Abstract
HIV-1 depends on many host factors for propagation. Other host factors, however, antagonize HIV-1 and may have profound effects on viral activation. Curing HIV-1 requires the reduction of latent viral reservoirs that remain in the face of antiretroviral therapy (ART). Using orthologous genetic screens, we identified bromodomain containing 4 (BRD4) as a negative regulator of HIV-1 replication. Antagonism of BRD4, via RNA interference or with a small molecule inhibitor, JQ1, both increased proviral transcriptional elongation and alleviated HIV-1 latency in cell line models. In multiple instances, JQ1 when used in combination with the NF-κB activators, Prostratin or PHA, enhanced the in vitro reactivation of latent HIV-1 in primary human T cells. These data are consistent with a model wherein BRD4 competes with the virus for HIV-1 dependency factors (HDFs) and suggests that combinatorial therapies that activate HDFs and antagonize HIV-1 competitive factors may be useful for curing HIV-1 infection.
Elucidation of the host factors that modulate HIV-1 propagation has been a long-term goal of the HIV research community (Friedrich et al., 2011). In principle, this knowledge should allow us to develop therapies that seek to eradicate this disease (Choudhary and Margolis, 2011; Yang et al., 2009). Current ART aimed at inhibiting viral enzymatic activities prevents HIV-1 replication and halts the virus’ destruction of the host’s immune system. However, patients develop viremia upon treatment interruption due to the reactivation of latent HIV-1 proviruses and long-term ART has been associated with some adverse effects (Hakre et al., 2012; Margolis, 2010; Siliciano and Greene, 2011). To cure HIV-1, such latent reservoirs must be purged, the most promising strategy for this being viral activation performed in unison with ART to prevent de novo infection (Archin et al., 2012; Siliciano and Greene, 2011).
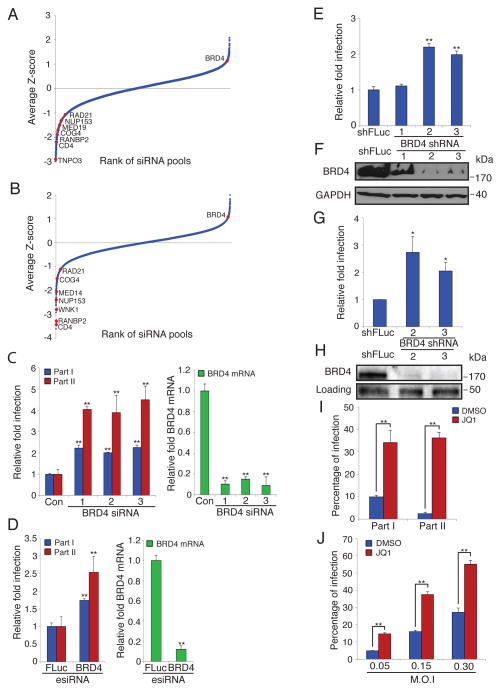
In an ongoing effort to saturate the HDF siRNA screen (Brass et al., 2008) we found that depletion of the transcriptional regulator BRD4 enhances HIV-1 infection using either of two orthologous large-scale siRNA libraries (Fig. 1A, B). BRD4 is a bromodomain protein that binds to acetylated histones and recruits transcription elongation factor b (PTEF-b), thereby increasing full-length mRNA expression (Jang et al., 2005; Yang et al., 2005). The establishment and maintenance of transcription of the HIV-1 genome is a critical step in the viral lifecycle (Karn, 2011; Ott et al., 2011). After initiating transcription from the HIV-1 long terminal repeat (LTR), the host’s RNA polymerase II complex pauses due to the influence of negative elongation factors (NELF and DSIF); this inhibition is overcome by the viral protein Tat, which binds to the viral mRNA’s TAR element and recruits PTEF-b, leading to phosphorylation of both the polymerase and the negative elongation factors, resulting in transcriptional elongation (Karn, 2011; Ott et al., 2011; Zhang et al., 2007). Because the Tat/PTEF-b complex regulates viral transcription in a feed-forward manner, subtle alterations in the availability of either component strongly influence viral gene expression and activation (Karn, 2011; Karn and Stoltzfus, 2012; Ott et al., 2011). Of note, the majority of PTEF-b is associated with either the host cell’s chromatin or its 7SK ribonucleoprotein (RNP) complex. The inaccessibility of PTEF-b presents a formidable barrier to viral activation that only Tat can overcome via its relocation of PTEF-b to the TAR element (Karn and Stoltzfus, 2012). BRD4 had also been previously implicated in HIV-1 biology. In different publications BRD4 has been suggested to be a positive or negative regulator of HIV-1 gene expression (Yang et al., 2005; Zhou et al., 2009). High level overproduction of a C-terminal portion of BRD4 has been observed to inhibit HIV-1 infection and compete with Tat for binding to PTEF-b in vitro (Bisgrove et al., 2007; Urano et al., 2008). The latter data suggest that BRD4 is necessary for basal viral transcription but might compete with Tat during active viral transcription (Bisgrove et al., 2007; Ott et al., 2011; Schroder et al., 2012). Together with