Summary
The ubiquitin proteasome system (UPS) is known to be responsible for the rapid turnover of many transcription factors, where half-life is held to be critical for regulation of transcriptional activity. However, the stability of key transcriptional regulators of development is often very poorly characterised. Neurogenin 3 (Ngn3) is a basic helix–loop–helix transcription factor that plays a central role in specification and differentiation of endocrine cells of the pancreas and gut, as well as spermatogonia and regions of the brain. Here we demonstrate that Ngn3 protein stability is regulated by the ubiquitin proteasome system and that Ngn3 can be ubiquitylated on lysines, the N-terminus and, highly unusually, on non-canonical residues including cysteines and serines/threonines. Rapid turnover of Ngn3 is regulated both by binding to its heterodimeric partner E protein and by the presence of cdk inhibitors. We show that protein half-life does appear to regulate the activity of Ngn3 in vivo, but, unlike the related transcription factor c-myc, ubiquitylation on canonical sites is not a requirement for transcriptional activity of Ngn3. Hence, we characterise an important new level of Ngn3 post-translational control, which may regulate its transcriptional activity.
Key words: Ubiquitylation, Neurogenin, Ngn3, Xenopus, bHLH, Proteolysis
Introduction
Development of the early embryo requires dynamic regulation of key regulatory molecules that drive and co-ordinate developmental processes. In particular, master regulatory transcription factors are often subject to stringent controls both at the level of their own transcription and by post-translational mechanisms. For example, ubiquitin-mediated proteolysis has been shown to be a requirement for transcriptional activity of the basic helix–loop–helix (bHLH) transcription factor c-myc (von der Lehr et al., 2003; Kim et al., 2003). Members of a related class of bHLH transcription factors include the proneural Neurogenin (Ngn) transcription factor family, which controls processes such as neuronal differentiation, endocrine pancreas formation and regulation of spermatogenesis (Massari and Murre, 2000; Yoshida et al., 2004).
While regulation of the level of proneural protein transcripts has been well documented, their post-translational regulation is much less well understood. However, recent studies have begun to shed light on several post-translational mechanisms that control Ngn2, a master regulator of neurogenesis. In particular, it has been demonstrated that Ngn2 is a very short-lived protein that is rapidly degraded by the ubiquitin proteasome system (UPS) (Vosper et al., 2007). This method of regulated protein degradation requires covalent linkages to the small ubiquitin (Ub) protein via a cascade of ubiquitin-conjugating and ubiquitin ligase enzymes: once an appropriate chain of at least 4 Ub moieties is attached to the substrate protein, it can be targeted for destruction by the proteasome (Thrower et al., 2000).
Ubiquitylation has classically been described to occur via an isopeptide linkage to substrate lysines (Hershko and Ciechanover, 1998), while ubiquitylation has also been demonstrated to occur by direct linkage to the free N-terminus of about a dozen proteins (Ciechanover and Ben-Saadon, 2004). These so-called canonical sites of ubiquitylation were originally thought to be the sole sites of Ub attachment during physiological ubiquitylation. However, recently it has become clear that non-canonical ubiquitylation can occur on a small number of proteins via thioester and ester linkage to substrate cysteines and/or serines and threonines, often using viral ubiquitin ligase enzymes and sometimes resulting in a change of substrate sub-cellular localisation (Cadwell and Coscoy, 2005; Carvalho et al., 2007; Williams et al., 2007). We have demonstrated recently that, highly unusually, Ngn2 can be ubiquitylated on non-canonical sites by cellular ubiquitin ligases and that these linkages alone can target Ngn2 for destruction (Vosper et al., 2009; McDowell et al., 2010). While ubiquitylation on cysteines can occur in interphase or mitosis of the cell cycle, cysteine ubiquitylation of Ngn2 is limiting for degradation only in mitosis (Vosper et al., 2009).
The related proneural transcription factor Neurogenin 3 (Ngn3) is required for cells to adopt an endocrine rather than an exocrine cell fate during development of both the pancreas and the gut (Schwitzgebel et al., 2000; Bertrand et al., 2002; Gu et al., 2002; Jenny et al., 2002). Tamoxifen-inducible Ngn3, under the control of the pdx1 promoter, has been used in mice to regulate Ngn3 activity at increasing developmental stages, and these studies show that, at defined developmental times, Ngn3 can drive differentiation towards all endocrine pancreatic lineages (Johansson et al., 2007). Moreover, at the appropriate stage, high levels of Ngn3 cause cells to exit the cell cycle and promote formation of insulin-expressing cells at the expense of exocrine pancreas (Johansson et al., 2007), indicating that manipulation of Ngn3 levels and/or activity may be highly beneficial for differentiation or transdifferentiation of cells to treat deficiencies of the endocrine pancreas such as diabetes.
Despite its central role in pancreatic development, the post-translational regulation of Ngn3 has barely been investigated. Ngn3 is 89% identical at the amino acid level to Ngn2 in the bHLH region, but only 43% similar outside it. Therefore, regulation of proteolytic degradation of Ngn3 may differ significantly from that of Ngn2. With this in mind, we set out to characterise the regulation of Ngn3 degradation using the highly tractable biochemical system afforded by the use of Xenopus egg extracts, identifying similarities and differences in regulation of ubiquitylation and proteolysis between the two proteins.
Here, we show that Ngn3 has a very short half-life and is degraded by ubiquitin-mediated proteolysis. We show that Ngn3 joins the very small number of proteins known to undergo extensive non-canonical ubiquitylation on cysteine, serine and/or threonine residues. However, in contrast to Ngn2, this non-canonical ubiquitylation does not contribute significantly to Ngn3's rapid turnover. We show that Ngn3 half-life is extended by binding to its heterodimeric E protein partner. In addition, Ngn3 is stabilised by the presence of a Cip/Kip family cyclin-dependent kinase inhibitor, indicating cell cycle regulation of proteolytic turnover. It has previously been proposed that ubiquitin-mediated proteolysis may be a requirement for activity of some transcription factors (Kim et al., 2003; von der Lehr et al., 2003; Collins and Tansey, 2006). Finally, we show that canonical ubiquitylation is not absolutely required for Ngn3's ability to drive transcription, but it may play a role in regulating the level of Ngn3 transcriptional activity in vivo.
Results
Ngn3 is an unstable protein degraded by the UPS
Cytoplasmic extract prepared from activated Xenopus eggs by centrifugation contain all the ubiquitin–proteasome system (UPS) components required to target Ngn2 for ubiquitylation and degradation, and provide a convenient system to investigate sites of ubiquitylation of targeted proteins. To determine whether Ngn3 was degraded by the UPS in these extracts, we prepared 35S-methionine-labeled in vitro translated Ngn3 protein in rabbit reticulocyte lysate (IVT Ngn3). IVT Ngn3 was incubated in interphase egg extract with and without the synthetic peptide proteasome inhibitor Mg132, at a dose previously shown to stabilise Ngn2 in egg extract (Vosper et al., 2007), and samples removed at increasing time-points for analysis by SDS Polyacrylamide Gel Electrophoresis (SDS PAGE).
