Abstract
Background
Carnitine Palmitoyltransferase-1c (CPT1c) is a neuron specific homologue of the carnitine acyltransferase family of enzymes. CPT1 isoenzymes transfer long chain acyl groups to carnitine. This constitutes a rate setting step for mitochondrial fatty acid beta-oxidation by facilitating the initial step in acyl transfer to the mitochondrial matrix. In general, neurons do not heavily utilize fatty acids for bioenergetic needs and definitive enzymatic activity has been unable to be demonstrated for CPT1c. Although there are studies suggesting an enzymatic role of CPT1c, its role in neurochemistry remains elusive.
Results
In order to better understand how CPT1c functions in neural metabolism, we performed unbiased metabolomic profiling on wild-type (WT) and CPT1c knockout (KO) mouse brains. Consistent with the notion that CPT1c is not involved in fatty acid beta-oxidation, there were no changes in metabolites associated with fatty acid oxidation. Endocannabinoids were suppressed in the CPT1c KO, which may explain the suppression of food intake seen in CPT1c KO mice. Although products of beta-oxidation were unchanged, small changes in carnitine and carnitine metabolites were observed. Finally, we observed changes in redox homeostasis including a greater than 2-fold increase in oxidized glutathione. This indicates that CPT1c may play a role in neural oxidative metabolism.
Conclusions
Steady-state metabolomic analysis of CPT1c WT and KO mouse brains identified a small number of metabolites that differed between CPT1c WT and KO mice. The subtle changes in a broad range of metabolites in vivo indicate that CPT1c does not play a significant or required role in fatty acid oxidation; however, it could play an alternative role in neuronal oxidative metabolism.
Background
Although the mammalian brain is lipid rich and mutations in lipid metabolizing enzymes result in debilitating neurological disease, neurons are generally not thought to rely on mitochondrial fatty acid beta-oxidation for bioenergetic requirements. Neurons instead mainly utilize the oxidation of glucose for most of their bioenergetic needs, although, during prolonged fasting, ketone bodies (i.e. acetoacetate and beta hydroxybutyrate) can also be used [1]. Most neurons have a low amount of the rate-setting enzymes in mitochondrial long chain fatty acid catabolism, namely, the malonyl-CoA sensitive Carnitine Palmitoyltransferase 1 (CPT1a and CPT1b) enzymes which limit most neurons potential for mitochondrial fatty acid beta-oxidation [2].
Carnitine acyltransferases are enzymes that catalyze the exchange of acyl groups between carnitine and Coenzyme A (CoA) to facilitate the transport acyl chains between the cytoplasm to the mitochondrial matrix [3]. CPT1 isoenzymes (EC 2.3.1.21) preferentially are positioned on the outer mitochondrial membrane and transfer long chain acyl groups from CoA to carnitine. CPT1a and CPT1b are malonyl-CoA sensitive and therefore inhibited when malonyl-CoA levels are high (e.g. during high glucose flux). The malonyl-CoA insensitive CPT2, on the other hand, is located in the mitochondrial matrix and reversibly transfers the acyl chain back to CoA to facilitate beta-oxidation. Although neurons have a relative dearth of CPT1a and CPT1b [2], they express a CPT1 homologue, CPT1c [4].
CPT1c has a high primary amino acid sequence similarity and identity to the canonical CPT enzymes. Therefore, it was surprising that definitive acyltransferase activity or enhanced oxidation of fatty acids could not be shown for CPT1c [4-6]. CPT1c KO mice exhibit both behavioral and metabolic deficits [6-9]. Over-expression of CPT1c in the brain of developing transgenic mice results in microencephaly [10]. Therefore, it is clear that CPT1c plays an important role in brain function. Although there were several metabolites identified that have been altered after over-expression [10,11] or knockout of CPT1c [7], the reaction that CPT1c catalyzes has remained elusive.
Here we used an unbiased metabolomic approach to broadly understand the consequence of CPT1c deletion to gain insight into the biochemical and physiological roles of CPT1c function. Similar to previous work in heterologous systems, we did not see changes consistent with a role for CPT1c in long chain fatty acid beta oxidation. However, there were changes in several fatty acid derived metabolites including endocannabinoids, which may explain the suppressed food intake in these models. Also, some of the most abundant changes were in redox biochemistry consistent with several models of CPT1c function recently proposed.
Methods
Animals
Mice with a targeted knockout of exons 1 and 2 of the cpt1c gene were propagated and genotyped as previously described [5,6]. Mice were fed a standard lab chow (Harlan 2018) after weaning. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and under the approval of the Johns Hopkins Medical School Animal Care and Use Committee.
Western blot analysis
A polyclonal rabbit antibody against CPT1c was used as a primary antibody for CPT1c detection in WT and CPT1c KO mice [5,6]. Anti-rabbit horseradish peroxidase (HRP) was used as a secondary antibody, and the blots for CPT1c were developed using ECL reagent. Mouse monoclonal anti-HSC70 (Santa Cruz biotech) and mouse monoclonal anti beta-actin (Sigma) was used as primary antibodies for loading control. Cy3 conjugated fluorescent secondary antibody was used for both HSC70 and beta-actin antibodies.
Metabolomic measurements and profiling
Unbiased metabolomics analysis of whole brain samples from WT and CPT1c KO mice (n=8/group) that were fasted overnight was performed using liquid chromatography/tandem mass spectrometry (HPLC/MS/MS2) and gas chromatography/mass spectrometry (GC/MS) platforms. The platform was able to screen and identify several metabolites in multiple classes, such as amino acids, lipids, and nucleotides. A complete list of the metabolites identified in this study is given in Tables 1, 2, 3 and 4. General platform methods about metabolomic measurements and profiling are described in the metabolomic study done by Eckel-Mahan et al. [12]
Table 1.