our loss-of-function data, these studies prompted us to further investigate BRD4’s role in the viral lifecycle and in particular evaluate it as an anti-latency target. To accomplish this, we individually transfected three unique siRNAs against BRD4 and monitored their effect on HIV-1 infection and the depletion of BRD4 mRNA levels (Fig. 1C, D). Similar experiments were done using a diverse pool of siRNAs against BRD4, esiRNAs. In all cases the level of HIV-1 infection correlated with the levels of BRD4 depletion suggesting that the effect on HIV-1 was attributable to the siRNA-mediated targeting of BRD4. We did routinely observe a greater difference in p24 staining in part 2 than in part 1 of these assays (Fig. 1D) and believe this may reflect the limitations of the image analysis to detect absolute levels of p24, in addition to the potential of a non-linear relationship existing between p24 levels detected in part 1 and percent infection scored in part 2. Two out of three short hairpins RNAs (shRNAs) against BRD4 also increased HIV-1 infection in a T cell line in accordance with their depletion levels (Fig. 1E, F). Importantly, we also detected higher levels of HIV-1 infection in primary human CD4+ T cells after BRD4 depletion (Fig. 1G, H). We then tested a small molecule, JQ1, which prevents BRD4-mediated transcriptional enhancement by interfering with its binding to acetylated histones (Delmore et al., 2011; Filippakopoulos et al., 2010). Consistent with BRD4 inhibiting HIV-1 infection and in keeping with a recent report (Banerjee et al., 2012), we found that JQ1 increased the replication of HIV-1 in both HeLa cells and in a T cell line (Fig. 1I, J, S1A), and that JQ1’s proviral effect was dependent on the presence of BRD4 (Fig. S1B).
Fig. 1. BRD4 depletion or inhibition increases HIV-1 replication.
(A, B) The results of the indicated RNAi library screens with the siRNA pools ranked in order of average relative fold infection, from lowest to highest. The positions of known HDFs as well as the newly identified HIV-1-competitive factor (HCF), BRD4, from the screen are indicated. (a) Ambion Library; (b) esiRNA Library
(C) HeLa MAGI cells were transfected with the indicated siRNAs for 72 hr, then infected with HIV-IIIB (Part I). Forty-eight hr later the viral supernatant was transferred to a new plate of HeLa MAGI cells for 48 hr (Part II). Part I and II cells were stained for p24. Relative fold infection is normalized to the non-targeting control siRNA (Con). Identically transfected cells were assessed for BRD4 mRNA knockdown using quantitative PCR (qPCR) and normalized to the control siRNA (right).
(D) Cells were treated as in (C) but were transfected with esiRNA pools as indicated. Values were normalized to the non-targeting firefly luciferase (FLuc) esiRNA control pool. Identically transfected cells were assessed for BRD4 mRNA knockdown using quantitative PCR (qPCR) and normalized to the control FLuc esiRNA pool (right).
(E) Jurkat T cells were stably transduced with the indicated shRNAs then infected with VSV-G NL4-3-GFP HIV-1. At 72 hr post infection the percentage of GFP positive cells was determined by flow cytometry, and the relative fold infection normalized to the shFLuc-expressing control line.
(F) Western blot for cells in (E). kDa = kilodaltons
(G) Primary Human CD4+ T cells were stably transduced with the indicated shRNAs then infected with VSV-G NL4-3-GFP HIV-1. At 72 hr post infection the percentage of GFP positive cells was determined by flow cytometry, and the relative fold infection normalized to the shFLuc-expressing control line.
(H) Western blot for cells in (G). The levels of a non-specific band (loading) show relative protein loading.
(I) HeLa MAGI cells were treated with either DMSO or JQ1 (500 nM) for 1 hr, then infected with HIV-IIIB as in (C).
(J) Jurkat T cells were treated with either DMSO or JQ1 (500 nM) for 1 hr, then infected with VSV-G NL4-3-GFP HIV-1 using the indicated multiplicity of infections (M.O.I). After 48 hr the percentage of GFP positive cells was determined by flow cytometry.
Values represent the mean +/− S.E.M. (standard error), N > 3 throughout. * P < 0.05, ** P < 0.01. Results were analyzed by unpaired t tests. See Fig. S1.
