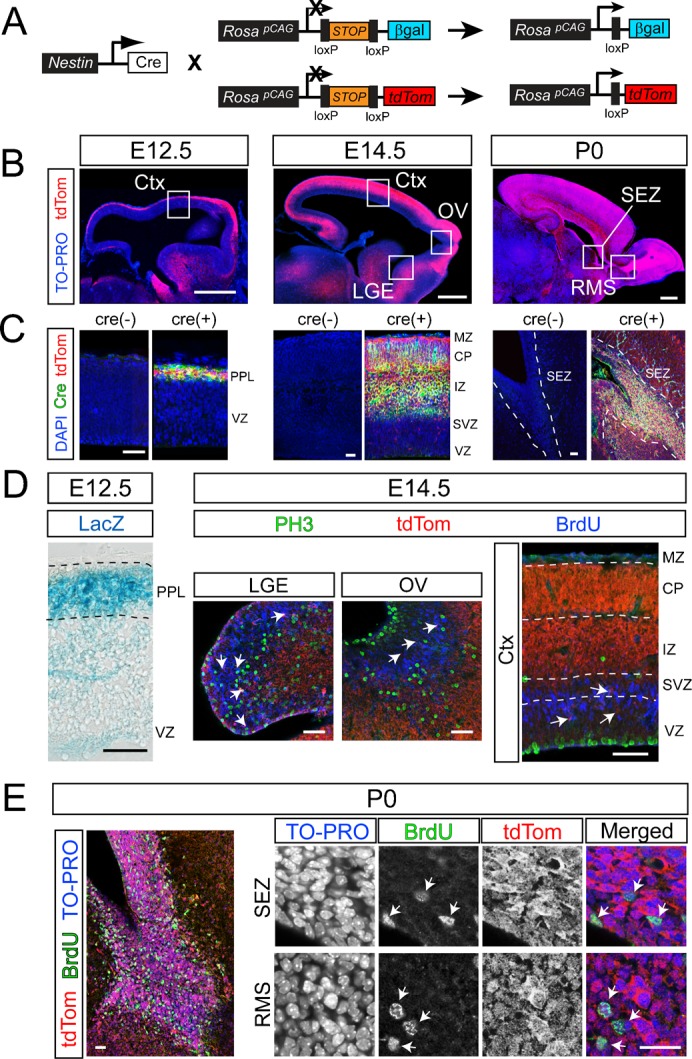
(A) Breeding schemes to generate Nestin-cre transgenic mice (Jax: B6.Cg-Tg(Nes-cre)1 Kln/J) on either a LacZ or tdTomato (tdTom) reporter background. LacZ or tdTom are conditionally expressed under the chicken β-actin promoter, which was knocked into the Rosa locus to track cre-mediated recombination. Two loxP sites flank a STOP codon upstream of the reporter cassettes, preventing reporter expression in the absence of cre-mediated recombination. Nestin-cre mediated recombination deletes the stop codon allowing the expression of the reporters. (B) Sagittal views of the developing CNS illustrating tdTom reporter expression mediated by Nestin-cre at embryonic stages E12.5, E14.5, and at birth (P0). (C) Analysis of cre expression at ages corresponding to sections in B, using a cre-specific antibody (green), in combination with tdTom and DAPI (blue). Cre(−) controls were obtained from brains of mice negative for Nestin-cre, but genetically positive for tdTom reporter. (D) Laminar pattern of Nestin-cre mediated recombination in NSCs and NPCs reported by LacZ expression in the E12.5 cerebral cortex. LacZ was predominantly expressed in the preplate (PPL), which consists of postmitotic cells, but not in the VZ, which consists of cycling NSCs. Similar pattern of expression was observed in the E14.5 lateral ganglionic eminence (LGE), olfactory ventricular zone (OV), and the cerebral cortices (Ctx). Each region is depicted in high magnification images of boxed regions marked in 1B. At E14.5, recombined tdTom+ cells in the VZ and SVZ (red, arrows) largely fail to co-label with the markers of cycling NSCs and NPCs in all three regions (PH3, green; BrdU, blue). Recombination in the E14.5 Ctx was dense in the intermediate zone (IZ) and the cortical plate (CP), which largely consists of post-mitotic neurons (MZ, marginal zone; devoid of recombined cells). (E) The rate of Nestin-cre mediated recombination in NSCs and NPCs reaches high levels at P0, revealed by expression of tdTom in nearly all cells (TO-PRO+, blue) and BrdU immunoreactive progenitors (green; arrows) of the subependymal zone (SEZ) and the rostral migratory stream (RMS). Scale bars: B, 500 µm; C, 30 µm; D, 40 µm; E, 20 µm.
