Abstract
Biological imaging is now a quantitative technique for probing cellular structure and dynamics, and increasingly for cell-based screens. However, the bioinformatics tools required for hypothesis-driven analysis of digital images are still immature. We are developing the Open Microscopy Environment (OME) as an informatics solution for the storage and analysis of optical microscope image data. OME aims to automate image analysis, modeling and mining of large sets of images and specifies a flexible data model, a relational database, and an XML-encoded file standard usable by potentially any software tool. With this design, OME provides a first step toward biological image informatics.
Introduction
Recent excitement in optical microscopy centers on the extraction of quantitative numerical information from digital images to generate and test specific scientific hypotheses. For example, combining computer vision and speckle microscopy makes it possible to test specific mechanistic models of actin flow during cell movement (1). The potential for automation in digital imaging is also driving interest in the use of microscopy for large-scale “screening by imaging” in which cells or organisms are treated with libraries of small molecules, banks of small inhibitory RNAs etc. to identify chemicals or genes that affect a particular biological process by virtue of a change in cellular behaviour or appearance (2, 3) (Fig. 1). However, the routine application of automated image analysis and large-scale screening is held back by significant limitations in the software used to store, process and analyze the large volumes of information generated by digital imaging. It is possible to interpret images only if we know the context in which they were acquired. Current software for microscopy automates image acquisition and provides hardware and software solutions for 3D imaging (using deconvolution, confocal and other methods) but does not keep track of image and analytical data in a rigorous way. It is usually possible to specify file name, date and experimenter, but few packages systematically record the identities of the genes being studied, the labels used, etc. (4). Interoperability between different software systems involves the exchange of TIFF files, which preserve none of the contextual information. In this Viewpoint, we describe the conceptual challenges faced by image informatics as applied to biological microscopy and describe some of the solutions incorporated in an open-source image informatics system currently under development in our laboratories, the Open Microscopy Environment (OME (5)).
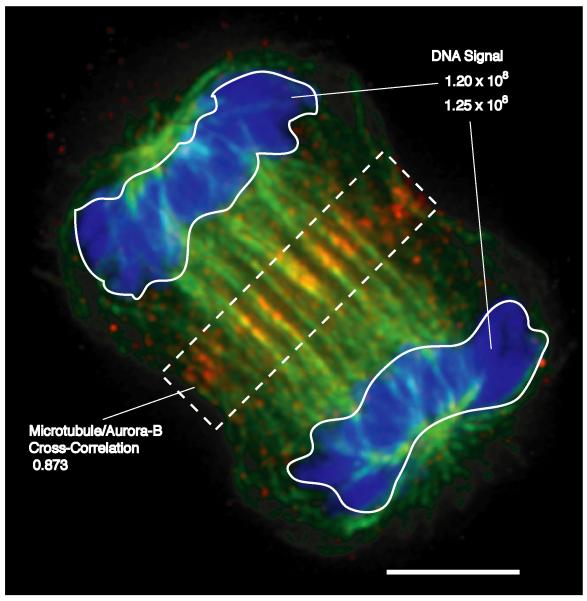
Figure 1. Applications for Quantitative Imaging.
The image shows an XlK2 cell during the process of cytokinesis stained for DNA (blue), microtubules (green) and the aurora-B protein kinase (red) (13). While the image demonstrates the relative localization of different cellular components and structures, quantitative analysis reveals specific characteristics that can be used to assay effects of inhibitors or expressed proteins. For example, integrating the signal from a DNA-specific fluorophore (top right) reveals defects in segregation of the genome in mitosis. Measuring the overlap of microtubules and aurora-B (e.g., using a cross correlation analysis (14)) within a sub-region of a dividing cell (dotted box) might be used to assess effectors of cytokinesis. Scale, 5 μm.
The primary goal of OME is to enable the automatic analysis, modelling and mining of large image sets with reference to specific biological hypotheses. OME aims to manage images from all optical microscopes, including confocal, wide-field and multi-photon systems but other image types (such as CT scans) are not necessarily supported. OME also aims to store – without loss or degradation – primary image data and the metadata that specifies the context and meaning of an image. Some metadata is devoted to describing the optics of the microscope, some to the experimental setup and sample, and some to information derived by analysis. Finally, OME aims to provide a flexible mechanism for incorporating new and existing image analysis routines and storing the output of these routines in a self-consistent and accessible manner.
The OME Data Model and Database
The OME data model is a formal description of the structure, meaning and behaviour of data stored and manipulated by the system, and is instantiated via both a database and a file format (Fig. 2). The OME data model has three parts: binary image data, data type semantics for managing modular image analysis and image metadata definitions for recording contextual information. Image data in OME is stored as time-lapse, three-dimensional, multi-spectral files (“5D images” (6, 7)). Data type semantics for OME are designed to allow analytic modules to be strung together in a flexible and simple fashion and are described in detail below. Image metadata describes the optics of the microscope, the filter sets, the objective lens etc. We hope that microscope manufacturers will agree (through OME or other projects) on a common format for metadata describing microscope hardware and image acquisition. Image metadata also describes the experimental setup, including genes under study, fluorophores, etc. Whenever possible, OME metadata definitions derive from pre-existing ontologies such as MESH and MGED and those being developed by the MIAME microarraying effort (8)).