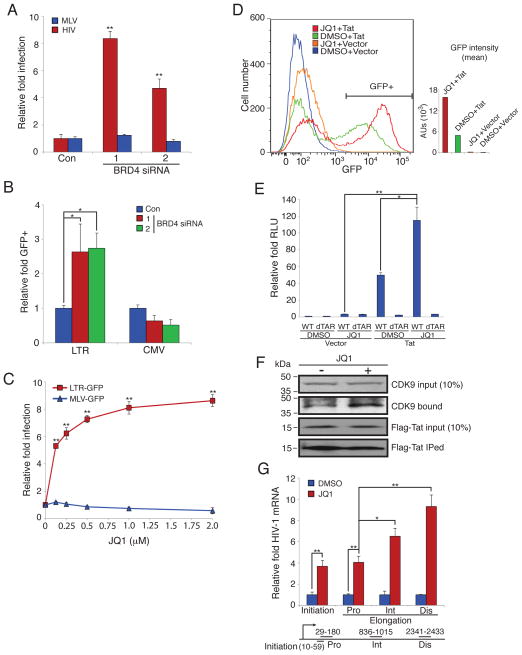
We next investigated the mechanism of BRD4’s inhibition of HIV-1. Based on previous studies together with our observed effect of JQ1 on pseudotyped viruses (Fig. 1E, G), we reasoned that inhibition of BRD4 may potentiate proviral transcription. In support of this notion, the depletion of BRD4 specifically increased the infection of HIV-1 proviruses in comparison to its effect on a γ-retrovirus (Moloney leukemia virus, MLV, Fig. 2A). BRD4’s loss also stimulated a stably integrated Tat-dependent reporter gene whose expression is driven by HIV-1’s LTR in contrast to the effect seen with CMV promoter-dependent gene expression (Fig. 2B). Similarly, the BRD4-inhibitor, JQ1, preferentially increased green fluorescent protein (GFP) expression from an HIV-1 LTR when compared to an MLV LTR (Fig. 2C (Delmore et al., 2011; Filippakopoulos et al., 2010)). Moreover, neither AP-1 nor NF-κB-dependent reporters were activated by JQ1 (Fig. S2). JQ1’s enhancement of LTR activity was predominantly seen to be both Tat- and TAR-dependent, suggesting that modulation of PTEF-b levels underlies the small molecule’s effects (Fig. 2D, E, Fig. S1B). In studies using a T cell line latently infected with an HIV-1 LTR-GFP provirus, we observed that the area under the GFP expression curve for cells stably expressing Tat and treated with JQ1 was 3.3 fold greater than seen with Tat and DMSO, with the percent of GFP positive cells increasing from 39% with Tat and DMSO to 54% with Tat and JQ1. Together these data suggest that JQ1 in combination with Tat resulted in more cells reactivating latent viruses, and doing so with greater magnitude. However, we did note a more modest stimulation of the HIV-LTR reporter in the absence of Tat (Fig. 2D, E) and this was found to be TAR-independent as well (Fig. 2D). Consistent with the assumption that JQ1 enhanced the available levels of PTEF-b, we found that JQ1 treatment increased the association of Tat with PTEF-b (Fig. 2F). Notably, JQ1 improved polymerase processivity along HIV-1 proviruses and increased all lengths of viral messenger RNAs (vmRNAs) tested (initial and elongated vmRNAs), with the most substantial enhancement seen for full-length vmRNAs (Fig. 2G). Given the known interactions of both BRD4 and Tat with PTEF-b (Bisgrove et al., 2007), these data suggest a model wherein BRD4 competes with Tat for the localization of a limiting amount of PTEF-b and, therefore, upon BRD4’s loss and/or chemical inhibition, more PTEF-b can bind to Tat and stimulate proviral transcriptional elongation.
Fig. 2. BRD4 inhibition stimulates HIV-1 gene expression and increases Tat-dependent transcriptional elongation and Tat-PTEF-b association.
(A) HeLa-T4 cells were transfected with the indicated siRNAs for 72 hr then infected with either VSV-G HIV-YFP (HIV) or VSV-G MLV-GFP (MLV). After 48 hr the percentage of YFP of GFP positive cells was determined and normalized to the control siRNA.
(B) HeLa-Tat-III cells stably expressing either an LTR- or CMV-driven d1-EGFP reporter gene were transfected with the indicated siRNAs. After 48 hr, GFP positive cells were measured and normalized to the control siRNA.
(C) HeLa-T4 cells stably transduced with HIV-1 LTR-GFP reporter (LTR-GFP) or a MLV LTR-GFP reporter (MLV-GFP) were treated with the indicated concentrations of JQ1 for 48 hr after which the relative fold infection was determined by imaging of GFP positive cells and cell nuclei (DNA-stained).
(D) Jurkat T cells stably expressing an HIV-1 LTR-GFP reporter gene (LTRG cells) were transduced with gamma retroviruses expressing either Tat or the empty vector alone (vector). 48 hr post transduction, cells were treated with DMSO or JQ1 (500 nM); 48 hr later the number of GFP expressing cells (GFP) was assessed using flow cytometry. The GFP+ % of cells: JQ1+Tat, 54.1%; DMSO+Tat, 39.0%; JQ1+Vector, 0.335%; DMSO+Vector, 0.0750%. The mean of GFP intensity was calculated for all sorted cells. Data are representative of three independent experiments.