Ngn3 is unstable in Xenopus egg extracts, with a half-life of 14.4±1.7 minutes in the absence of Mg132 (Fig. 1A), even shorter than the half-life of Ngn2 (approximately 22 minutes (Vosper et al., 2007)). Ngn3 is significantly stabilised by Mg132, averaging a half-life of 66.0±4.5 minutes (Fig. 1). Failure of Mg132 to completely stabilise Ngn3 may reflect only partial inhibition of the proteasome, or else could indicate that Ngn3 can also be degraded by non-proteasomal pathways. We also see retardation of Ngn3 protein after incubation in egg extract (compare t = 0 and t = 15 time-points). Ngn2 is highly phosphorylated in egg extracts (Ali et al., 2011; Hindley et al., 2012) and it is likely that Ngn3 is similarly modified. Xenopus interphase egg extracts can be driven into a stable mitotic state by addition of CycBΔ90, a non-degradable form of cyclin B (Glotzer et al., 1991), and cell cycle stage of the extract significantly alters half-life in a number of proteins including Ngn2 (Vosper et al., 2007). We determined the relative half-life of Ngn3 in mitotic extract using the method described above. Unlike Ngn2, whose half-life is significantly shortened in mitosis, the stability of Ngn3 in mitotic extract is similar to that in interphase, with a half-life of 15.0±1.4 minutes in the absence of Mg132 and 70.0±2.0 minutes in the presence of Mg132 (Fig. 1).
Fig. 1. Ngn3 is degraded by ubiquitin-mediated proteolysis in interphase and mitosis.
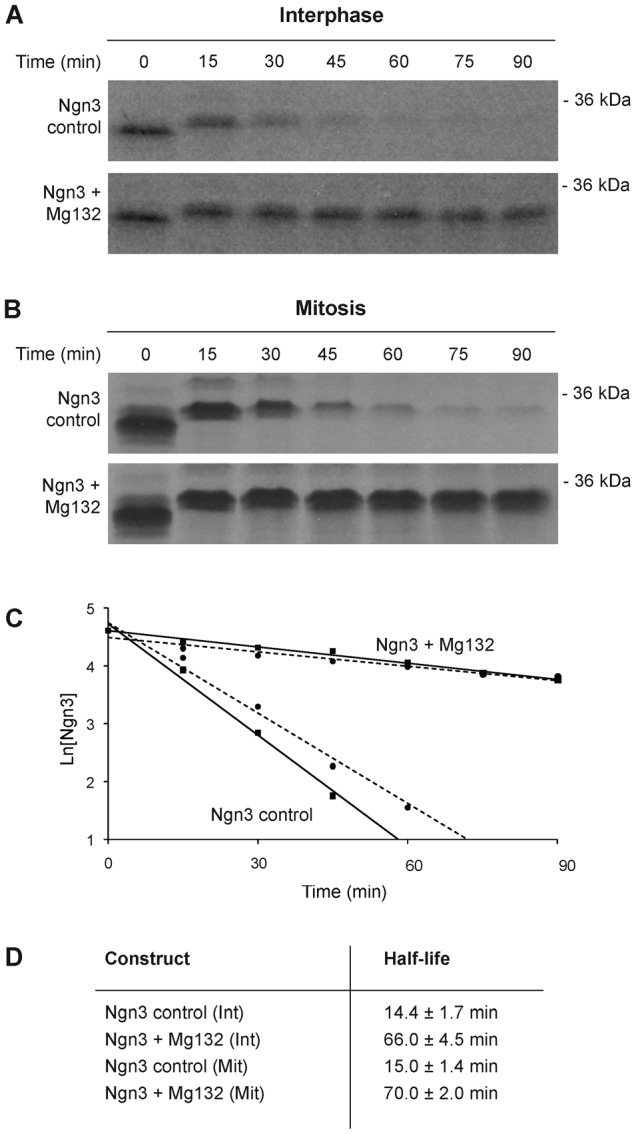
Degradation assays were performed in interphase (A) and mitotic (B) Xenopus egg extract using 35S-IVT Ngn3 in either the presence or absence of 100 µM Mg132 proteasome inhibitor. Samples were run on SDS-PAGE gels, analysed by autoradiography (A,B) and quantified by phosphorimaging (C). Degradation half-lives were calculated using first-order rate kinetics (D). In C, the solid line shows degradation in interphase extracts, and the dotted line shows degradation in mitotic extracts.
Identification of sites of ubiquitylation in Ngn3
The vast majority of ubiquitylated cellular proteins undergo modification of lysine residues via an isopeptide linkage to Ub. A much smaller number of proteins have been shown to be ubiquitylated directly onto the N-terminus, and only a handful have been demonstrated to undergo ubiquitylation on non-canonical sites including cysteine, serine and/or threonine (Ciechanover and Ben-Saadon, 2004; Carvalho et al., 2007; Cadwell and Coscoy, 2005; Williams et al., 2007). Perhaps uniquely, Ngn2 has been shown to use all three of these modes of ubiquitylation to direct protein degradation (McDowell et al., 2010). Therefore, in this study, we set out to determine the sites of ubiquitylation that can target Ngn3 for UPS-mediated degradation in interphase egg extracts. To this end, mutant forms of Ngn3 were created to remove putative sites of canonical and non-canonical ubiquitylation.
To block ubiquitylation on lysine residues we generated a mutant form of Ngn3 in which all 7 lysines are mutated to arginine (Ngn3KO). Co-translational N-terminal acetylation is incompatible with N-terminal ubiquitylation (Kuo et al., 2004). The N-terminal acetylation prediction tool TermiNator (available from the CNRS) predicts that the likelihood for co-translational acetylation of wild-type Ngn3 on its N-terminus is 22%, so there is a good likelihood that this site would be available for ubiquitylation. To prevent this, we mutated the first 5 amino acids to a sequence that is predicted to be N-terminally acetylated with an 83% likelihood of producing acetylated Ngn3 (AcNgn3). A similar mutational strategy was used to block N-terminal ubiquitylation on Ngn2 (Vosper et al., 2009). We then combined both types of mutation to generate AcNgn3KO, which contains no canonical sites of ubiquitylation. We then tested the ability of these mutated proteins to be ubiquitylated.
We have previously shown that wild-type Ngn2 is ubiquitylated on cysteines even when lysines are present (Vosper et al., 2009). In the present study, we set out to determine whether wild-type Ngn3 is also ubiquitylated on non-canonical residues and, if so, whether this modification occurs in the native protein or only in the absence of lysines. To this end, His-tagged ubiquitin precipitation assays were performed in interphase egg extract on wild-type Ngn3 and mutants thereof, as described above. IVT Ngn3 protein was incubated in interphase egg extract along with His-tagged ubiquitin or untagged ubiquitin, as a negative control, in the presence of the proteasome inhibitor Mg132. After ubiquitylation was allowed to occur, ubiquitylated proteins were isolated by binding to Ni-NTA agarose beads, and washed in buffer containing 8M urea to remove non-covalently bound proteins. Ubiquitylated proteins were then eluted into SDS PAGE loading buffer under either non-reducing conditions (pH 6.8) designed to retain weaker thioester/ester Ub linkages or high pH, highly reducing conditions (10% (v/v) β-mercaptoethanol, pH 10.5), which should both reduce thioester bonds and hydrolyse ester linkages efficiently. All samples were heated at 95°C for 5 minutes prior to separation by SDS PAGE.