Biochemicals involved in lipid metabolic pathways
| PATHWAY | SUB PATHWAY | BIOCHEMICAL NAME | KEGG | CPT1c KOCPT1c WT | Welch's Two-Samplet-Test | CAS | PUBCHEM |
|---|---|---|---|---|---|---|---|
| Lipid |
Essential fatty acid |
linoleate (18:2n6) |
C01595 |
0.93 |
0.4643 |
60-33-3; |
5280450 |
| linolenate [alpha or gamma; (18:3n3 or 6)] |
C06427 |
1.04 |
0.4808 |
|
|
||
| dihomo-linolenate (20:3n3 or n6) |
C03242 |
0.81 |
0.0608 |
|
5312529 |
||
| eicosapentaenoate (EPA; 20:5n3) |
C06428 |
0.72 |
0.0236 |
10-2005-9;10417-94-4; |
446284 |
||
| docosapentaenoate (n3 DPA; 22:5n3) |
C16513 |
0.80 |
0.0662 |
2234-74-4; |
|
||
| docosapentaenoate (n6 DPA; 22:5n6) |
C06429 |
0.77 |
0.3030 |
25182-74-5; |
6441454 |
||
| docosahexaenoate (DHA; 22:6n3) |
C06429 |
0.89 |
0.2879 |
6217-54-5; |
445580 |
||
|
Medium chain fatty acid |
caproate (6:0) |
C01585 |
0.98 |
0.5408 |
142-62-1; |
8892 |
|
| caprylate (8:0) |
C06423 |
0.99 |
0.9309 |
124-07-2; |
379 |
||
| pelargonate (9:0) |
C01601 |
0.89 |
0.1531 |
112-05-0; |
5461016 |
||
| laurate (12:0) |
C02679 |
1.01 |
0.9051 |
143-07-7; |
3893 |
||
|
Long chain fatty acid |
myristate (14:0) |
C06424 |
0.99 |
0.8942 |
544-63-8; |
11005 |
|
| myristoleate (14:1n5) |
C08322 |
1.26 |
0.1786 |
544-64-9; |
5281119 |
||
| palmitate (16:0) |
C00249 |
0.85 |
0.0781 |
57-10-3; |
985 |
||
| palmitoleate (16:1n7) |
C08362 |
0.93 |
0.3794 |
373-49-9; |
445638 |
||
| margarate (17:0) |
|
0.86 |
0.2288 |
506-12-7; |
10465 |
||
| 10-heptadecenoate (17:1n7) |
|
0.81 |
0.1051 |
29743-97-3; |
5312435 |
||
| stearate (18:0) |
C01530 |
0.94 |
0.4536 |
57-11-4; |
5281 |
||
| oleate (18:1n9) |
C00712 |
0.88 |
0.2434 |
112-80-1; |
445639 |
||
| 10-nonadecenoate (19:1n9) |
|
0.72 |
0.0470 |
73033-09-7; |
5312513 |
||
| eicosenoate (20:1n9 or 11) |
|
0.78 |
0.1453 |
|
|
||
| dihomo-linoleate (20:2n6) |
C16525 |
0.75 |
0.0804 |
2091-39-6; |
6439848 |
||
| arachidonate (20:4n6) |
C00219 |
0.94 |
0.4832 |
506-32-1; |
444899 |
||
| docosadienoate (22:2n6) |
C16533 |
0.84 |
0.3185 |
7370-49-2; |
5282807 |
||
| adrenate (22:4n6) |
C16527 |
0.81 |
0.1467 |
2091-25-0; |
5282844 |
||
|
Fatty acid, ester |
n-Butyl Oleate |
|
0.96 |
0.7046 |
142-77-8; |
5354342 |
|
|
Fatty acid, dicarboxylate |
2-hydroxyglutarate |
C02630 |
1.00 |
0.8616 |
40951-21-1; |
43 |
|
|
Fatty acid, amide |
oleamide |
|
1.22 |
0.7962 |
301-02-0; |
5283387 |
|
| stearamide |
C13846 |
1.19 |
0.6546 |
124-26-5; |
31292 |
||
|
Eicosanoid |
prostaglandin D2 |
C00696 |
1.20 |
0.1092 |
41598-07-6; |
448457 |
|
| prostaglandin E2 |
C00584 |
0.93 |
0.3928 |
363-24-6; |
5280360 |
||
| 5-HETE |
|
0.99 |
0.8472 |
73307-52-5; |
9862886 |
||
| 15-HETE |
C04742 |
0.83 |
0.9669 |
54845-95-3; |
5280724 |
||
|
Endocannabinoid |
palmitoyl ethanolamide |
|
0.64 |
0.0331 |
|
4671 |
|
|
Fatty acid & BCAA metabolism |
propionylcarnitine |
C03017 |
1.08 |
0.4494 |
17298-37-2; |
107738 |
|
|
Carnitine metabolism |
carnitine |
C00487 |
0.88 |
0.0084 |
461-05-2; |
288 |
|
| 3-dehydrocarnitine* |
C02636 |
1.22 |
0.0103 |
10457-99-5; |
6991982 |
||
| acetylcarnitine |
C02571 |
0.89 |
0.2172 |
5080-50-2; |
7045767 |
||
| oleoylcarnitine |
|
0.83 |
0.3694 |
|
|
||
|
Fatty alcohol, long chain |
1-octadecanol |
D01924 |
1.01 |
0.8513 |
112-92-5; |
8221 |
|
|
Glycerolipid metabolism |
choline phosphate |
C00588 |
0.97 |
0.4914 |
72556-74-2; |
1014 |
|
| ethanolamine |
C00189 |
1.10 |
0.4703 |
141-43-5; |
|
||
| phosphoethanolamine |
C00346 |
1.06 |
0.5812 |
1071-23-4; |
52,323,241,015 |
||
| glycerol |
C00116 |
0.97 |
0.6203 |
56-81-5; |
753 |
||
| glycerol 3-phosphate (G3P) |
C00093 |
0.98 |
0.6926 |
29849-82-9; |
754 |
||
| glycerophosphorylcholine (GPC) |
C00670 |
0.96 |
0.9071 |
28319-77-9; |
657272 |
||
| cytidine 5'-diphosphocholine |
C00307 |
1.25 |
0.0583 |
33818-15-4; |
13805 |
||
|
Inositol metabolism |
myo-inositol |
C00137 |
0.94 |
0.0882 |
87-89-8; |
|
|
| chiro-inositol |
|
0.77 |
0.1568 |
643-12-9; |
|
||
| inositol 1-phosphate (I1P) |
|
1.01 |
0.8432 |
106032-59-1; |
|
||
| scyllo-inositol |
C06153 |
0.90 |
0.1635 |
488-59-5; |
|
||
|
Ketone bodies |
3-hydroxybutyrate (BHBA) |
C01089 |
1.23 |
0.2197 |
625-72-9; |
441 |
|
|
Lysolipid |
1-palmitoylglycerophosphoethanolamine |
|
1.12 |
0.8591 |
|
9547069 |
|
| 2-palmitoylglycerophosphoethanolamine* |
|
0.83 |
0.1926 |
|
|
||
| 1-stearoylglycerophosphoethanolamine |
|
1.20 |
0.7882 |
69747-55-3; |
9547068 |
||
| 1-oleoylglycerophosphoethanolamine |
|
1.14 |
0.8654 |
|
9547071 |
||
| 2-oleoylglycerophosphoethanolamine* |
|
1.07 |
0.9602 |
|
|
||
| 1-arachidonoylglycerophosphoethanolamine* |
|
1.06 |
0.8488 |
|
|
||
| 2-arachidonoylglycerophosphoethanolamine* |
|
0.45 |
0.2213 |
|
|
||
| 2-docosahexaenoylglycerophosphoethanolamine* |
|
0.48 |
0.3141 |
|
|
||
| 1-palmitoylglycerophosphocholine |
|
0.47 |
0.1450 |
17364-16-8; |
86554 |
||
| 2-palmitoylglycerophosphocholine* |
|
0.59 |
0.2106 |
|
|
||
| 1-stearoylglycerophosphocholine |
|
0.51 |
0.1452 |
19420-57-6; |
497299 |
||
| 2-stearoylglycerophosphocholine* |
|
1.00 |
|
|
10208382 |