(E) 293T cells were treated with JQ1 (500 nM) or DMSO for 1 hr, then transfected with the indicated combinations of plasmids: wild-type (WT) or TAR-deleted (dTAR) HIV-1 LTR-luciferase reporter vectors, and a HIV-1 Tat expression vector (Tat) or the matching empty vector (Vector). A dual luciferase assay was performed at 48 hr post transfection and the relative light units (RLU) were normalized to the DMSO-treated cells transfected with the WT HIV-1 LTR-luciferase and empty vector plasmids.
(F) Hela cells stably transduced with a FLAG-Tat were incubated with JQ1 (500 nM) or DMSO for 24 hr. Cells were then lysed and subjected to immunoprecipitation using anti-FLAG antibodies. An anti-CDK9 antibody was used to detect the endogenous CDK9 levels. An anti-FLAG antibody was used to detect the FLAG-Tat level.
(G) Jurkat T cells were treated with JQ1 (500 nM) or DMSO for 1 hr, then infected with VSV-G NL43-GFP HIV-1. After 48 hr cDNA was prepared and vmRNA length assessed using the indicated primers. Initiation: 10–59bp. Elongation: proximal (Pro, 29–180bp), intermediate (Int, 836–1015bp) and distal (Dis, 2341–2433), with the numbers denoting the amplification region of the NL4–3 HIV-1 transcript as determined from the transcriptional start site. Data was normalized to the DMSO control.
Values represent the mean +/− S.E.M., N > 3 throughout. * P < 0.05, ** P < 0.01. Results were analyzed by unpaired t tests. See Fig. S1, 2.
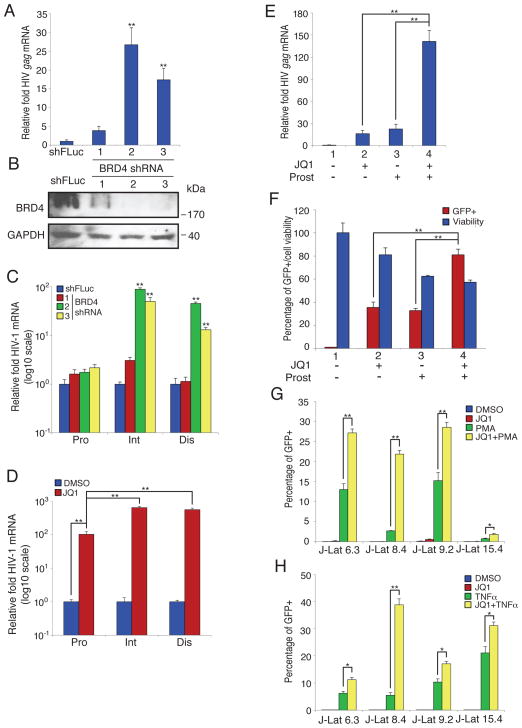
Since the depletion or inhibition of BRD4 results in the production of 2 to 3-fold or greater viral gene expression this suggests that a significant amount of proviruses, 50% to 75%, are not fully active. These quiescent viruses are reminiscent of latent proviruses present in HIV-1 infected individuals treated with long term ART. To explore this similarity, we investigated BRD4’s contribution to viral latency. Depletion of BRD4 in a promonocyte latency model, U1 cells, strongly increased both gag vmRNA expression and transcriptional processivity (Fig. 3A–C, (Folks et al., 1987; Pomerantz et al., 1990)). Chemical inhibition of BRD4 by JQ1 also enhanced transcriptional elongation in U1 cells (Fig. 3D). U1 cells are impaired for vmRNA transcription because they express Tat containing a point mutation that reduces transcriptional activation (Cannon et al., 1994; Emiliani et al., 1998). In contrast when we decreased BRD4 in another latency cell line, ACH2, which contains a provirus with a mutant TAR, we saw more moderate increases in vmRNA production as compared to the U1 cells (Fig. S4A, (Cannon et al., 1994; Emiliani et al., 1996)). This is consistent with BRD4 antagonizing Tat’s recruitment of PTEF-b.
Fig. 3. BRD4 inhibition alleviates HIV-1 latency.
(A) U1 cells were stably transduced with the indicated shRNAs, then HIV-1 gag mRNA levels were measured by qPCR and normalized to the shFLuc control.
(B) Western blot of cells in (A).
(C) A transcriptional elongation assay was performed as above using cells in (A). The level of transcripts was normalized to the shFLuc control. Note log scale on the Y axis.
(D) A transcriptional elongation assay was performed as above using U1 cells treated with either JQ1 (500 nM) or DMSO for 1 hr. The level of transcripts was normalized to the values of the DMSO-treated controls. Note log scale on the Y axis.
(e) U1 cells were treated with either JQ1 (500 nM) or Prostratin (1 uM) alone, or the combination. After 72 hr cDNA was prepared and levels of gag mRNA determined.
(f) J-Lat A2 cells were treated with JQ1 (500 nM) or Prostratin (1 μM) for 72h. Cells were then assessed for GFP expression and viability. For the cell viability assays the relative luminescence units (RLU) of DMSO treated cells was set at 100%.