Ngn3 becomes extensively ubiquitylated after incubation in interphase extract, resulting in a ladder of poly-Ub forms of the protein with retarded migration on SDS PAGE, the most prominent rapidly migrating form running higher than the approx. 35 KDa of the unconjugated protein (Fig. 2A,B). More highly conjugated forms run as a ladder rising up the gel with the most poly-ubiquitylated forms running together above the 70 kDa molecular weight marker. In contrast, no Ngn3 is pulled down using untagged Ub in this assay (data not shown), demonstrating that all protein detected is a result of covalent attachment to Ub. AcNgn3 shows a similar pattern of ubiquitylation to that of the wild-type protein (Fig. 2), with a poly-Ub ladder still rising to the top of the gel. However, the lysineless variants Ngn3KO and AcNgn3KO show both a ladder of poly-Ub forms and a prominent band running at approximately 35 kDa, the size at which native unconjugated Ngn3 migrates on SDS PAGE (Fig. 2A,C, arrow). Highly reducing conditions have little effect on Ub ladders of Ngn3 and AcNgn3 (Fig. 2C compared to Fig. 2B) demonstrating covalent ubiquitylation. In contrast, ubiquitylated forms of lysineless variants Ngn3KO and AcNgn3KO are essentially lost in reducing conditions (see loss of retarded bands between 35KDa and the top of the gel in these samples), although the prominent band of unconjugated Ngn3 (Fig. 2C, arrow) remains. These results are consistent with the predominant Ub linkages in the wild-type protein occurring via stable isopeptide bonds to lysines, whereas Ub on lysineless variants occurs via labile ester and thioester linkages to non-canonical Ub acceptor sites, disrupted by high pH and reducing conditions. However, we noted that ubiquitylated Ngn3 and AcNgn3 do release a detectable band of unconjugated protein running at approx. 35 kDa, particularly after reducing treatment (Fig. 2C, arrow), indicating that a subset of even these proteins that retain lysines are ubiquitylated via labile linkages. These labile linkages are likely to include cysteine residues (Vosper et al., 2007). To test this, we generated further Ngn3 mutants in which cysteines were mutated to alanines either in a wild-type background to generate Ngn3CO or in the presence of additional mutations of canonical ubiquitylation sites to generate Ngn3KOCO and AcNgn3KOCO. We then tested the ability of each mutant to be ubiquitylated.
Fig. 2. Ubiquitylation of Ngn3 and mutants on canonical and non-canonical sites.
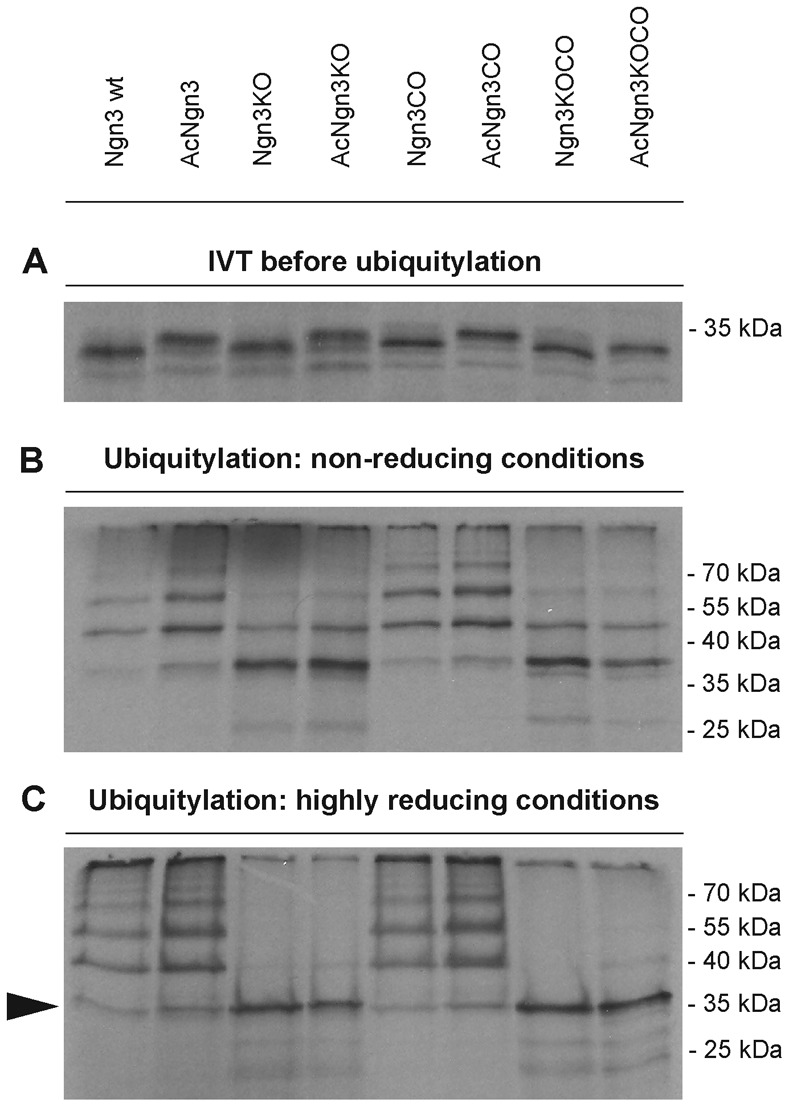
Ubiquitylation assays were performed in interphase Xenopus egg extract using 35S-labeled IVT Ngn3 and mutants as labelled, in the presence of His-tagged ubiquitin (B,C). IVT protein levels were normalised, as demonstrated by the levels of starting material shown in A. Proteins were eluted from NTA-agarose beads using either non-reducing conditions (pH 6.8) (B) or highly reducing conditions (pH 10, 10% (v/v) β-mercaptoethanol) (C).
The patterns of ubiquitylated forms of Ngn3CO and Ac3Ngn3CO were indistinguishable from those of Ngn3 and AcNgn3 in either non-reducing or reducing conditions (Fig. 2). Perhaps more surprisingly, there is also no difference in the pattern of ubiquitylation of Ngn3KO compared to Ngn3KOCO or of AcNgn3KO compared to AcNgn3KOCO; all these samples show poly-ubiquitylated forms in non-reducing conditions releasing unconjugated Ngn3 protein (arrow) under reducing conditions. These results indicate again that non-canonical ubiquitylation can occur on Ngn3, but that these Ub events can occur readily on serines/threonines, regardless of the presence or absence of cysteines.