||
| 1-oleoylglycerophosphocholine |
|
0.56 |
0.1923 |
19420-56-5; |
16081932 |
||
| 2-oleoylglycerophosphocholine* |
|
0.65 |
0.3441 |
|
|
||
| 1-arachidonoylglycerophosphocholine* |
C05208 |
1.00 |
|
|
|
||
| 2-arachidonoylglycerophosphocholine* |
|
0.89 |
0.4485 |
|
|
||
| 1-docosahexaenoylglycerophosphocholine* |
|
1.00 |
|
|
|
||
| 2-docosahexaenoylglycerophosphocholine* |
|
0.86 |
0.4614 |
|
|
||
| 1-palmitoylglycerophosphoinositol* |
|
0.85 |
0.2160 |
|
|
||
| 1-stearoylglycerophosphoinositol |
|
0.77 |
0.1315 |
|
|
||
| 1-arachidonoylglycerophosphoinositol* |
|
0.87 |
0.3521 |
|
|
||
| 1-oleoylglycerophosphoserine |
|
0.92 |
0.6515 |
|
9547099 |
||
| 2-oleoylglycerophosphoserine* |
|
0.80 |
0.1921 |
|
|
||
| 1-palmitoylplasmenylethanolamine* |
|
1.23 |
0.5225 |
|
|
||
|
Monoacylglycerol |
1-palmitoylglycerol (1-monopalmitin) |
|
0.83 |
0.1685 |
542-44-9; |
14900 |
|
| 1-stearoylglycerol (1-monostearin) |
D01947 |
0.92 |
0.3625 |
123-94-4; |
24699 |
||
| 2-stearoylglycerol (2-monostearin) |
|
0.75 |
0.1774 |
621-61-4; |
79075 |
||
| 1-oleoylglycerol (1-monoolein) |
|
0.80 |
0.1139 |
111-03-5; |
5283468 |
||
| 2-oleoylglycerol (2-monoolein) |
|
0.59 |
0.0769 |
3443-84-3; |
5319879 |
||
|
Sphingolipid |
sphingosine |
C00319 |
0.71 |
0.3009 |
123-78-4; |
5353955 |
|
| palmitoyl sphingomyelin |
|
0.84 |
0.1297 |
|
9939941 |
||
| stearoyl sphingomyelin |
C00550 |
1.07 |
0.2147 |
85187-10-6;85187-10-6; |
6453725 |
||
|
Mevalonate metabolism |
3-hydroxy-3-methylglutarate |
C03761 |
1.07 |
0.4426 |
503-49-1; |
5459993 |
|
| Sterol/Steroid | cholesterol |
C00187 |
1.00 |
0.9987 |
57-88-5; |
6432564 |
|
| 7-alpha-hydroxycholesterol |
C03594 |
1.24 |
0.2998 |
566-27-8; |
107722 |
||
| 7-beta-hydroxycholesterol |
C03594 |
1.11 |
0.2969 |
566-27-8; |
473141 |
||
| 24(S)-hydroxycholesterol |
C13550 |
0.94 |
0.5728 |
2140-46-7; |
|
||
| corticosterone | C02140 | 0.59 | 0.2402 | 50-22-6; | 5753 |
Table 2.
Biochemicals in the amino acid and peptide pathways
| PATHWAY | SUB PATHWAY | BIOCHEMICAL NAME | KEGG | CPT1c KOCPT1c WT | Welch's Two-Samplet-Test | CAS | PUBCHEM |
|---|---|---|---|---|---|---|---|
|
Amino acid |
Glycine, serine and threonine metabolism |
glycine |
C00037 |
0.91 |
0.1984 |
56-40-6; |
5,257,127,750 |
| serine |
C00065 |
0.98 |
0.6400 |
56-45-1; |
59,516,857,581 |
||
| N-acetylserine |
|
1.16 |
0.2513 |
97-14-3; |
65249 |
||
| homoserine |
C00263,C02926 |
1.04 |
0.5460 |
672-15-1; |
126,476,971,022 |
||
| 3-phosphoserine |
C01005 |
1.06 |
0.4516 |
407-41-0; |
|
||
| threonine |
C00188 |
0.98 |
0.8340 |
72-19-5; |
69,710,196,288 |
||
| allo-threonine |
C05519 |
0.98 |
0.7264 |
28954-12-3; |
992,896,995,276 |
||
| betaine |
C00719 |
0.62 |
0.0393 |
107-43-7; |
247 |
||
|
Alanine and aspartate metabolism |
alanine |
C00041 |
0.99 |
0.8540 |
56-41-7; |
59,507,311,724 |
|
| beta-alanine |
C00099 |
0.95 |
0.7707 |
56-41-7;107-95-9; |
2,394,755,801 |
||
| N-acetylalanine |
C02847 |
0.96 |
0.7172 |
97-69-8; |
88064 |
||
| aspartate |
C00049 |
1.02 |
0.5759 |
56-84-8; |
5960 |
||
| N-acetylaspartate (NAA) |
C01042 |
0.98 |
0.7849 |
997-55-7;997-55-7; |
65065 |
||
|
Glutamate metabolism |
glutamate |
C00025 |
1.10 |
0.1218 |
56-86-0; |
611 |
|
| glutamine |
C00064 |
0.96 |
0.3866 |
56-85-9; |
69,920,865,961 |
||
| gamma-aminobutyrate (GABA) |
C00334 |
1.07 |
0.4581 |
56-12-2; |
6,992,099,119 |
||
| N-acetylglutamate |
C00624 |
1.21 |
0.1108 |
5817-08-3; |
1549099 |
||
| N-acetyl-aspartyl-glutamate (NAAG) |
C12270 |
1.04 |
0.6033 |
3106-85-2; |
5255 |
||
| N-acetylglutamine |
C02716 |
0.79 |
0.1871 |
2490-97-3; |
182230 |
||
|
Histidine metabolism |
histidine |
C00135 |
1.11 |
0.1815 |
5934-29-2; |
7,733,651,426 |
|
|
Lysine metabolism |
lysine |
C00047 |
0.81 |
0.0655 |
56-87-1; |
5962 |
|
| 2-aminoadipate |
C00956 |
0.99 |
0.9856 |
542-32-5;1118-90-7; |
469 |
||
| pipecolate |
C00408 |
0.91 |
0.4383 |
4043-87-2; |
849 |
||
| glutaroyl carnitine |
|
0.77 |
0.0244 |
102636-82-8; |
|
||
|
Phenylalanine & tyrosine metabolism |
phenylalanine |
C00079 |
0.93 |
0.0731 |
63-91-2; |
69,256,656,140 |
|
| tyrosine |
C00082 |
1.10 |
0.1569 |
60-18-4; |
60,576,942,100 |
||
| 3-(4-hydroxyphenyl)lactate |
C03672 |
1.28 |
0.1580 |
6482-98-0; |
9378 |
||
|
Tryptophan metabolism |
tryptophan |
C00078 |
1.10 |
0.1009 |
73-22-3; |
69,235,166,305 |
|
| C-glycosyltryptophan* |
|
1.00 |
0.9578 |
|
|
||
| 5-hydroxyindoleacetate |
C05635 |
0.99 |
0.9982 |
54-16-0; |
1826 |
||
|
Valine, leucine and isoleucine metabolism |
isoleucine |
C00407 |
0.99 |
0.7705 |
73-32-5; |
791 |
|
| leucine |
C00123 |
0.92 |
0.1061 |
61-90-5; |
70,457,986,106 |
||
| valine |
C00183 |
1.00 |
0.9896 |
72-18-4; |
69,710,186,287 |
||
| alpha-hydroxyisovalerate |
|
0.92 |
0.9081 |
600-37-3; |
99823 |
||
| 2-methylbutyroylcarnitine |
|
0.98 |
0.7985 |
31023-25-3; |
6426901 |
||
| isovalerylcarnitine |
|
0.90 |
0.1479 |
|
6426851 |
||
| hydroxyisovaleroyl carnitine |
|
0.90 |
0.0807 |
99159-87-2; |
|
||
|
Cysteine, methionine, SAM, taurine metabolism |
cysteine |
C00097 |
1.13 |
0.0835 |