In both reporter backgrounds, the majority of recombined cells were situated in the preplate layer (PPL) of the E12.5 cortex as well as intermediate zone (IZ) and cortical plate (CP) of the E14.5 cortex, suggesting that recombination became sufficient during postmitotic stages of neuronal differentiation (Fig. 1B–D). In contrast, the rate of recombination in NSCs and NPCs increased dramatically in the subependymal zone (SEZ) and the rostral migratory stream (RMS) of newborn (P0) mice (Fig. 1B,E). Nearly every BrdU-labeled NSC and NPC expressed tdTom (93±5% of BrdU+) (Fig. 1E), suggesting sufficient Nestin-cre mediated recombination in these cells. These findings suggest a gradual increase in the rate of Nestin-cre mediated recombination during perinatal development in NSCs and NPCs.
For comparison we utilized a knock-in Emx1cre line of mice (Fig. 2A) that has been reported to drive sufficient recombination in dorsal telencephalic NSCs and NPCs (Gorski et al., 2002). In contrast to the Nestin-cre line, Emx1cre mediated recombination was sufficient in the VZ/SVZ of the developing cortex at both E12.5 and E14.5 (Fig. 2B), which we confirmed again using antibody staining for cre (Fig. 2C). In addition, robust recombination was found in the majority (83±8%) of PH3+ progenitors in the E12.5 VZ and SVZ of the cortex (Fig. 2D). The efficiency increased to nearly 100% by E14.5 (Fig. 2D). Thus, the low degree of recombination in Nestin-cre brains is not due to potential issues associated with cre-loxP interactions in embryonic progenitors, but is most likely related to the transgene in this specific strain.
Fig. 2. Sufficient rate of recombination in embryonic NSCs and NPCs of Emx1cre knock-in mice.
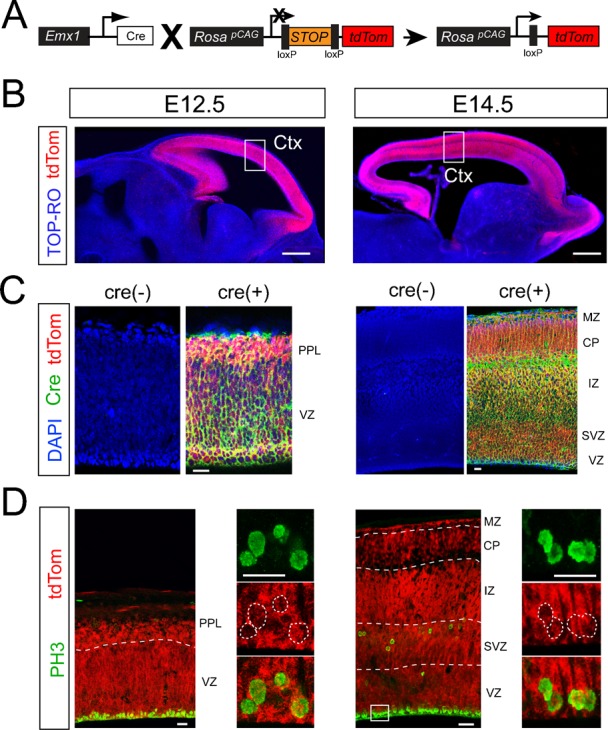
(A) Breeding schemes to generate mice with the Emx1cre locus on the tdTom reporter background. (B) tdTom reporter expression (red) in the E12.5 and E14.5 Emx1cre brains. Robust tdTom expression was detected in the entire cortex at both E12.5 and E14.5. (C) Analysis of cre expression at ages corresponding to sections in B, using a cre-specific antibody (green), in combination with tdTom and DAPI (blue). Cre(−) controls were obtained from brains of mice negative for Emx1cre, but genetically positive for tdTom reporter. (D) Emx1cre recombination in neural progenitors of E14.5 cortex (high magnification for the box region indicated in 2B). Emx1cre mediated tdTom expression (red) was found in the entire cortex including the proliferative VZ and SVZ. High magnification of the VZ regions (box) revealed robust co-localization of PH3+ mitotic NSCs (green) with tdTom (red). Scale bars: B, 300 µm; C, 30 µm; D, low mag (left), 20 µm; high mag (right panels), 10 µm.