Thus, non-canonical ubiquitylation of Ngn3 occurs extensively in the absence of lysines, but less prominently when lysines are available. To what extent do canonical and non-canonical ubiquitylation at distinct types of acceptor residues contribute to the rapid degradation of Ngn3? To test this we undertook degradation assays of Ngn3 and of cysteineless Ngn3 derivatives, with or without mutation of canonical ubiquitylation sites, first in interphase egg extract.
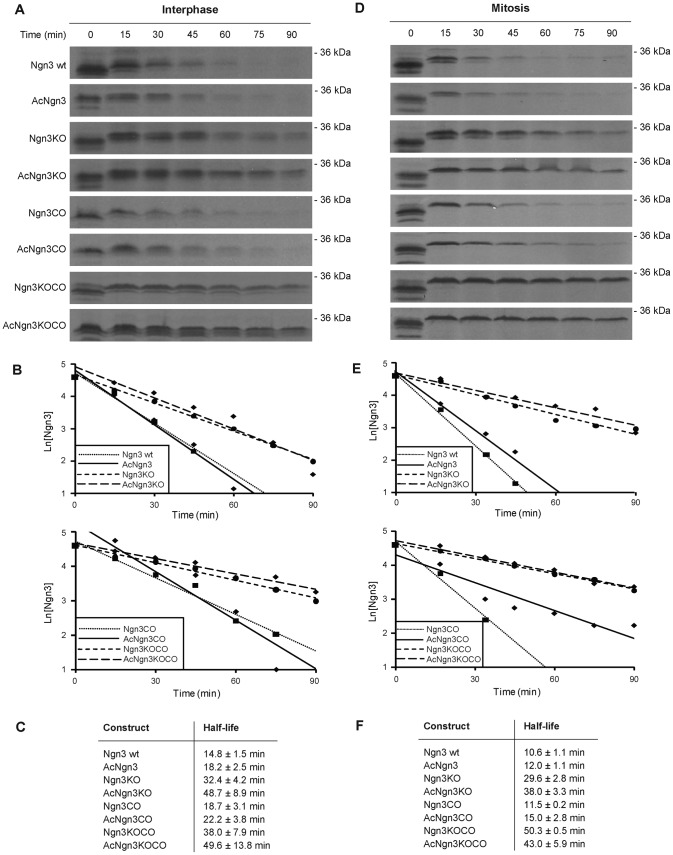
In interphase egg extract, mutation of lysines approximately doubled Ngn3 protein half-life (Ngn3 14.8±1.5 minutes, Ngn3KO 32.4±4.2 minutes) (Fig. 3A–C). Blocking N-terminal ubiquitylation alone had only a very small effect on half-life (AcNgn3 18.2±2.5 minutes), indicating that the N-terminus is not readily used as a site of ubiquitylation to target for destruction in the otherwise native protein. However, we saw that the half-life of AcNgn3KO (48.7±8.9 minutes) was significantly longer than that of Ngn3KO (32.4±4.2 minutes), indicating that N-terminal ubiquitylation is able to direct Ngn3KO degradation when lysines are no longer available for ubiquitylation.
Fig. 3. Identification of Ngn3 ubiquitylation sites involved in degradation.
Degradation assays were performed in interphase (A,B,C) and mitotic (D,E,F) Xenopus egg extract using 35S-IVT Ngn3 and mutants (including N-terminal acetylation (Ac), lysine to arginine (KO), and cysteine to alanines (CO) mutations). Samples were run on SDS-PAGE gels, analysed by autoradiography (A,D) and quantified by phosphorimaging (B,E). Degradation half-lives were calculated using first-order rate kinetics (C,F).
Nevertheless, AcNgn3KO, which cannot be ubiquitylated on canonical ubiquitylation sites, is still unstable in interphase (and less stable than Ngn3 incubated in the presence of Mg132) (Fig. 1), indicating that non-canonical ubiquitylation can target Ngn3 for degradation. To test the contribution of ubiquitylation on cysteine residues to the degradation of Ngn3, we tested the half-life of various mutants of Ngn3 in which all 3 cysteines had been mutated to alanines (Ngn3CO mutants).
Ngn3CO was found to have a similar half-life to wild-type Ngn3 (Ngn3CO 18.7±3.1 minutes, Ngn3 14.8±1.5 minutes). Likewise, AcNgn3CO (22.2±3.8 minutes) has a similar half-life to AcNgn3 (18.2±2.5 minutes). We also saw that, even when lysines of Ngn3 have been mutated, further mutation of cysteines has little effect on half-life. Ngn3KOCO (half-life 38.0±7.9 minutes) and AcNgn3KOCO (half-life 49.6±13.8 minutes) are not significantly stabilised in comparison with their counterparts with cysteines intact (Ngn3KO 32.4±4.2 minutes, AcNgn3KO 48.7±8.9 minutes). Thus, ubiquitylation on cysteines of Ngn3 appears not to contribute significantly to its degradation in interphase. Moreover, when the lysines are mutated and the N-terminus is blocked, it is clear that potential ubiquitylation on cysteines alone is not sufficient to bring about efficient degradation of Ngn3.
Cysteine ubiquitylation plays a more prominent role in driving degradation of Ngn2 in mitosis as compared to interphase (Vosper et al., 2009). To determine whether non-canonical ubiquitylation of Ngn3 was contributing to rapid protein turnover in mitosis, we also assayed the half-lives of lysine and cysteine mutants of Ngn3 in mitotic egg extract (Fig. 3D–F). As in interphase, ubiquitylation on the N-terminus was not seen to be a significant contributor to ubiquitin-mediated degradation (Ngn3 10.6±1.1 minutes, AcNgn3 12.0±1.1 minutes), except in the absence of internal lysines (Ngn3KO 29.3±2.8 minutes, AcNgn3KO 38.0±3.3 minutes), when it appears that N-terminal ubiquitylation can target Ngn3KO for degradation. The mutation of cysteines to alanines does not result in significant stabilisation of the otherwise native Ngn3 protein in mitotic extract (Ngn3 10.6±1.1 minutes, Ngn3CO 11.5±0.2 minutes). These half-life data suggest that the majority of ubiquitin-mediated degradation of Ngn3 in mitosis, as in interphase, can be accounted for by ubiquitylation at internal lysines.
Thus, in contrast to Ngn2, which was seen to require cysteines for its rapid rate of degradation in mitosis (Vosper et al., 2009), ubiquitylation of cysteines appears to play no role in the rapid turnover of Ngn3 protein when lysines are available for ubiquitylation. However, in the absence of viable canonical ubiquitylation sites, the half-life data presented in Fig. 3F suggest that ubiquitylation on internal cysteines is capable of targeting Ngn3 for degradation in mitosis. To illustrate this, in the absence of internal lysines, Ngn3KO half-life is 29.3±2.8 minutes, while the further mutation of cysteines to alanines results in an increased half-life for Ngn3KOCO of 50.3±0.5 minutes.