52-90-4;56-89-3; |
58,626,419,722 |
|
| cystine |
C00491 |
0.86 |
0.5529 |
56-89-3; |
595 |
||
| taurine |
C00245 |
1.03 |
0.7783 |
107-35-7; |
11,234,068,592 |
||
| S-adenosylhomocysteine (SAH) |
C00021 |
0.96 |
0.4778 |
979-92-0; |
|
||
| methionine |
C00073 |
0.95 |
0.1654 |
63-68-3; |
69,920,876,137 |
||
| N-acetylmethionine |
C02712 |
0.88 |
0.1362 |
65-82-7; |
448580 |
||
| 2-hydroxybutyrate (AHB) |
C05984 |
1.23 |
0.5077 |
3347-90-8; |
440864 |
||
|
Urea cycle; arginine-, proline-, metabolism |
arginine |
C00062 |
0.95 |
0.0964 |
1119-34-2; |
5,246,487,232 |
|
| ornithine |
C00077 |
0.90 |
0.2453 |
3184-13-2; |
6262 |
||
| urea |
C00086 |
0.71 |
0.2913 |
57-13-6; |
117,616,150,869 |
||
| proline |
C00148 |
0.97 |
0.6099 |
147-85-3; |
1,457,426,971,047 |
||
| N-acetylornithine |
C00437 |
1.26 |
0.3497 |
6205-08-9; |
6,992,102,439,232 |
||
| trans-4-hydroxyproline |
C01157 |
1.03 |
0.6431 |
51-35-4; |
58,106,971,053 |
||
| argininosuccinate |
C03406 |
0.86 |
0.3803 |
156637-58-0; |
828 |
||
|
Creatine metabolism |
creatine |
C00300 |
1.03 |
0.2564 |
57-00-1; |
586 |
|
| creatinine |
C00791 |
1.20 |
0.1694 |
60-27-5; |
588 |
||
|
Butanoate metabolism |
2-aminobutyrate |
C02261 |
1.03 |
0.8503 |
1492-24-6; |
4,396,916,971,251 |
|
|
Polyamine metabolism |
5-methylthioadenosine (MTA) |
C00170 |
1.08 |
0.2023 |
2457-80-9; |
439176 |
|
| putrescine |
C00134 |
0.83 |
0.4688 |
110-60-1; |
|
||
| spermidine |
C00315 |
1.04 |
0.6645 |
124-20-9; |
1102 |
||
| spermine |
C00750 |
0.99 |
0.4470 |
71-44-3; |
1103 |
||
|
Guanidino and acetamido metabolism |
4-guanidinobutanoate |
C01035 |
0.98 |
0.7911 |
463-003;463-00-3; |
500 |
|
|
Glutathione metabolism |
glutathione, reduced (GSH) |
C00051 |
1.53 |
0.1024 |
70-18-8; |
124886 |
|
| 5-oxoproline |
C01879 |
0.86 |
0.0291 |
98-79-3; |
7405 |
||
| glutathione, oxidized (GSSG) |
C00127 |
2.15 |
0.0307 |
103239-24-3; |
6,535,911,215,652 |
||
| cysteine-glutathione disulfide |
|
1.33 |
0.0802 |
13081-14-6; |
4247235 |
||
| Peptide |
Dipeptide derivative |
carnosine |
C00386 |
0.98 |
0.8057 |
305-84-0; |
4,392,246,992,100 |
| homocarnosine |
C00884 |
1.00 |
0.9807 |
3650-73-5; |
10243361 |
||
| gamma-glutamyl | gamma-glutamylleucine |
|
0.91 |
0.1529 |
2566-39-4; |
151023 |
|
| gamma-glutamylglutamate |
|
1.24 |
0.1880 |
1116-22-9; |
92865 |
||
| gamma-glutamylglutamine |
|
0.93 |
0.4450 |
10148-81-9; |
150914 |
||
| gamma-glutamylphenylalanine | 0.93 | 0.5544 | 7432-24-8; | 111299 |
Table 3.
Biochemicals from the carbohydrate and energy pathways
| PATHWAY | SUB PATHWAY | BIOCHEMICAL NAME | KEGG | CPT1c KOCPT1c WT | Welch's Two-Samplet-Test | CAS | PUBCHEM |
|---|---|---|---|---|---|---|---|
|
Carbohydrate |
Aminosugars metabolism |
N-acetylglucosamine |
C00140 |
1.03 |
0.7477 |
7512-17-6; |
24139 |
| erythronate* |
|
0.98 |
0.7434 |
13752-84-6; |
2781043 |
||
| N-acetylneuraminate |
C00270 |
1.03 |
0.4494 |
131-48-6; |
|
||
|
Fructose, mannose, galactose, starch, and sucrose metabolism |
fructose |
C00095 |
0.98 |
0.8393 |
57-48-7; |
5984 |
|
| mannose |
C00159 |
0.94 |
0.6417 |
3458-28-4; |
161658 |
||
| mannose-6-phosphate |
C00275 |
0.97 |
0.7187 |
70442-25-0;104872-94-8; |
|
||
| sorbitol |
C00794 |
0.92 |
0.5926 |
6706-59-8; |
107428 |
||
|
Glycolysis, gluconeogenesis, pyruvate metabolism |
1,5-anhydroglucitol (1,5-AG) |
C07326 |
0.95 |
0.7426 |
154-58-5; |
|
|
| glycerate |
C00258 |
0.96 |
0.4928 |
600-19-1; |
752 |
||
| glucose-6-phosphate (G6P) |
C00668 |
0.96 |
0.5074 |
103192-55-8; |
|
||
| glucose |
C00293 |
0.86 |
0.1984 |
50-99-7; |
79025 |
||
| fructose-6-phosphate |
C05345 |
0.83 |
0.1261 |
103213-47-4; |
|
||
| Isobar: fructose 1,6-diphosphate, glucose 1,6-diphosphate |
|
0.98 |
0.8050 |
|
|
||
| 3-phosphoglycerate |
C00597 |
0.80 |
0.1220 |
80731-10-8; |
|
||
| dihydroxyacetone phosphate (DHAP) |
C00111 |
1.02 |
0.6910 |
102783-56-2; |
4643300 |
||
| 1,3-dihydroxyacetone |
C00184 |
1.12 |
0.4601 |
62147-49-3; |
670 |
||
| pyruvate |
C00022 |
0.83 |
0.0193 |
127-17-3; |
107735 |
||
| lactate |
C00186 |
1.06 |
0.3677 |
79-33-4; |
612 |
||
|
Nucleotide sugars, pentose metabolism |
arabitol |
C00474 |
1.30 |
0.0435 |
488-82-4; |
94154 |
|
| ribitol |
C00474 |
0.86 |
0.1732 |
488-81-3; |
|
||
| sedoheptulose-7-phosphate |
C05382 |
0.91 |
0.4130 |
2646-35-7; |
616 |
||
| ribose 5-phosphate |
C00117 |
1.39 |
0.0353 |
18265-46-8;108321-05-7; |
447634 |
||
| Isobar: ribulose 5-phosphate, xylulose 5-phosphate |
|
1.06 |
0.5400 |
|
|
||
| arabinose |
C00181 |
1.08 |
0.5432 |
28697-53-2; |
66308 |
||
| Energy |
Krebs cycle |
citrate |
C00158 |
1.02 |
0.5785 |
77-92-9; |
311 |
| alpha-ketoglutarate |
C00026 |
0.79 |
0.2702 |
305-72-6;328-50-7;22202-68-2; |
51 |
||
| succinate |
C00042 |
0.88 |
0.5010 |
110-15-6; |
1110 |
||
| fumarate |
C00122 |
0.94 |
0.5055 |
100-17-8; |
|
||
| malate |
C00149 |
1.11 |
0.2256 |
6915-15-7; |
525 |
||
| Oxidative phosphorylation | phosphate |
C00009 |
0.98 |
0.3284 |
7664-38-2; |
1061 |
|
| pyrophosphate (PPi) | C00013 | 0.84 | 0.4801 | 1466-09-3; | 644102 |
Table 4.