(g) The indicated J-Lat cell lines were treated with JQ1 (500 nM), PMA (200 nM), or the both for 72 hr. GFP positive cells were assessed using flow cytometry, and the percentage of positive cell was calculated.
(h) The indicated J-Lat cell lines were treated with the noted compounds (JQ1 (500 nM), TNF-α (10 ng/ml) alone or in combination. At 72 hr the percentage of GFP positive cells was determined by flow cytometry.
Values represent the mean +/− S.E.M., N > 3 throughout. * P < 0.05, ** P < 0.01. Results were analyzed by unpaired t tests. See Fig. S3.
Overcoming insufficient transcription is important for activating latent HIV-1 (Dahl and Palmer, 2009; Karn and Stoltzfus, 2012). Therefore, given JQ1’s effect on transcriptional elongation we evaluated its impact on latent HIV-1 when paired with compounds that boost transcriptional initiation by stimulating HDFs (HDF-activators). Two such HDF-activators are the natural product, Prostratin (12-deoxyphorbol 13-acetate, (Gulakowski et al., 1997; Williams et al., 2004)), and the cytokine, TNF-α (Duh et al., 1989; Folks et al., 1987), both of which stimulate the HDF, NF-κB, a transcription factor that is essential for HIV-1 LTR activity (Chan and Greene, 2012; Williams et al., 2004). Interestingly, when we tested JQ1 together with Prostratin in U1 cells, we discovered that this combined treatment synergistically increased latent viral gene expression >100-fold, likely by enhancing both transcriptional initiation and elongation (Fig. 3E, (Barboric et al., 2001; Chan and Greene, 2012)). Using several T cell J-Lat cell lines (Jordan et al., 2003), we observed increased HIV-1 LTR expression using JQ1 and Prostratin in combination (Fig. 3F, S3C). In addition, we found that stimulation of HIV-1 reporter gene expression in J-Lat T cell lines by either TNF-α or PMA was also enhanced by JQ1 (Fig. 3G, H). It is unclear why these clonal lines do not achieve full activation under these conditions but possible explanations include cell to cell variability and genetic drift. We note, however, that JQ1 alone had less effect in the J-Lat T cell lines compared to the other cells tested, perhaps reflecting cell line specific activation states or requirements. In addition, when we evaluated JQ1 together with another latent HIV-1 activator, SAHA (suberoylanilide hydroxamic acid), a bifunctional cell-differentiating agent and non-specific histone deacetylase inhibitor, we saw a modest improvement in HIV-1 gene expression in the J-Lat cell line A2, when compared to JQ1 treatment alone (Fig. S3D (Archin et al., 2009; Richon et al., 1998)).
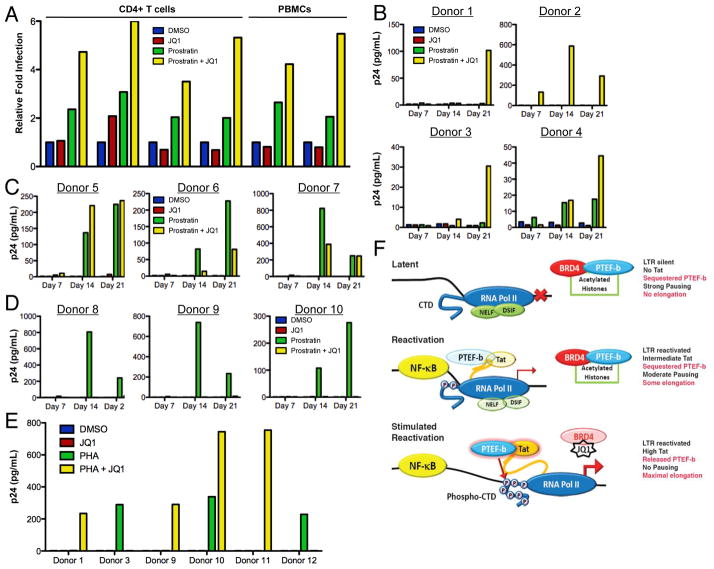
Based on the enhanced viral activation produced by JQ1 alone or together with the HDF-activators in the cell line latency models, we next tested these molecules in the setting of newly infected primary human T cells. Peripheral blood mononucleocytes (PBMCs) from uninfected volunteers were isolated, then either selected for CD4+ expression (donors 1–4), or depleted of CD8+ T cells (donors 5, 6). The cells were stimulated with plate bound CD3 and CD28 antibodies together with IL-2. Pretreatment of these activated T cells with JQ1 together with either PMA (donors 1, 2) or Prostratin (donors 3–6), led to substantial increases in HIV-1-encoded reporter gene expression in all cases (Fig. 4A). Interestingly, under these conditions, JQ1 alone had little to no effect on HIV-1 gene expression similar to what was seen with the J-Lat T cell lines.
Figure 4. BRD4 inhibition in combination with HIV-1 dependency factor (HDF)-activators enhances both HIV-1 replication in primary human T cells, as well as HIV-1 reactivation in latently infected T cells from patients on long term ART.