To conclusively confirm our surprising findings on the developmental patterns of cre-mediated recombination in NSCs and NPCs in the Nestin-cre lines, and to rule out possible recombinogenic issues related to the Rosa locus in the reporter lines in the first part of the study, we crossed both cre lines into the Mosaic Analysis with Double Markers on chromosome 11 (MADM-11) line of mice (Hippenmeyer et al., 2010). In this line, cre-dependent mitotic recombination results in expression of GFP or tdTom independently or together resulting in various cells with three potential expression patterns (Fig. 3A, red, green, yellow). Recombination can also occur in postmitotic cells during the G0 phase, but in this case only cells expressing both GFP and tdTom (yellow cells) will be obtained (Fig. 3A). Using this strategy we found recombined cells with all three colors (red, green, yellow) throughout all layers of the cerebral cortices in Emx1cre MADM11 brains at E12.5 (Fig. 3B). In addition, recombined NSCs and NPCs were co-labeled with the mitosis marker PH3 in the VZ region (Fig. 3B). Thus, the Emx1cre knock-in allele is sufficient for driving mitotic recombination in NSCs and NPCs as early as E12.5.
Fig. 3. Analysis of mitotic and post-mitotic specificity of recombination in the cerebral cortices of Emx1cre and Nestin-cre mice using the MADM-11 genetic system.
(A) Scheme of cre-induced reporter expression in Mosaic Analysis with Double Markers on chromosome 11 (MADM-11) mice. In this line, cre-dependent mitotic recombination results in expression of GFP or tdTom independently or together yielding various cells with three potential expression patterns (tdTom+, red; GFP+, green; tdTom+/GFP+, yellow). Recombination can also occur in postmitotic cells during G0, but only yellow cells will be obtained due to the presence of only one allele for each reporter system. (B) MADM-11 recombined cells in E12.5 Nestin-cre and Emx1cre cerebral cortices. Only a few Nestin-cre:MADM11 cells were detected predominantly in the preplate (PPL) away from the VZ. Almost all recombined Nestin-cre:MADM-11 cells were negative for the mitotic marker PH3 (blue). In contrast, the Emx1cre knock-in allele was sufficient for driving mitotic recombination in the cerebral cortices at E12.5 as evidenced by the presence of cells expressing a single reporter. Co-localization of PH3 (blue) with recombined cells was detectable in the VZ (arrows). (C) Nestin-cre:MADM-11 recombined cells in E14.5 and E17.5 cerebral cortices. By E14.5, a few mitotically recombined cells were found in the IZ and CP, but very few clones could be detected in the VZ and SVZ. In addition, the majority of recombined cells in the VZ and SVZ were largely devoid of PH3 immunolabeling. By E17.5, mitotic recombination became sufficient in the entire cerebral cortex and PH3+ recombined cells could be readily detected in both the VZ and SVZ (arrows). Scale bars: B, 30 µm; C, 60 µm.
In contrast to the Emx1cre line, Nestin-cre mediated recombination in the MADM-11 background was extremely infrequent in the cerebral cortices at E12.5, and the few recombined cells failed to co-label with PH3 in the VZ. Moreover, the few MADM-11 cells were predominantly situated in the preplate (PPL) and all of these cells expressed both GFP and tdTom, suggesting that recombination had occurred during postmitotic stages of their development (Fig. 3B). At E14.5, a few more recombined cells could be found in Nestin-cre:MADM-11 cerebral cortices, but fewer clones could be detected in the VZ/SVZ compared to IZ/CP situated cells. Moreover, the majority of VZ/SVZ recombined cells were largely devoid of PH3 immunostaining (Fig. 3C). By E17.5, recombination became sufficient in the entire cerebral cortex and PH3+ recombined cells in both VZ and SVZ regions could be readily detected (Fig. 3C, arrows). These data confirm our finding that mitotic recombination (i.e., recombination in NSCs and NPCs) only becomes sufficient during perinatal development in the specific Nestin-cre line we obtained from the Jackson laboratory.