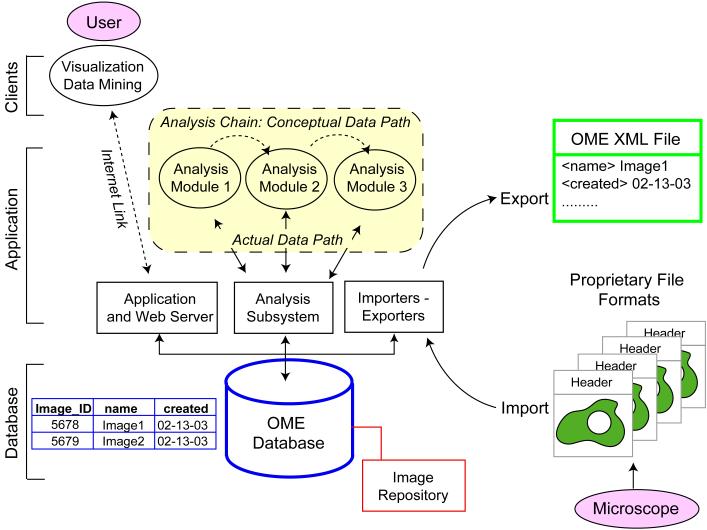
Figure 2. The Database is the Interface: OME Architecture.
OME is constructed as a standard “three-tier” application with a relational database that stores information in a table-based structure (blue), an application server that processes data and a client that lives on the users desktop and communicates via the internet (that is, via IP). Multiple clients can communicate with OME including Web browsers, commercial microscopy software and data mining applications. The OME data model is instantiated via a relational database (“OME database;” blue) in which metadata is stored in tables as specified by the schema and binary image data is stored in a trusted file system (the “image repository;” red). When data is transported between databases, or stored in a flat file, metadata and image data in the database are translated into XML (“OME XML File;” green). The OME database communicates with analysis modules via a subsystem (“analysis subsystem”) that ensures the consistent treatment of semantic datatypes. The analysis modules also calculate and store the history of the analysis chain (see online supplemental material). When analysis modules are chained together, each communicates independently with the database (“actual data path;” yellow block) even though the conceptual path appears is from one module to the next.(“conceptual data path”). Existing commercial or independent software tools can read OME data without substantial modification. OME itself is open source and available through a LGPL license but applications that talk to it can be either open or proprietary.
OME is designed to connect a desktop computer to an Oracle or PostgreSQL relational database using a standard client-server paradigm (Fig. 2; blue). The relational structure of OME makes it easy to access images on the basis of content and meaning: “Find all images of HeLa cells recorded by Jason in 2002.” Queries of this type are accomplished via an application layer comprising import and export routines, interfaces for analytic and visualization tools, and ancillary software. As an aid to performance, binary image data is stored in a file system (a repository) accessible only to OME (Fig. 2; red). Images from commercial file formats are imported into OME using a translator that reads the image data and converts it into a multi-dimensional image repository format. Any metadata stored with the image (usually, in a “header” that precedes the pixel data) is extracted from an input file and stored in the appropriate database tables (Fig 2; blue table) The net result is the conversion of a polyglot of commercial file formats into a single database representation. The OME file format is used when image data and metadata must be translated into a file for transport between OME databases, or for storage outside of a database. In OME files, each piece of data is associated with a tag (e.g. <filter_wavelength>), that defines its meaning in extensible markup language (XML), providing a vendor-neutral file format that conforms to public web-compliant standards (Fig. 2; green). We anticipate that commercial software tools will eventually be able to interact with the OME database as clients or possibly even directly read and write OME files.
Data Semantics for Image Analysis
Searchable image archives are useful, but we really require informatic systems that can extract and store quantitative information derived from images. Typically, image analysis involves several processing steps but the precise steps and their sequence necessarily depends on properties of the image and on scientific goals. We therefore require an extensible tool box of algorithms (including fourth-generation languages such as MatLab) that can be applied in different combinations to different images. Consider, the problem of tracking labeled vesicles in a time-lapse movie. A segmentation algorithm finds the vesicles and produces a list of centroids, volumes, signal intensities, etc; a tracker then defines trajectories by linking centroids at different timepoints according to predetermined set of rules; and finally a viewer displays the analytic results overlaid on the original movie. As designers of OME, we cannot anticipate exactly which tools work best for vesicle tracking. Instead, we must build general mechanisms for linking an analysis toolbox to images and storing analysis results.