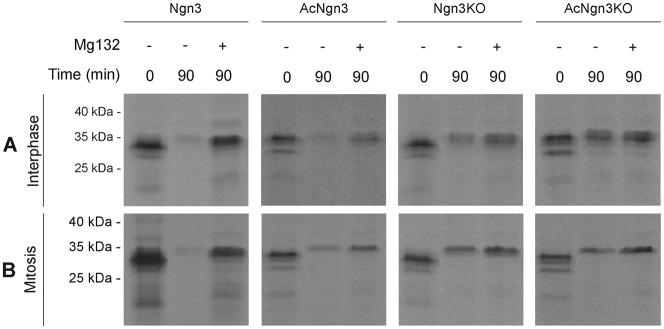
To confirm that the degradation we are seeing in interphase and mitosis is due to the proteasome, we compared degradation of Ngn3, AcNgn3, Ngn3KO and AcNgn3KO in the presence and absence of Mg132 (Fig. 4). In each case, the protein appears stabilised in the presence of Mg132 and, as expected, stabilisation is more dramatic for the Ngn3 constructs containing lysines that have the fastest degradation rate.
Fig. 4. Forms of Ngn3 lacking lysines are stabilised by Mg132.
Degradation assays at a fixed point of 90 minutes were performed in interphase (A) and mitotic (B) Xenopus egg extract using 35S-IVT Ngn3 and mutants thereof, as labelled, in either the presence or absence of 100 µM Mg132 proteasome inhibitor. Samples were run on SDS-PAGE gels, analysed by autoradiography (A,B).
Taken together, these results demonstrate that ubiquitylation and proteasomal degradation can occur via both canonical and non-canonical cysteine sites in Ngn3, even in the wild-type protein, but only canonical sites of ubiquitylation, primarily lysines, have a significant effect on Ngn3 half-life. This is in contrast to Ngn2, which requires ubiquitylation of both canonical and non-canonical cysteine sites for rapid turnover of the wild-type protein. Having established both similarities and differences in regulation of sites of ubiquitylation and their effect on degradation between Ngn2 and Ngn3 as described above, we went on to investigate further potential mechanisms of control of Ngn3 proteolytic degradation.
The stability of proneural bHLH transcription factors such as Ngn2 and MASH1 has been shown to be additionally regulated by association with their heterodimerisation E protein partners, E12 or E47 (Vosper et al., 2007). We therefore investigated whether Ngn3 protein is stabilised by the addition of E12. We mixed IVT Ngn3 and E12 in interphase egg extracts and investigated Ngn3 stability in our degradation assay. Binding to E12 resulted in an approximately 4-fold increase in Ngn3 half-life (Fig. 5A,C).
Fig. 5. Ngn3 is stabilised by E12 and Xic1 in interphase Xenopus egg extract.
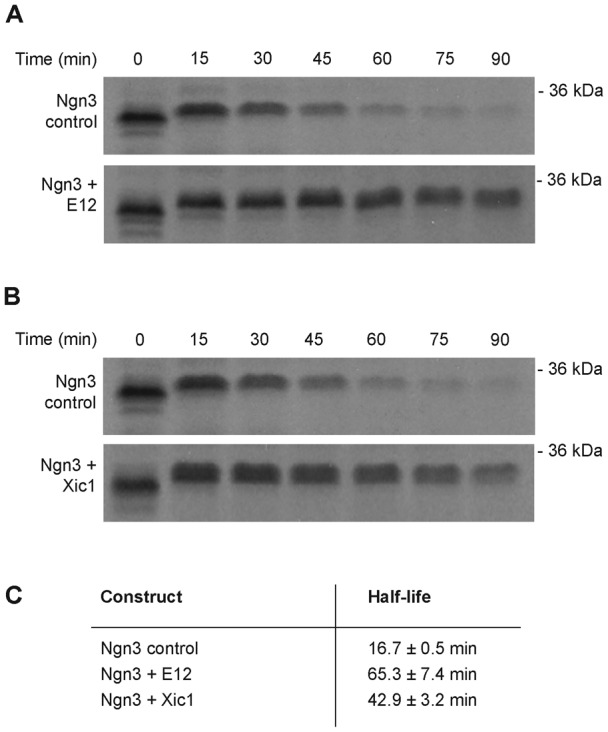
Degradation assays were performed in the presence of either (A) IVT E12 (at a 1:1 volume ratio to Ngn3) or IVT mix alone, or (B) 15 µM Xic1-MBP or 15 µM MBP as a control. Samples were run on SDS-PAGE gels, analysed by autoradiography (A,B) and quantified by phosphorimaging. Degradation half-lives were calculated using first-order rate kinetics (C).
Cyclin-dependent kinase inhibitors (cdkis) have also been shown to be required for Ngn2 function, independent of their ability to inhibit the cell cycle and overall cdk kinase activity. In vivo, cdkis promote Ngn2 protein accumulation, and may act by inhibiting ubiquitin-mediated proteolysis (Vernon et al., 2003; Nguyen et al., 2006). We investigated whether the Xenopus cdki, Xic1, which shows homology to the mammalian cdkis p21Cip1, p27Kip1 and p57Kip2, was able to stabilise Ngn3 protein directly in egg extract. Addition of recombinant Xic1 protein to egg extract resulted in a 2.5-fold increase in the half-life of IVT Ngn3 (Fig. 5B,C).
It has been suggested that ubiquitylation is important for activity of some transcription factors, where their rapid proteolytic turnover may be a prerequisite for promoter activity (Kim et al., 2003; von der Lehr et al., 2003; Collins and Tansey, 2006). Conversely, one might hypothesise that increasing the half-life of this highly unstable protein by inhibiting UPS-mediated turnover might enhance Ngn3's transcriptional activity. We investigated whether ubiquitylation of Ngn3 on canonical sites, the mode of Ngn3 ubiquitylation that regulates its stability as demonstrated above, was absolutely required for Ngn3 activity in vivo in developing Xenopus embryos. DNA binding is a prerequisite for transcriptional activation, so first we investigated the ability of AcNgn3KO to bind to E box DNA by electrophoretic mobility shift assay (EMSA) (Fig. 6).
Fig. 6. AcNgn3KO binds DNA less tightly than Ngn3.
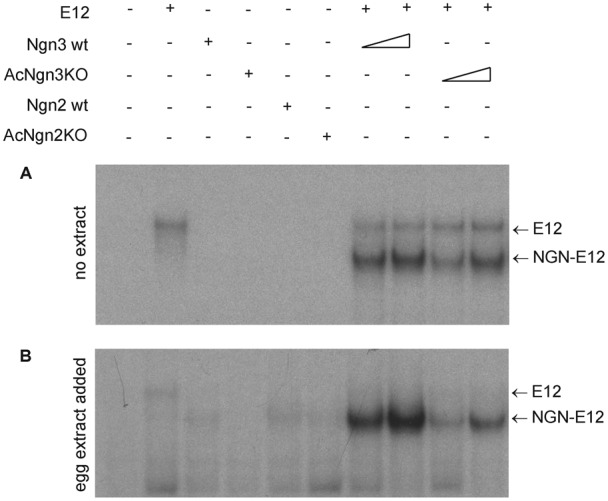
Ngn3, AcNgn3KO, Ngn2 and AcNgn2KO binding to E box-containing probe, with or without added E protein as indicated, were assayed by EMSA either in buffer alone (A) or in the presence of interphase egg extract (B), triangle representing two different amounts of each IVT (see Materials and Methods).