Biochemicals in nucleotide, cofactors and vitamins, and xenobiotic Pathways
| PATHWAY | SUB PATHWAY | BIOCHEMICAL NAME | KEGG | CPT1c KOCPT1c WT | Welch's Two-Samplet-Test | CAS | PUBCHEM |
|---|---|---|---|---|---|---|---|
|
Nucleotide |
Purine metabolism, (hypo)xanthine/inosine containing |
xanthine |
C00385 |
1.02 |
0.7727 |
69-89-6; |
1188 |
| hypoxanthine |
C00262 |
0.98 |
0.4343 |
68-94-0; |
790 |
||
| inosine |
|
1.00 |
0.8754 |
58-63-9; |
|
||
|
Purine metabolism, adenine containing |
adenine |
C00147 |
1.11 |
0.0801 |
73-24-5; |
190 |
|
| adenosine |
C00212 |
0.86 |
0.1407 |
58-61-7; |
60961 |
||
| N1-methyladenosine |
C02494 |
0.94 |
0.3601 |
15763-06-1; |
5460178 |
||
| adenosine 2'-monophosphate (2'-AMP) |
C00946 |
1.00 |
|
130-49-4; |
|
||
| adenosine 5'-monophosphate (AMP) |
C00020 |
0.89 |
0.2000 |
149022-20-8; |
15938965 |
||
|
Purine metabolism, guanine containing |
guanosine |
C00387 |
1.01 |
0.9130 |
118-00-3; |
6802 |
|
|
Purine metabolism, urate metabolism |
urate |
C00366 |
1.06 |
0.4983 |
69-93-2;120K5305; |
|
|
| allantoin |
C02350 |
0.76 |
0.1685 |
97-59-6; |
204 |
||
|
Pyrimidine metabolism, cytidine containing |
cytidine |
C00475 |
0.94 |
0.1562 |
65-46-3; |
6175 |
|
| cytidine 5'-monophosphate (5'-CMP) |
C00055 |
1.01 |
0.8988 |
63-37-6; |
7058165 |
||
|
Pyrimidine metabolism, orotate containing |
orotate |
C00295 |
0.86 |
0.2325 |
50887-69-9; |
967 |
|
|
Pyrimidine metabolism, uracil containing |
uracil |
C00106 |
0.97 |
0.5212 |
66-22-8; |
1174 |
|
| uridine |
C00299 |
0.91 |
0.0141 |
58-96-8; |
6029 |
||
| pseudouridine |
C02067 |
0.99 |
0.7648 |
1445-07-4; |
|
||
|
Purine and pyrimidine metabolism |
methylphosphate |
|
0.85 |
0.1460 |
7023-27-0; |
13130 |
|
|
Cofactors and vitamins |
Ascorbate and aldarate metabolism |
ascorbate (Vitamin C) |
C00072 |
0.87 |
0.1924 |
134-03-2; |
|
| dehydroascorbate |
C05422 |
1.70 |
0.2338 |
490-83-5; |
835 |
||
| threonate |
C01620 |
0.96 |
0.5529 |
70753-61-6; |
151152 |
||
|
Hemoglobin and porphyrin |
heme* |
C00032 |
0.69 |
0.3695 |
14875-96-8; |
|
|
|
Nicotinate and nicotinamide metabolism |
nicotinamide |
C00153 |
1.00 |
0.9275 |
98-92-0; |
936 |
|
| nicotinamide adenine dinucleotide (NAD+) |
C00003 |
0.87 |
0.0469 |
53-84-9; |
1,089,765,158,925,280,000 |
||
|
Pantothenate and CoA metabolism |
pantothenate |
C00864 |
0.94 |
0.7951 |
137-08-6; |
6613 |
|
| phosphopantetheine |
C01134 |
0.85 |
0.0841 |
NA; |
115254 |
||
|
Pyridoxal metabolism |
pyridoxal |
C00250 |
1.05 |
0.5803 |
65-22-5; |
1050 |
|
|
Riboflavin metabolism |
flavin adenine dinucleotide (FAD) |
C00016 |
0.93 |
0.1085 |
146-14-5;84366-81-4; |
643975 |
|
| riboflavin (Vitamin B2) |
C00255 |
0.93 |
0.2187 |
83-88-5; |
493570 |
||
| flavin mononucleotide (FMN) |
C00061 |
0.96 |
0.7167 |
130-40-5; |
710 |
||
|
Tocopherol metabolism |
alpha-tocopherol |
C02477 |
1.04 |
0.6234 |
59-02-9;10191-41-0; |
14985 |
|
| Xenobiotics |
Chemical |
glycolate (hydroxyacetate) |
C00160 |
1.06 |
0.7194 |
79-14-1; |
3,698,251,757 |
| glycerol 2-phosphate |
C02979,D01488 |
1.02 |
0.9683 |
819-83-0; |
2526 |
||
| 2-phenoxyethanol |
|
0.94 |
0.9231 |
122-99-6; |
|
||
| 2-pyrrolidinone |
|
0.84 |
0.6590 |
616-45-5; |
12025 |
||
|
Food component/Plant |
ergothioneine |
C05570 |
0.88 |
0.0968 |
58511-63-0; |
3032311 |
|
| Sugar, sugar substitute, starch | erythritol | C00503 | 0.89 | 0.0966 | 149-32-6; |
Statistical analysis
Pair-wise comparisons between CPT1c WT and KO were performed using Welch’s two-sample t-tests. From the p-values, any value below the significance level of 0.05 was interpreted as statistically significant.
Results
Carnitine Palmitoyltransferase-1c KO mice
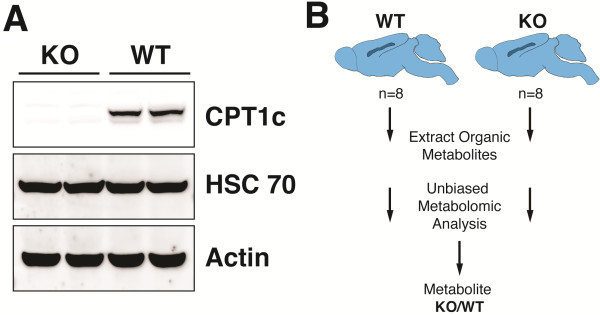
Although CPT1c is widely expressed in transformed cells and tumors [13], we have only been able to reliably detect CPT1c in neurons in vivo. To understand the endogenous function of CPT1c, we performed metabolomic profiling on brains of CPT1c KO mice and their littermate controls. Therefore, we collected and snap froze the brains of CPT1c KO and WT littermate sex matched adult mice after an overnight fast. Western blot analysis of WT and CPT1c KO mice showed that KO mice were indeed completely deficient of CPT1c (Figure 1A). These samples were then homogenized and the small organic metabolites were extracted and analyzed by a mixture of GC-MS and LC-MS/MS by a commercial supplier of metabolomic analyses (Figure 1B). Below, we detail the changes in steady-state biochemicals between WT and KO brains that were identified through an unbiased metabolomic screen.