(A) Primary CD4+ T cells or PBMCs were isolated from HIV-1 negative donors and treated with the indicated compounds (JQ1 (500 nM), PMA (200 nM), or Prostratin (1 μM)) alone or in combination for 1 hr, then infected with VSV-G NL4-3-GFP HIV-1. After 72 hr the number of GFP expressing cells was determined by flow cytometry and normalized to the DMSO control. Values represent the mean, these data are representative of 2 to 4 independent experiments.
(B, C, D) CD8-depleted PBMCs isolated from ART-treated patients were rested for 3d in the presence of Nevaripine (600 nM), followed by reactivation with DMSO or Prostratin (250 nM) for 48h in the presence or absence of JQ1 (250 nM). Viral replication was assessed by p24 ELISA on day 7, 14, and 21. Donors were numbered by whether JQ1 had a beneficial effect on Prostratin reactivation (B), or a non-beneficial or inhibitory effect (C, D).
(e) As above but assessing reactivation with DMSO or PHA (2 μg/mL) in the presence or absence of JQ1 (250 nM) for 48 h.
(f) Model of BRD4-inhibition reactivating latent HIV-1 transcription. Latent HIV-1 (top panel) is initially reactivated by the binding of transcription factors (i.e. NF-κB) to the LTR, resulting in the production of sufficient Tat to modestly compete for some of the PTEF-b associated with BRD4/chromatin (“Acetylated Histones”) or 7SK-RNP (reactivation, middle panel). Upon antagonism of BRD4 by either chemical inhibition with JQ1 or RNAi, additional PTEF-b is made available from the chromatin-sequestered pool, thus lowering the barrier for interaction with Tat and producing maximal proviral transcription (stimulated reactivation, bottom panel). Enhanced viral transcription leads to the amplification of the feed-forward regulatory loop producing higher levels of Tat that in turn recruit the now more readily available PTEF-b (denoted as more intensely colored Tat and PTEF-b components). RNA pol II = RNA polymerase II, CTD = carboxy terminal domain of RNApol, AC = histone acetylation, NELF and DSIF = negative elongation factors removed after phosphorylation by PTEF-b. See Fig. S4, Tables S1, S2.
To explore the ability of JQ1 to both reactivate and stimulate the replication of HIV-1 within latently infected cells, we tested its effect on human CD4 T cells isolated from 19 ART-treated patients, all of whom had achieved durable viral suppression (plasma HIV RNA levels <75 copies/mL) for a minimum of 52 weeks (Table S1). At analysis, patients had a median period of stable viral suppression of 226 weeks (interquartile range 149 – 390 weeks) and a median CD4+ T cell count of 698 cells/mm3 (interquartile range 595 – 1027). Cryopreserved PBMCs were rested for 3 d in media containing Nevaripine to prevent viral infection (600 nM), then washed and stimulated with either Prostratin (250 nM), Phytohaemagglutinin (PHA, 2 μg/mL), or SAHA (500 nM) for 6–48 h (methods), in the presence or absence of JQ1 (250 nM).
Treatment with Prostratin, PHA, or SAHA resulted in detectable HIV replication in 12 of 19 patient samples (Table S2). Of these 12 samples, 7 showed enhanced viral replication when JQ1 treatment was used in combination with either Prostratin or PHA (Fig. 4B, E), with 3 of the 8 patient samples having a negligible response to Prostratin or PHA treatment in the absence of JQ1 (Donors 1, 2, 11). Interestingly, JQ1 also inhibited the viral replication induced by either Prostratin or PHA in 5 patient samples in the absence of cytotoxicity (Fig. 4B–D, S4A, B). Indeed in two instances, treatment with JQ1 enhanced HIV-1 reactivation with one compound (either Prostratin or PHA) while inhibiting reactivation by the other (Donors 3, 9). A 6h treatment with SAHA reactivated latent HIV-1 in three samples (Donors 2, 5, 7), but in each instance JQ1’s addition suppressed SAHA’s effect. In summary, we found that a heterogeneic response to latent HIV-1 reactivation occurs in T cells from different donors and that in multiple instances JQ1 could augment or synergize with Prostratin or PHA to reactivate latent viruses.
Tat-mediated transcription serves as a critical regulator of HIV-1 activation (Karn and Stoltzfus, 2012; Ott et al., 2011). Our data support the notion that PTEF-b levels are rate-limiting for HIV-1’s transcription in both transformed and primary cells (Isel and Karn, 1999; Kao et al., 1987). In this model, latent HIV-1 (Fig. 4E, top panel) is initially reactivated by the binding of transcription factors (i.e. NF-κB) to the LTR (middle panel). This early event produces sufficient Tat to recruit the limited amount of free PTEF-b, the majority of which is sequestered on chromatin by BRD4. Upon inhibition or depletion of BRD4 additional PTEF-b is released and can interact with Tat, thereby producing maximal transcription (stimulated reactivation, bottom panel). This PTEF-b-dependent stimulation is most significant when BRD4 is inhibited under conditions of high level LTR activity (Prostratin, PMA, PHA or TNF-α), which increases transcriptional initiation and elevate Tat levels. Based on the increase in viral replication which occurs when NF-κB is activated and BRD4 is inhibited, our data demonstrate that in some instances the pairing of JQ1 with HDF-activators more effectively alleviates latency.