IVT Ngn3 and AcNgn3KO at two different concentrations were tested for E box DNA binding ability by EMSA. Ngn3 binds strongly to E box DNA probe dependent on the presence of E protein, but binding of AcNgn3KO appears somewhat less efficient (Fig. 6A). This likely reflects an intrinsic difference in the ability of the mutated AcNgn3KO protein to bind to DNA, rather than ubiquitylation or other potential modification of lysines, as this semi-purified system containing only IVT protein and radiolabelled DNA probe. To recapitulate the environment of the Xenopus egg and embryo, where ubiquitylation of lysines and the N-terminus occurs, we then conducted a similar EMSA analysis in the presence of interphase egg extract, which was permissive for ubiquitylation but where degradation had been prevented by addition of Mg132. AcNgn3KO showed substantially weaker binding to DNA than Ngn3, indicating that lysines play a greater role in contributing to stable DNA association in the presence of egg extract when these residues could be modified (Fig. 6B). Thus, in Xenopus cytoplasm AcNgn3KO has a considerably longer half-life than Ngn3 (Fig. 1), but has a significantly lower affinity for E-box-containing DNA (Fig. 6). If both strong DNA binding and ubiquitylation are required for transcriptional activity, one might predict that AcNgn3KO would show a significantly reduced capacity to drive cellular differentiation. However, conversely, an increased Ngn3 half-life could promote the ability of AcNgn3KO to drive transcription by increasing promoter occupancy time and this may compensate for weaker DNA binding. To test this, we compared the ability of Ngn3 and AcNgn3KO to drive a downstream transcriptional program in vivo, in Xenopus embryos.
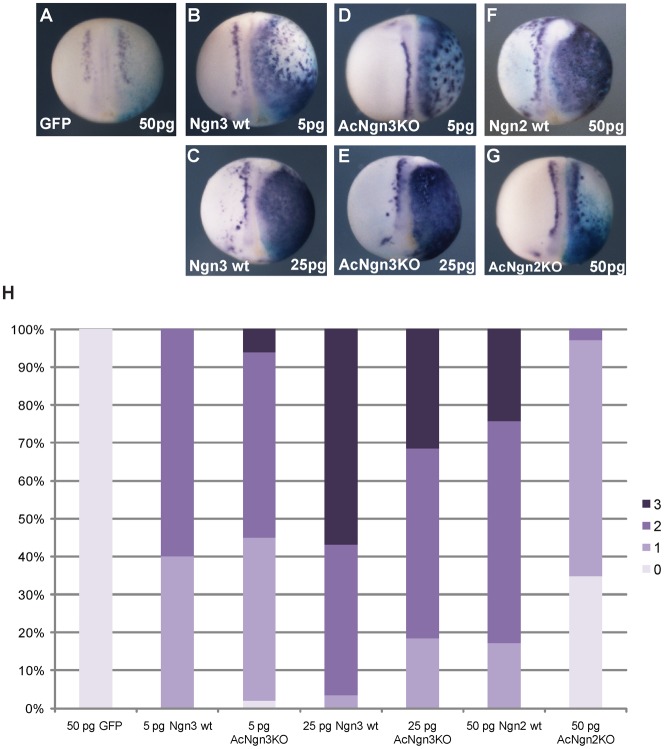
Ngn3 upregulates a large number of direct downstream targets to control differentiation of endocrine cells of the gut and pancreas, among other tissues (Gradwohl et al., 2000; Juhl et al., 2008). These differentiation pathways have several key downstream effectors in common with those required to induce neurogenesis by Ngn2, such as NeuroD and Delta. Indeed, experimentally, Ngn3 overexpression in early Xenopus embryos results in upregulation of NeuroD and Delta, converting epidermis to a neural fate (Huang et al., 2000) (Fig. 7; data not shown), providing a convenient assay for Ngn3 transcriptional activity in vivo. Using this assay, we compared the abilities of Ngn3 and AcNgn3KO to induce ectopic neurogenesis in Xenopus after mRNA micro-injection, using in situ hybridisation for neural β tubulin to detect ectopic differentiated neurons.
Fig. 7. Ngn3 and AcNgn3KO show similar activity in vivo.
1 cell of a 2 cell-stage Xenopus embryo was injected with mRNA encoding GFP (as a control, A), Ngn3 (B,C), AcNgn3KO (D,E), Ngn2 (F) or AcNgn2KO (G) in the amounts as indicated, along with βgal mRNA as a lineage tracer. Expression of neural β tubulin at stage 15 was assayed by in situ hybridisation. A semi-quantitative scoring method for neural β tubulin expression was devised (H), where 0 = no ectopic neurogenesis, 1 = low levels of ectopic neurogenesis, 2 = moderate ectopic neurogenesis and 3 = extensive ectopic neurogenesis (number of embryos scored per condition: between 49 and 69).
Ngn3 induced formation of ectopic primary neurons in a dose-dependent manner, where 25 pg micro-injected mRNA produced substantially more extensive neurogenesis than 5 pg. Surprisingly, 5 pg AcNgn3KO showed very similar activity to 5 pg Ngn3, and at 25 pg, AcNgn3KO was only marginally less active than the wild-type protein. This contrasts with the effect of a lysineless mutant of Ngn2 (Vosper et al., 2009); AcNgn2KO showed very low levels of ectopic neurogenesis compared to its wild-type counterpart, Ngn2. From these results, it is clear that rapid ubiquitin-mediated proteolysis is not a prerequisite for Ngn3 transcriptional activity, unlike the related bHLH protein c-myc (Kim et al., 2003; von der Lehr et al., 2003; Collins and Tansey, 2006). Moreover, these results indicate that the increased half-life of AcNgn3KO may compensate for reduced DNA binding, allowing the protein to retain considerable activity in vivo.
Discussion
Xenopus egg extracts provide an ideal system for detailed biochemical analysis of mechanisms regulating Ngn3 degradation, as it contains stockpiles of E1, E2 and E3 enzymes and proteasomal components required for its ubiquitin-mediated destruction. Xenopus also provides a versatile system for measuring Ngn3 activity; while found in the hypothalamus and hippocampus (Sommer et al., 1996; Simon-Areces et al., 2010), Ngn3 is not endogenously expressed in most tissues of the brain during development. However, when exogenously expressed, Ngn3 can activate neuronal differentiation in Xenopus ectoderm primed for this fate in a dose-dependent manner by activating NeuroD (Huang et al., 2000) (data not shown), on a par with neuronal activation by Ngn2. Hence, overexpression by mRNA microinjection provides a convenient system to semi-quantitatively monitor Ngn3's transcriptional activity. Here we have used both Xenopus egg extracts and expression in developing Xenopus embryos to undertake a detailed study determining modes of Ngn3 ubiquitylation and their relationship with rates of degradation. We have also investigated the role of co-factors in control of Ngn3 protein half-life and the relationship between ubiquitylation and Ngn3 transcriptional activity.