Figure 1.
CPT1c KO mice and metabolomic profiling. (A) CPT1c protein from homogenized brains of WT and CPT1c KO mice were analyzed by western blot using the anti-CPT1c antibody. Hsc70 and Actin were used for loading controls. (B) A schematic pathway of metabolomic profiling for KO and WT brains. A commercial supplier of metabolic analysis homogenized 8 brain samples from independent mice to extract organic metabolites for performing unbiased metabolomic analysis using a mixture of GC-MS and LC-MS/MS.
Fatty acid oxidative metabolites show no difference in overall trend in CPT1c KO mice
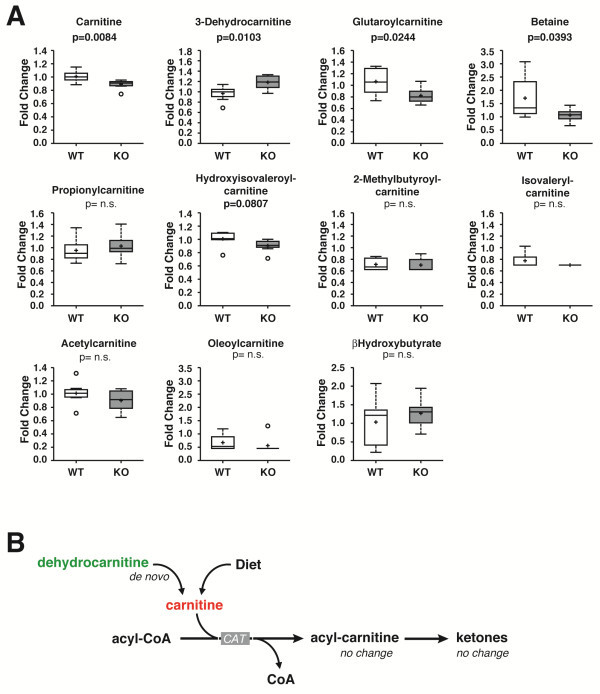
Given the high primary amino acid homology of CPT1c to other CPTs, it would follow that CPT1c may be involved in fatty acid beta oxidation or at least in long chain acyl-CoA metabolism. If CPT1c was involved in fatty acid oxidation, we would expect that the deletion of CPT1c would decrease the level of acyl-carnitines and potentially increase the levels of other long chain acyl-CoA dependent biosyntheses. A broad range of lipid species were identified in the metabolomic screen (Table 1). No changes were seen in oleoyl-carnitine, beta-hydroxybutyrate, or acetyl-carnitine, as we would have expected (Figure 2A). However, the metabolomic analysis did show that free carnitine, 3-dehydrocarnitine, glutaroylcarnitine, and betaine were significantly changed (Figure 2A).
Figure 2.
Loss of CPT1c results in decreased free carnitine and no change in fatty acid oxidative metabolites in the brain. (A) Biochemicals involved in carnitine, amino acid, and fatty acid metabolism from WT and CPT1c KO brains were compared through metabolomic analyses, revealing a statistically significant change in levels of free carnitine (p=0.084), 3-dehydrocarnitine (p=0.0103), glutaroylcarnitine (p=0.0244) and betaine (p=0.0383). (B) Schematic of biochemical pathways altered in CPT1c KO mice. Based on this schematic pathway, glutaroyl carnitine and betaine may affect the level of free carnitine, since these biochemicals play a role in carnitine biosynthesis.
Among the metabolites that showed a statistically significant difference, only 3-dehydrocarnitine increased in CPT1c KO mice while glutaroyl carnitine, betaine and free carnitine decreased. Glutaroyl carnitine and betaine are biochemicals that are involved in carnitine biosynthesis (Figure 2B; Table 2). Glutaroyl carnitine is involved in lysine metabolism, which is one of the amino acids that is used to synthesize carnitine. In the carnitine biosynthesis pathway, betaine takes the form of butyrobetaine to synthesize L-carnitine [14]. As a result, it is possible that the decrease in glutaroyl carnitine and betaine could have caused free carnitine levels to decrease in CPT1c KO mice. Previous studies also tested hypothalamic and cortical explants from WT and CPT1c KO mice for their ability to oxidize fatty acids, but there was no evidence that unique properties in neurons existed to allow activation of fatty acid oxidation by CPT1c [5]. CPT1c over-expressed in heterologous cells in vitro also did not show a change in fatty acid oxidation [5]. Therefore, our results remain consistent with previous findings that CPT1c, although it is highly homologous with its isoforms CPT1a and CPT1b, does not participate substantially in neuronal mitochondrial fatty acid oxidation.
Loss of CPT1c results in decreased levels of endogenous endocannabinoids
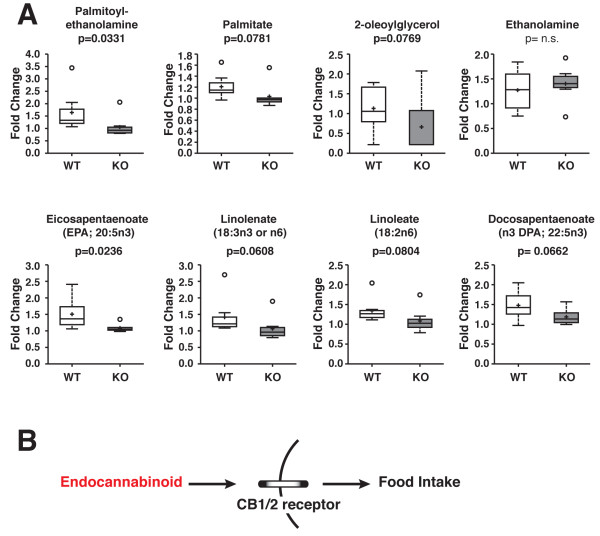
Several studies have investigated the neurological role of endocannabinoids on food intake [15]. A study investigated the role of endocannabinoids in regulating food intake in the tongue, gut and different brain regions, suggesting that the cannabinoid system plays a role in modulating the activity of neural pathways that regulate food intake and energy expenditure [15]. The brain cannabinoid system, as shown in Figure 3B, regulates food intake through the interaction of endogenous ligands and cannabinoid receptors. From our metabolomic analyses, there was a significant decrease in palmitoylethanolamine and a trend for a decrease in 2-oleolylglycerol in CPT1c KO mouse brains compared to WT mouse brains (Figure 3). There was no significant difference between WT and CPT1c KO mice for free nonesterified fatty acids (Table 1). Among the metabolites shown in Figure 3A, eicosapentaenoate and palmitoylethanolamine showed a significant decrease in CPT1c KO mice with a p-value of 0.0236 and 0.0331, respectively. There was also a slight increase in ethanolamine between WT and CPT1c KO mice, and decrease in 2-oleoylglycerol (p=0.0769), an endogenous cannabinoid (CB) CB-1 agonist (Figure 3A).
Figure 3.
Loss of CPT1c results in decreased endocannabinoids in the brain. (A) Biochemicals involved in fatty acid biochemistry from WT and CPT1c KO mouse brains were compared to determine if metabolomic analyses showed any statistically significant changes. There was an overall decreasing trend in endocannabinoids in CPT1c KO mice. Specifically, eicosapentaenoate (p=0.0236) and palmitoylethanolamine (p=0.0331) significantly decreased in CPT1c KO mice. (B) A schematic of how a decrease in endocannabinoids can induce a decrease in food intake by interacting with CB1 and CB2 cannabinoid receptors.