Beginning with orthologous large-scale genetic screening for host factors that modulate HIV-1 replication in vitro, we discovered that depletion of BRD4 increased viral replication in transformed cells and enhanced HIV infection in primary human CD4+ T cells. This work significantly extends elegant previous studies demonstrating that BRD4 modulates HIV-1 infection (Bisgrove et al., 2007; Urano et al., 2008). Indeed, we now show that BRD4’s depletion/inhibition stimulated viral gene expression from both newly integrated and latent proviruses. Furthermore, we revealed that BRD4’s loss enhanced HIV-1 gene expression by increasing Tat/PTEF-b association and host polymerase transcriptional elongation.
We found that in multiple instances a small molecule inhibitor of BRD4, JQ1, when used in combination with the transcriptional activators, Prostratin or PHA, could trigger and/or markedly enhance the in vitro reactivation of latent HIV-1 in primary human T cells, thereby identifying a new first-in-class anti-latency small molecule. While this manuscript was in preparation, Banerjee et al. also reported that JQ1 alone could stimulate HIV-1 replication in a T cell line and in the primary T cells from one of three patients receiving ART (Banerjee et al., 2012). Our study now significantly extends those observations by providing mechanistic insight into how BRD4, and thus JQ1, impact the HIV-1 lifecycle. Importantly, we also newly describe that using JQ1 in combination with compounds known to alleviate viral latency, (Prostratin or PHA), greatly increased reactivation of HIV-1 in latently infected primary T cells from 7 of 19 patients on long term ART. Of note, our work shows that JQ1 alone does not reactivate HIV-1 in latently infected primary T cells, which differs from the single example of modest viral reactivation by JQ1 alone recently reported (Banerjee et al., 2012). Our data instead reveal that JQ1 potentiates the actions of several known transcriptional HIV-1 reactivating compounds, PMA, PHA, TNF-α and Prostratin. Therefore in comparison our study demonstrates an enhanced potential for the use of agents that improve transcriptional elongation, i.e. JQ1, in reactivating latent HIV-1 reservoirs, via their pairing with small molecules that mobilize LTR-enhancing HDFs (i.e. NF-κB). Furthermore by using such combinations to create synergies it may be possible to improve the efficacy and minimize the toxicity of anti-latency therapy (Choudhary and Margolis, 2011; Siliciano and Greene, 2011). A common mechanism of HIV-LTR activation shared by these compounds is the stimulation of NF-κB. In certain instances NF-κB’s activity is dependent on interaction with BRD4 (Sharma et al., 2007). However Huang et al. determined that NF-κB transcriptionally activates a subset of genes in a BRD4-independent manner and our data are consistent with the HIV-LTR being one such example (Huang et al., 2009).
In contrast to data generated using several transformed cell lines or newly infected primary T cells, we observed a marked heterogeneity in the ability of the tested compounds to reactivate latent HIV-1 in primary T cells from patients on long term ART. We also noted that the addition of JQ1 along with these HDF-activating compounds could either promote or inhibit viral reactivation. While the dichotomous effects of JQ1 are likely attributable to many factors, insight may come from earlier work showing that BRD4 is needed for basal HIV-1 LTR transcription, suggesting that interfering with this early action of BRD4 may prevent further increases in replication (Ott et al., 2011); additional possibilities include cell lineage-dependent variations of BRD4 or PTEF-b expression or availability. Together these results suggest that it will be useful to carry out personalized in vitro testing to identify efficacious combinations of HIV-1 reactivators, including JQ1.
Our work also demonstrates that in addition to HDFs that are needed by HIV-1 (Brass et al., 2008), and HIV-1 restriction factors (HRFs) that have evolved to specifically inhibit the virus (Yan and Chen, 2012), there exists a third functional class of host factors, HIV-1 competitive factors (HCFs), i.e. BRD4. Such factors have not been selected to be anti-viral per se, but none-the-less can blunt infections and promote latency by competing with HIV-1 for limiting cellular resources, i.e. PTEF-b. It follows then that anti-latency strategies might be improved by combining HDF-activating therapies together with small molecules that transiently inactivate specific HRFs and/or HCFs. Because HIV-1 latency exists via multiple mechanisms, we deem it likely that continuing to improve and integrate our basic understanding of how host proteins modulate latency will provide additional insights and in so doing help guide the way to a cure.
Experimental Procedures
Small molecules
Prostratin and SAHA were from Santa Cruz (sc-203422, sc-220139). PMA was from Sigma (P1585). Compounds were used at optimized concentrations: JQ1 (500 nM), Prostratin (1 μM), PMA (200 nM), SAHA (0.5 μM). Please see Supplement.