The results presented here suggest that, in both interphase and mitosis, the N-terminus of Ngn3 is a viable site of ubiquitylation in the absence of lysines, but that it is not significantly involved in ubiquitin-mediated degradation of wild-type Ngn3. Our results also show that Ngn3 can join the very small number of identified proteins that can undergo non-canonical ubiquitylation via labile thioester or ester bonds between ubiquitin and residues such as cysteines, serines, or threonines.
Although Ngn2 and Ngn3 share important downstream targets, and although they are both regulated by the UPS, our results reveal major differences between the ubiquitin-mediated regulation of Ngn2 and Ngn3. We previously saw that wild-type Ngn2 was targeted for degradation in mitosis by ubiquitylation not only on internal lysines and the N-terminus, but also on internal cysteines and potentially also serines and/or threonines (Vosper et al., 2009; McDowell et al., 2010). Unlike Ngn2, we see that Ngn3 is targeted for proteasome-mediated degradation overwhelmingly by ubiquitylation at internal lysines (Fig. 3) in interphase and mitosis. One reason for this difference between Ngn2 and Ngn3 may be that Ngn3 has only 3 cysteines, compared to 7 cysteines in Ngn2. These experiments do, however, show evidence of ubiquitylation on sites other than lysines, cysteines, and the N-terminus (most likely on serines/threonines) of Ngn3 leading to degradation, as the mutation of lysines, cysteines, and N-terminus (resulting in AcNgn3KOCO) is less stabilising than the addition of proteasome inhibitor Mg132 (Figs 1, 2).
These data also point to a significant promiscuity in the ubiquitylation machinery that results in Ub moieties being passed to any available acceptor in the absence of available lysines. Sites of ubiquitylation need to be near unstructured protein regions to target for proteasome-mediated unfolding and degradation (Prakash et al., 2004). It seems likely that Ngn3, like its relative Ngn1 (Aguado-Llera et al., 2010), may be largely unstructured. This could potentially give considerable flexibility in the use of different sites of ubiquitylation to target for destruction, and provide few intrinsic structural constraints for specifying precise sites of ubiquitylation.
Boosting transcript levels of Ngn3, as a central regulator of endocrine pancreas formation, has been extensively investigated as a means of potentiating islet formation in cell replacement therapies for diabetes (for a review, see Miyatsuka et al., 2008). However, enhancing protein activity post-translationally may be more amenable to practical manipulation by extrinsic factors than introduction or overexpression of the Ngn3 gene, when designing stem cell differentiation protocols. A full understanding of the relationship between Ngn3 protein stability and activity will be essential for this. We see that Ngn3 is ubiquitylated on both canonical and non-canonical ubiquitin acceptor sites. However, our results indicate that non-canonical ubiquitylation plays a minimal role in rapid degradation of the native Ngn3 protein. A mutant Ngn3 protein, in which all lysines have been mutated, shows much reduced DNA binding, yet retains substantial activity in vivo, indicating that increased protein half-life can promote transcriptional activity of Ngn3 even when its DNA binding ability is compromised. In addition, Ngn3 half-life can be increased by cdki expression.
Taken together, our results could have significant implications for control of Ngn3 availability and activity for in vitro manipulation of stem cell fate; our results indicate that increasing half-life of Ngn3 in cells might be achieved either by increasing cdki expression, which would have the additional effect of slowing or stopping cell cycling, and/or by identification and manipulation of the E3 ligase(s) that target Ngn3 for ubiquitylation.
The findings presented here, along with other recent reports of non-canonical ubiquitylation, indicate that ubiquitin-mediated protein turnover is far more complex, and the ubiquitylation machinery far more flexible, than initially recognised. Although Ngn2 and Ngn3 are members of the same family and can activate many of the same downstream targets, regulation of their proteolysis differs significantly, demonstrating the need to take care in extrapolating findings from one protein family member to another.
Materials and Methods
Plasmid constructs
Mouse Ngn3 was cloned into pCS2+. AcNgn3 was created by PCR as previously described (Vosper et al., 2009). Ngn3KO and AcNgn3KO were synthesised commercially (GeneCust) and cloned into pCS2+. From these constructs, CO mutants Ngn3CO, AcNgn3CO, Ngn3KOCO, and AcNgn3KOCO were created by site-directed mutagenesis, using the QuikChange® multisite-directed mutagenesis kit (Stratagene).
Degradation assays
Activated interphase and mitotic Xenopus egg extracts were prepared and supplemented with ubiquitin as previously described (Vosper et al., 2007; Vosper et al., 2009). 35S-labeled IVT Ngn3 was generated in rabbit reticulocyte lysate (Promega) containing 35S-methionine (Perkin Elmer). 35S-IVT Ngn3 (or mutant derivatives thereof) was added to egg extract (18% (v/v) 35S-IVT Ngn3) and incubated at 20°C. At indicated time-points, aliquots were removed and mixed with Laemmli sample buffer supplemented with 100 mM β-mercaptoethanol. Samples were heated at 95°C for 5 minutes and run out on SDS-PAGE gels (15% Tris-glycine). Gels were analysed visually by autoradiography and quantitatively by phosphorimaging (Fujifilm). Degradation half-lives were calculated using first-order rate kinetics, using a natural log (ln) scale (Vosper et al., 2007). Three repeats of each degradation assay were performed and analysed.
Ubiquitylation assays
His-ubiquitin precipitation assays were performed, under both non-reducing and highly reducing conditions, as previously described with minor alterations (Vosper et al., 2009). Interphase Xenopus egg extract was supplemented with 20 µM Mg132 (BioMol) and 2.5 mg/ml of either His-ubiquitin or untagged ubiquitin (both Sigma). IVT Ngn3 (or mutant) (20% (v/v)) was added to this supplemented extract and incubated at 20°C for 90 minutes. Each reaction was then diluted 10-fold in His buffer (8 M urea, 600 mM NaCl, 20 mM imidazole, 100 mM Tris pH 7.4, 10% ethanol, 1% (v/v) IgePal), supplemented with 15 µl Ni-NTA agarose (Qiagen), and incubated at 20°C for 90 minutes (12 rpm). Following this incubation, beads were washed 3 times with His buffer. His-conjugated proteins were eluted from nickel beads by the addition of 15 µl 2× Laemmli buffer and heating at 95°C for 5 minutes. For non-reducing conditions, the Laemmli buffer was prepared at pH 6.8 with no β-mercaptoethanol. For highly reducing conditions, the Laemmli buffer was prepared at pH 10.0 with 10% (v/v) β-mercaptoethanol. Eluted proteins were separated on SDS-PAGE gels (15% Tris-glycine), which were analysed by autoradiography.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were undertaken as previously described (Ali et al., 2011). Either IVT E12 (25% (v/v)) and wild-type or mutant Ngn3 (as indicated) (12.5% or 25% (v/v)) were incubated in activated interphase Xenopus laevis egg extract (50% (v/v)) supplemented with 20 µM Mg132 for 20 minutes at RT (Fig. 5B) before dilution to 40% (v/v) in binding buffer. Alternatively, IVT E12 (10% (v/v)) and wild-type or mutant Ngn3 (as indicated) (5% or 10% (v/v)) (Fig. 6A) were immediately incubated in binding buffer. Incubations in binding buffer took place at RT for 20 minutes with synthetic non-specific competitor poly(dI–dC) and 32P-labeled probe consisting of the E1 E box domain of the NeuroD promoter. Samples were then separated on a non-denaturing 5% Bis–Tris gel, which was dried and analyzed by autoradiography.