In summary, our study unraveled an important technical issue that has significant implications for neurodevelopmental investigators using the cre-lox technology as a genetic approach. We found that a readily utilized, and the only commercially available, Nestin-cre line of mice is a poor cre-driver for directing recombination to embryonic neural stem- and progenitor- cells. Recombination commences in the embryonic brain as reported in the past, but is largely confined to postmitotic neurons such as those occupying the preplate and cortical plate of the developing cortex. We think this discrepancy previously led many investigators to assume that the high rate of recombination includes progenitors in the VZ and SVZ of the developing central nervous system. However, we find that the rate of Nestin-cre driven recombination reaches nearly 100% in neural and glial progenitors during perinatal development. Whether or not this temporal gradient of recombination efficiency is a transgenic issue in the specific line of Nestin-cre mice used in our study, or if the endogenous Nestin promoter also exhibits a similar expression pattern remains to be determined. Temporal and spatial patterns of transgene expression have inherent problems including uncontrollable random copy number and integration site of the transgene (reviewed by Haruyama et al., 2009). For example, a transgene may be integrated into a locus that contains repressor sites or one with epigenetic modifications that do not favor transcription. It is also known that dynamic alterations occur in both transcriptional and epigenetic machinery in maturing NSCs (Juliandi et al., 2010). Hence, randomly integrated transgenes may be transcriptionally repressed in stem cells, but become active upon differentiation, or vice versa, as may be the case in the Nestin-cre line tested in this study. Thus, investigators planning to use the only Nestin-cre line of transgenic mice distributed by the Jackson Laboratory should take this technical limitation into consideration prior to utilizing it in their studies.
Acknowledgments
This work was supported by NIH grant R01NS062182, a grant from the American Federation for Aging Research, and generous institutional funds to H.T.G.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Bouvier J., Cheng J.–G. (2009). Recombineering-based procedure for creating Cre/loxP conditional knockouts in the mouse. Curr. Protoc. Mol. Biol. Chapter 23, Unit 23.13 10.1002/0471142727.mb2313s85 [DOI] [PubMed] [Google Scholar]
- Branda C. S., Dymecki S. M. (2004). Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 10.1016/S1534-5807(03)00399-X [DOI] [PubMed] [Google Scholar]
- Dubois N. C., Hofmann D., Kaloulis K., Bishop J. M., Trumpp A. (2006). Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis 44, 355–360 10.1002/dvg.20226 [DOI] [PubMed] [Google Scholar]
- Gavériaux–Ruff C., Kieffer B. L. (2007). Conditional gene targeting in the mouse nervous system: Insights into brain function and diseases. Pharmacol. Ther. 113, 619–634 10.1016/j.pharmthera.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Gorski J. A., Talley T., Qiu M., Puelles L., Rubenstein J. L., Jones K. R. (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruyama N., Cho A., Kulkarni A. B. (2009). Overview: engineering transgenic constructs and mice. Curr. Protoc. Cell Biol. Chapter 19, Unit 19.10 10.1002/0471143030.cb1910s42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S., Youn Y. H., Moon H. M., Miyamichi K., Zong H., Wynshaw–Boris A., Luo L. (2010). Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709 10.1016/j.neuron.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka F., Ishibashi M., Taki W., Hashimoto N., Nakanishi S., Kageyama R. (1999). Ectopic expression of the bHLH gene Math1 disturbs neural development. Eur. J. Neurosci. 11, 2582–2588 10.1046/j.1460-9568.1999.00699.x [DOI] [PubMed] [Google Scholar]
- Juliandi B., Abematsu M., Nakashima K. (2010). Epigenetic regulation in neural stem cell differentiation. Dev. Growth Differ. 52, 493–504 10.1111/j.1440-169X.2010.01175.x [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R.et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2007). Staining whole mouse embryos for β-galactosidase (lacZ) activity. CSH Protoc. pdb.prot4725 10.1101/pdb.prot4725 [DOI] [PubMed] [Google Scholar]
- Petersen P. H., Zou K., Hwang J. K., Jan Y. N., Zhong W. (2002). Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 10.1038/nature01124 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., Schütz G. (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Trumpp A., Depew M. J., Rubenstein J. L., Bishop J. M., Martin G. R. (1999). Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 13, 3136–3148 10.1101/gad.13.23.3136 [DOI] [PMC free article] [PubMed] [Google Scholar]