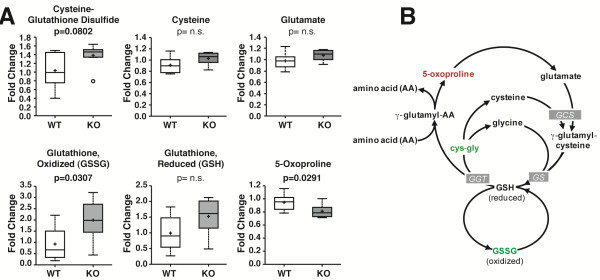
Loss of CPT1c results in increased levels of glutathione
The oxidized form of GSH (GSSG) and 5-oxoproline, biochemicals involved in the gamma-glutamyl redox cycle, resulted in a statistically significant difference in CPT1c KO mice (Table 2). GSSG and cysteine-glutathione disulfide levels increased while 5-oxoproline levels decreased in CPT1c KO mice (Figure 4A). Based on the schematic redox pathway shown in Figure 4B, our results suggest that CPT1c may play a role in oxidative metabolism. This is consistent with findings in cancer metabolism. Zaugg et al. depleted the levels of CPT1c in MCF-7 cells to determine whether these cells were sensitive to oxidative stress. Hypoxia was used as a stress inducer, and they found that CPT1c depletion caused an increased sensitivity to oxidative stress, implying that CPT1c may play a crucial role in protecting the cells from stress from the environment [13]. Furthermore, the loss of CPT1c resulted in an increase in ceramides [7,8], a key mediator of oxidative stress [16,17]. However, the mechanism and role of CPT1c in oxidative metabolism remains unknown.
Figure 4.
Loss of CPT1c results in elevated oxidative demands in the brain. (A) In a comparison of biochemicals involved in redox homeostasis in WT and CPT1c KO mouse brains, GSSG and 5-oxoproline were statistically significant. GSSG levels increased in CPT1c KO mice with a p-value of 0.0307, while 5-oxoproline decreased in KO mice (p=0.0291). The biochemicals shown displayed an overall increasing trend in CPT1c KO mice. (B) A schematic of the gamma-glutamyl redox cycle. Based on the pathway, an increase in the biochemicals from Figure 4A may cause the cells to become more sensitive to oxidative stress.
Discussion
Role of CPT1c in behavior and physiology
Carnitine acyltransferases are enzymes that catalyze the exchange of acyl groups between carnitine and CoA to facilitate the transport of acyl groups from the cytoplasm to the mitochondrial matrix. Carnitine acetyltransferase (CRAT) and carnitine octonyltransferase (CROT) facilitate transport short- and medium-chain acyl-CoA, while CPT1 facilitate transports long chain acyl-CoA to the mitochondria. CPT1 enzymes are encoded by three genes in mammals that are localized in different tissues and have different properties. CPT1a, which is enriched in the liver, has been heavily studied due to its crucial role in β-oxidation and human fatty oxidation disorders (OMIM #255120) and is lethal when knocked out in mice [18]. CPT1b is localized mainly in the muscle and is a regulator for the use of fatty acids in muscle and is also lethal when knocked out in mice [19]. These two enzymes, which are present on the outer mitochondrial membrane, play a critical role in regulating and facilitating fatty acid beta-oxidation.
The brain specific CPT1c is highly homologous to its closely related genes, CPT1a and CPT1b [4]. However, despite its high homology, CPT1c does not catalyze acyl transfer from long chain acyl-CoA to carnitine [4-6]. Other distinguishing properties of CPT1c include a longer C-terminus and localization in the endoplasmic reticulum (ER) instead of the mitochondria [11]. Although it does not facilitate acyl transfer in the cell, CPT1c most likely remains sensitive to the endogenous allosteric CPT1 inhibitor, malonyl-CoA, binding with a similar affinity as CPT1a [4,6]. Moreover, while other isoenzymes are expressed in a broad range of organisms, CPT1c seems to have risen late in evolution, raising the question whether CPT1c has a specific role in mammalian brain function.
Several studies used CPT1c knockout (KO) and CPT1c transgenic mice to investigate the role of CPT1c in the CNS. Knockout studies showed that loss of CPT1c did not affect the viability or fertility of the mice, but resulted in a suppression in food intake and decrease in body weight when they were fed a normal or low-fat diet [6,9]. Paradoxically, when high fat diet was given to CPT1c KO mice, they exhibited diet-induced obesity which ultimately resulted in a diabetic phenotype [5,6]. Even though fatty acid oxidative metabolites showed no significant change based on the metabolomic analysis, due to a decrease in peripheral energy expenditure CPT1c KO mice were more susceptible to obesity and diabetes when fed a high fat diet. This suggests that CPT1c has a hypothalamic function in protecting the body from adverse weight gain when the mice were fed a high fat diet. Transgenic CPT1c mice (CPT1c-TgN), on the other hand, which allowed conditional expression of CPT1c in a tissue-specific manner via cre-lox recombination, showed enhanced expression of CPT1c and they were protected from diet-induced obesity even on a high-fat diet [10].
CPT1c KO mice also showed impaired spatial learning [7]. Cpt1c deficiency was shown to alter dendritic spine morphology by increasing immature filopodia and reducing mature mushroom and stubby spines. Compared to WT mice, CPT1c KO mice showed a higher escape latency, implying that they had a delay in the acquisition phase [7]. Based on this study, CPT1c deficiency interfered with consolidating new information but did not affect retaining information or motor behavior. As a result, there may be other physiological roles of CPT1c in addition to regulating food intake and energy expenditure consistent with its broad expression throughout the nervous system [7].
Endocannabinoid regulation of food intake
Endocannabinoids are endogenous ligands that bind to cannabinoid receptors to regulate many aspects of physiology and behavior. Specifically, the brain endocannabinoid system regulates food intake via the hypothalamus, where it activates necessary mediators to induce appetite after a short-term food deprivation. CB1 receptor KO mice showed reduced food intake, similar to CPT1c KO mice [20,21]. Based on our results, CPT1c could be interacting with the cannabinoid system, causing an overall decreasing trend in endocannabinoids in CPT1c KO mice. In this context, the loss of CPT1c could have influenced the endocannabinoid system and its function to regulate food intake and body weight, which may explain the suppressed food intake in CPT1c KO mice [5,9]. Therefore, a decrease in endocannabinoids based on metabolomic profiling may suggest a putative role of the endocannabinoid system in suppressing food intake in CPT1c KO mice. However, it is unclear if CPT1c affects endocannabinoid metabolism directly or more likely indirectly by altering neuronal specific fatty acid metabolism.