Xenopus laevis extracts and embryos
Acquisition of Xenopus laevis eggs and embryos, preparation and injection of synthetic mRNA, staging of embryos, in situ hybridisation and egg extracts preparation have been described previously (Vernon et al., 2003; Vosper et al., 2007; Vosper et al., 2009).
Acknowledgments
We would like to thank Alison Jones, Gary McDowell, Chris Hurley, Chris Hindley, Jon Vosper, Helen Wise and Xana Almeida for technical support, helpful discussions and preliminary data leading up to this study. This work was supported by Medical Research Council (UK) [research grants G0700758 (AP) and G1002329]. R.R. was funded by the Marshall Aid Commemoration Commission, St. John's College, and the American National Science Foundation Graduate Research Fellowship Program. Any findings or opinions expressed in this work are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Aguado–Llera D., Goormaghtigh E., de Geest N., Quan X. J., Prieto A., Hassan B. A., Gómez J., Neira J. L. (2010). The basic helix-loop-helix region of human neurogenin 1 is a monomeric natively unfolded protein which forms a “fuzzy” complex upon DNA binding. Biochemistry 49, 1577–1589 10.1021/bi901616z [DOI] [PubMed] [Google Scholar]
- Ali F., Hindley C., McDowell G., Deibler R., Jones A., Kirschner M., Guillemot F., Philpott A. (2011). Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277 10.1242/dev.067900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Castro D. S., Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Cadwell K., Coscoy L. (2005). Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 10.1126/science.1110340 [DOI] [PubMed] [Google Scholar]
- Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá–Miranda C., Azevedo J. E. (2007). Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 10.1074/jbc.M706325200 [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Ben–Saadon R. (2004). N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14, 103–106 10.1016/j.tcb.2004.01.004 [DOI] [PubMed] [Google Scholar]
- Collins G. A., Tansey W. P. (2006). The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16, 197–202 10.1016/j.gde.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. (2000). neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 97, 1607–1611 10.1073/pnas.97.4.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D. A. (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hindley C., Ali F., McDowell G., Cheng K., Jones A., Guillemot F., Philpott A. (2012). Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 139, 1718–1723 10.1242/dev.077552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. P., Liu M., El–Hodiri H. M., Chu K., Jamrich M., Tsai M. J. (2000). Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell. Biol. 20, 3292–3307 10.1128/MCB.20.9.3292-3307.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M., Uhl C., Roche C., Duluc I., Guillermin V., Guillemot F., Jensen J., Kedinger M., Gradwohl G. (2002). Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21, 6338–6347 10.1093/emboj/cdf649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson K. A., Dursun U., Jordan N., Gu G., Beermann F., Gradwohl G., Grapin–Botton A. (2007). Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev. Cell 12, 457–465 10.1016/j.devcel.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Juhl K., Sarkar S. A., Wong R., Jensen J., Hutton J. C. (2008). Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes 57, 2755–2761 10.2337/db07-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003). Skp2 regulates Myc protein stability and activity. Mol. Cell 11, 1177–1188 10.1016/S1097-2765(03)00173-4 [DOI] [PubMed] [Google Scholar]
- Kuo M. L., den Besten W., Bertwistle D., Roussel M. F., Sherr C. J. (2004). N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18, 1862–1874 10.1101/gad.1213904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E., Murre C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell G. S., Kucerova R., Philpott A. (2010). Non-canonical ubiquitylation of the proneural protein Ngn2 occurs in both Xenopus embryos and mammalian cells. Biochem. Biophys. Res. Commun. 400, 655–660 10.1016/j.bbrc.2010.08.122 [DOI] [PubMed] [Google Scholar]
- Miyatsuka T., Matsuoka T. A., Kaneto H. (2008). Transcription factors as therapeutic targets for diabetes. Expert Opin. Ther. Targets 12, 1431–1442 10.1517/14728222.12.11.1431 [DOI] [PubMed] [Google Scholar]
- Nguyen L., Besson A., Heng J. I., Schuurmans C., Teboul L., Parras C., Philpott A., Roberts J. M., Guillemot F. (2006). p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 20, 1511–1524 10.1101/gad.377106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004). An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 10.1038/nsmb814 [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V. M., Scheel D. W., Conners J. R., Kalamaras J., Lee J. E., Anderson D. J., Sussel L., Johnson J. D., German M. S. (2000). Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127, 3533–3542. [DOI] [PubMed] [Google Scholar]
- Simon–Areces J., Membrive G., Garcia–Fernandez C., Garcia–Segura L. M., Arevalo M. A. (2010). Neurogenin 3 cellular and subcellular localization in the developing and adult hippocampus. J. Comp. Neurol. 518, 1814–1824 10.1002/cne.22304 [DOI] [PubMed] [Google Scholar]
- Sommer L., Ma Q., Anderson D. J. (1996). neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8, 221–241 10.1006/mcne.1996.0060 [DOI] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon A. E., Devine C., Philpott A. (2003). The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development 130, 85–92 10.1242/dev.00193 [DOI] [PubMed] [Google Scholar]
- von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I.et al. (2003). The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11, 1189–1200 10.1016/S1097-2765(03)00193-X [DOI] [PubMed] [Google Scholar]
- Vosper J. M., Fiore–Heriche C. S., Horan I., Wilson K., Wise H., Philpott A. (2007). Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochem. J. 407, 277–284 10.1042/BJ20070064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosper J. M., McDowell G. S., Hindley C. J., Fiore–Heriche C. S., Kucerova R., Horan I., Philpott A. (2009). Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284, 15458–15468 10.1074/jbc.M809366200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C., van den Berg M., Sprenger R. R., Distel B. (2007). A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Takakura A., Ohbo K., Abe K., Wakabayashi J., Yamamoto M., Suda T., Nabeshima Y. (2004). Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev. Biol. 269, 447–458 10.1016/j.ydbio.2004.01.036 [DOI] [PubMed] [Google Scholar]