Although we typically think of databases as storage systems, databases also represent an ideal mechanism for linking independent pieces of software together in a modular fashion. Conceptually, the data path in OME is from one analytical module to the next (Fig. 2; dashed box), but in practice, each module communicates independently with the database. The advantage of this architecture is that the problem of building links between analysis modules written in different computer languages is simplified to the task of linking each module independently to the database using known methods. For this to work however, the output of one module must match the input of the next module. The OME data model therefore includes a set of semantic data types that describe analytic results such as “centroid,” “trajectory,” “maximum signal,” etc. Semantic typing defines the types of relationships a data type can participate in, and thus, determines which analytic modules can use the data as inputs and outputs. However, the process of data analysis is tightly tied to prior knowledge of the biological system, the experiment, and the properties of the analytic routines. It is simply not possible to create a standards body that will rule on which definitions of centroid are valid and which are not. However, a database can solve this problem by linking each result to an operational record of the data processing steps that produced it, including the algorithm used and the states of any settings or variables. Thus, semantic data types, like centroid, can be defined broadly and then given specific meaning by the recorded history of their derivation. In this way we can judge each result in light of the methods that generated it and determine the accuracy of measurements a posteriori given the known operation of the analytic algorithms and characteristics of the data.
A final challenge for OME is providing a mechanism to add new analysis modules. In some cases, the inputs and outputs of the new module correspond to existing OME semantic data types (supporting online text). A more complex situation arises if OME lacks the necessary data types for a new module to interact with the system. In this case, the database must be augmented with tables to store the new data and present the changes to the user interface. This is a challenging problem in database design and represents a type of extensibility that is absolutely critical, but usually absent, in most bioinformatics software. Our solution is to specify the data requirements (the inputs and outputs) of an analytical module in an XML description written to the OME XML specification. OME is designed to then create the necessary tables on the fly. The net result is an analytic system that is extensible, modular and language-independent.
Image Informatics in Practice
The OME system is being developed as an open-source collaboration between academic labs and commercial hardware and software imaging companies (9). OMEv1.0 (10) demonstrated the utility of general-purpose image informatics software (11). A tutorial demonstrating this system is available (12). A system with features necessary for general use is being developed as OMEv2.0 for release late in 2003 (10). Our software is open-source, but commercial code plays a vital role in modern digital microscopy and image analysis. OME is therefore designed to integrate effectively with commercial code. Like DNA and protein sequence analysis, biological imaging must develop features of an information science to meet the demands of screening and hypothesis-driven analysis. Macromolecular and imaging bioinformatics have many things in common, but the complexity and unstructured nature of biological images presents a unique set of challenges in data analysis and interpretation. We are confident that if commercial and academic microscopists can solve the informatic problems that currently make quantitative analysis of microscope images difficult, quantitative image analysis will assume an important position in the future of bioinformatics.
Supplementary Material
Acknowledgements
We gratefully acknowledge helpful discussions with our academic and commercial partners(9), G. Danuser, and members of the Swedlow, Goldberg and Sorger groups. Research in the authors’ laboratories is supported by grants from the Wellcome Trust and Cancer Research UK (to J. R. S.), and the National Institutes of Health and the Harvard Institute of Chemistry and Cell Biology (to P. K. S and I. G.), J. R. S. is a Wellcome Trust Senior Research Fellow.
Footnotes
References
- 1.Waterman-Storer CM, Danuser G. Curr Biol. 2002;12:R633–40. doi: 10.1016/s0960-9822(02)01139-9. [DOI] [PubMed] [Google Scholar]
- 2.Mayer TU, et al. Science. 1999;286:971–4. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 3.Gonczy P, et al. Nature. 2000;408:331–6. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 4.Huang K, Lin J, Gajnak JA, Murphy RF. Proc IEEE Symp Biomed Imaging. 2002:325–328. [Google Scholar]
- 5. http://www.openmicroscopy.org.
- 6.Chen H, Sedat JW, Agard DA. In: Handbook of Biological Confocal Microscopy. Pawley JB, editor. Plenum; New York: 1989. pp. 141–150. [Google Scholar]
- 7.Andrews PD, Harper IS, Swedlow JR. Traffic. 2002;3:29–36. doi: 10.1034/j.1600-0854.2002.30105.x. [DOI] [PubMed] [Google Scholar]
- 8.Brazma A, et al. Adv Biochem Eng Biotechnol. 2002;77:113–39. doi: 10.1007/3-540-45713-5_7. [DOI] [PubMed] [Google Scholar]
- 9. http://www.openmicroscopy,org/participants2.htm.
- 10. http://www.openmicroscopy.org/technology6.htm.
- 11.Platani M, Goldberg I, Lamond AI, Swedlow JR. Nature Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- 12. http://www.openmicroscopy.org/OMEV1/OME_V1.html.
- 13.Adams RR, Carmena M, Earnshaw WC. Trends Cell. Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 14.Bracewell RN. The Fourier Transform and its Applications. McGraw-Hill; New York: 1986. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.