Glutathione and redox metabolism
Neurons are particularly sensitive to oxidative stress and damage caused by reactive oxygen species (ROS). On the cellular level, there are many endogenous metabolic stress inducers, such as ROS produced from the mitochondria and cytosolic enzymes, such as cyclooxygenase and lipoxygenase. There are also various exogenous conditions that can also promote the level of ROS species to increase, such as H2O2 and hypoxia, that induces irreversible cellular damage or cell death. As shown by the pathway in Figure 4B, reduced glutathione (GSH) and oxidized glutathione (GSSG) are tightly regulated in order to maintain cellular redox homeostasis and to protect the cells from oxidative damage [17]. Carrasco et al. showed that CPT1c expression correlated with ceramide production and loss of CPT1c resulted in reduced ceramide levels. [7]. A recent study on the role of CPT1c in cancer cells in response to metabolic stress showed that CPT1c could participate in protecting cells from stress. In addition, they postulated that metabolic stress could alter regulation of the CPT1c gene, reducing ATP production and increasing sensitivity towards metabolic stress [13]. Here, we showed that CPT1c deficiency results in an increased oxidative environment. This may indicate that although CPT1c does not contribute in large part to beta-oxidation, it may be involved in other neuron specific oxidative metabolism. Alternatively, CPT1c may need to be activated in a yet to identified stress-induced manner. Barger et al. [22] showed that CPT1c was required for leukemia growth under low glucose conditions. Therefore, CPT1c may have a context dependent role in fatty acid catabolism. Although here we show that CPT1c could play a role in oxidative stress, the precise role of CPT1c in relation to oxidative stress remains unknown.
Conclusion
Unbiased metabolomic profiling of steady-state metabolites in WT and CPT1c KO brains revealed subtle changes in a broad range of metabolites in vivo. The metabolic alterations are not consistent with CPT1c playing a role in beta-oxidation or a large non-redundant role in bioenergetics.
Abbreviations
WT: Wild-type; KO: Knockout; CPT1: Carnitine Palmitoyltransferase 1; CPT2: Carnitine Palmitoyltransferase 2; CoA: Coenzyme A; CB: Cannabinoids; GC: Gas chromatography; MS: Mass spectrometry.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MJW conceived of the project, collected samples and aided in writing. JL interpreted results and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jieun Lee, Email: jlee425@jhmi.edu.
Michael J Wolfgang, Email: mwolfga1@jhmi.edu.
Acknowledgments
We would like to thank Amanda Reamy for technical assistance. This work was supported in part by the American Heart Association (SDG2310008 to M.J.W.) and NIH NINDS (NS072241 to M.J.W.).
References
- Cahill GF Jr. Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Lane MD. The role of hypothalamic malonyl-CoA in energy homeostasis. J Biol Chem. 2006;281(49):37265–37269. doi: 10.1074/jbc.R600016200. [DOI] [PubMed] [Google Scholar]
- Price N, van der Leij F, Jackson V, Corstorphine C, Thomson R, Sorensen A, Zammit V. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 2002;80(4):433–442. doi: 10.1006/geno.2002.6845. [DOI] [PubMed] [Google Scholar]
- Wolfgang MJ, Cha SH, Millington DS, Cline G, Shulman GI, Suwa A, Asaumi M, Kurama T, Shimokawa T, Lane MD. Brain-specific carnitine palmitoyl-transferase-1c: role in CNS fatty acid metabolism, food intake, and body weight. J Neurochem. 2008;105(4):1550–1559. doi: 10.1111/j.1471-4159.2008.05255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Kurama T, Dai Y, Suwa A, Asaumi M, Matsumoto S, Cha SH, Shimokawa T, Lane MD. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci USA. 2006;103(19):7282–7287. doi: 10.1073/pnas.0602205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco P, Sahun I, McDonald J, Ramirez S, Jacas J, Gratacos E, Sierra AY, Serra D, Herrero L, Acker-Palmer A. et al. Ceramide levels regulated by carnitine palmitoyltransferase 1C control dendritic spine maturation and cognition. J Biol Chem. 2012;287(25):21224–21232. doi: 10.1074/jbc.M111.337493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Zhu G, Gao X, Wu D, Carrasco P, Casals N, Hegardt FG, Moran TH, Lopaschuk GD. Important roles of brain-specific carnitine palmitoyltransferase and ceramide metabolism in leptin hypothalamic control of feeding. Proc Natl Acad Sci USA. 2011;108(23):9691–9696. doi: 10.1073/pnas.1103267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XF, Chen W, Kong XP, Xu AM, Wang ZG, Sweeney G, Wu D. Enhanced susceptibility of Cpt1c knockout mice to glucose intolerance induced by a high-fat diet involves elevated hepatic gluconeogenesis and decreased skeletal muscle glucose uptake. Diabetologia. 2009;52(5):912–920. doi: 10.1007/s00125-009-1284-0. [DOI] [PubMed] [Google Scholar]
- Reamy AA, Wolfgang MJ. Carnitine palmitoyltransferase-1c gain-of-function in the brain results in postnatal microencephaly. J Neurochem. 2011;118(3):388–398. doi: 10.1111/j.1471-4159.2011.07312.x. [DOI] [PubMed] [Google Scholar]
- Sierra AY, Gratacos E, Carrasco P, Clotet J, Urena J, Serra D, Asins G, Hegardt FG, Casals N. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J Biol Chem. 2008;283(11):6878–6885. doi: 10.1074/jbc.M707965200. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B. et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25(10):1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Black SM. Carnitine homeostasis, mitochondrial function, and cardiovascular disease. Drug Discov Today Dis Mech. 2009;6(1–4):e31–e39. doi: 10.1016/j.ddmec.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipatrizio NV, Piomelli D. The thrifty lipids: endocannabinoids and the neural control of energy conservation. Trends Neurosci. 2012;35(7):403–411. doi: 10.1016/j.tins.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvicens N, Cotter TG. Ceramide is the key mediator of oxidative stress-induced apoptosis in retinal photoreceptor cells. J Neurochem. 2006;98(5):1432–1444. doi: 10.1111/j.1471-4159.2006.03977.x. [DOI] [PubMed] [Google Scholar]
- Andrieu-Abadie N, Gouaze V, Salvayre R, Levade T. Ceramide in apoptosis signaling: relationship with oxidative stress. Free Radic Biol Med. 2001;31(6):717–728. doi: 10.1016/S0891-5849(01)00655-4. [DOI] [PubMed] [Google Scholar]
- Nyman LR, Cox KB, Hoppel CL, Kerner J, Barnoski BL, Hamm DA, Tian L, Schoeb TR, Wood PA. Homozygous carnitine palmitoyltransferase 1a (liver isoform) deficiency is lethal in the mouse. Mol Genet Metab. 2005;86(1–2):179–187. doi: 10.1016/j.ymgme.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Ji S, You Y, Kerner J, Hoppel CL, Schoeb TR, Chick WS, Hamm DA, Sharer JD, Wood PA. Homozygous carnitine palmitoyltransferase 1b (muscle isoform) deficiency is lethal in the mouse. Mol Genet Metab. 2008;93(3):314–322. doi: 10.1016/j.ymgme.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal P, Bellocchio L, Clark S, Cannich A, Klugmann M, Lutz B, Marsicano G, Cota D. Hypothalamic CB1 cannabinoid receptors regulate energy balance in mice. Endocrinology. 2012;153(9):4136–4143. doi: 10.1210/en.2012-1405. [DOI] [PubMed] [Google Scholar]
- Cota D, Marsicano G, Tschop M, Grubler Y, Flachskamm C, Schubert M, Auer D, Yassouridis A, Thone-Reineke C, Ortmann S. et al. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J Clin Invest. 2003;112(3):423–431. doi: 10.1172/JCI17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JF, Gallo CA, Tandon P, Liu H, Sullivan A, Grimes HL, Plas DR. S6K1 determines the metabolic requirements for BCR-ABL survival. Oncogene. 2012. [Epub ahead of print] [DOI] [PMC free article] [PubMed]