Genetic screening for HIV-1 modulators using orthologous RNAi libraries
To identify host factors which modulate HIV-1 replication, we performed RNAi screens using two genome-wide siRNA resources (Silencer Select, Ambion; Library 2, Mission esiRNAs, Sigma) following a previous protocol (Brass et al., 2008).
siRNAs
BRD4 siRNAs and shRNAs. All siRNAs were used at 50 nM final concentration using Oligofectamine transfection lipid (Invitrogen).
Study Subjects
HIV-infected adults and HIV-seronegative controls were recruited from outpatient clinics at Massachusetts General Hospital and affiliated hospitals. The respective institutional review boards approved this study, and all subjects gave written consent.
Supplementary Material
Acknowledgments
We thank the ICCB-L; C. Shamu, J. Smith, S. Chang, S. Rudnicki, S. Johnston, K. Rudnicki, and D. Wrobel; Ragon Institute (RI); M. Boyarina, K. Donnelly, P. Richtmeyer. Brigham and Women’s Hospital; M. Hinely. We thank X. Yu (RI), and M. Lichterfeld (MGH) for helpful discussions. We thank J. Rohrbach and D. Kavanagh (RI) for help with primary T cell experiments. This research was funded by a generous donation from the Bill and Melinda Gates Foundation. A.L.B. is grateful to the Charles H. Hood Foundation, the Phillip T. and Susan M. Ragon Foundation, the Harvard and UMass CFARs, and the NIH (1R01AI091786) for their generous support; S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Author contributions:
J.Z., G.D.G., S.J.E., and A.L.B designed the study; J.Z., G.D.G., S.J.E., and A.L.B wrote the manuscript; J.Z., S.P.J., G.D.G., G.G., C.R.C., H.Q., T.P., S.J.E. and A.L.B performed experiments and analyzed data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012 doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Cannon P, Kim SH, Ulich C, Kim S. Analysis of Tat function in human immunodeficiency virus type 1-infected low-level-expression cell lines U1 and ACH-2. J Virol. 1994;68:1993–1997. doi: 10.1128/jvi.68.3.1993-1997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JK, Greene WC. Dynamic roles for NF-kappaB in HTLV-I and HIV-1 retroviral pathogenesis. Immunol Rev. 2012;246:286–310. doi: 10.1111/j.1600-065X.2012.01094.x. [DOI] [PubMed] [Google Scholar]
- Choudhary SK, Margolis DM. Curing HIV: Pharmacologic approaches to target HIV-1 latency. Annu Rev Pharmacol Toxicol. 2011;51:397–418. doi: 10.1146/annurev-pharmtox-010510-100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl V, Palmer S. Establishment of drug-resistant HIV-1 in latent reservoirs. J Infect Dis. 2009;199:1258–1260. doi: 10.1086/597760. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Ott M, Van Lint C, Amella CA, Verdin E. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J Virol. 1998;72:1666–1670. doi: 10.1128/jvi.72.2.1666-1670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Van Lint C, Fischle W, Paras P, Jr, Ott M, Brady J, Verdin E. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc Natl Acad Sci U S A. 1996;93:6377–6381. doi: 10.1073/pnas.93.13.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. Host factors mediating HIV-1 replication. Virus Res. 2011;161:101–114. doi: 10.1016/j.virusres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Gulakowski RJ, McMahon JB, Buckheit RW, Jr, Gustafson KR, Boyd MR. Antireplicative and anticytopathic activities of prostratin, a non-tumor-promoting phorbol ester, against human immunodeficiency virus (HIV) Antiviral Res. 1997;33:87–97. doi: 10.1016/s0166-3542(96)01004-2. [DOI] [PubMed] [Google Scholar]
- Hakre S, Chavez L, Shirakawa K, Verdin E. HIV latency: experimental systems and molecular models. FEMS Microbiol Rev. 2012;36:706–716. doi: 10.1111/j.1574-6976.2012.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J, Stoltzfus CM. Transcriptional and Posttranscriptional Regulation of HIV-1 Gene Expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM. Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep. 2010;7:37–43. doi: 10.1007/s11904-009-0033-9. [DOI] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RJ, Trono D, Feinberg MB, Baltimore D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci U S A. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnolzer M, Verdin E, et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, George AA, Singh BN, Sahoo NC, Rao KV. Regulation of transcript elongation through cooperative and ordered recruitment of cofactors. J Biol Chem. 2007;282:20887–20896. doi: 10.1074/jbc.M701420200. [DOI] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV Latency. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano E, Kariya Y, Futahashi Y, Ichikawa R, Hamatake M, Fukazawa H, Morikawa Y, Yoshida T, Koyanagi Y, Yamamoto N, et al. Identification of the P-TEFb complex-interacting domain of Brd4 as an inhibitor of HIV-1 replication by functional cDNA library screening in MT-4 cells. FEBS Lett. 2008;582:4053–4058. doi: 10.1016/j.febslet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Fenard D, Bisgrove D, Verdin E, Greene WC. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279:42008–42017. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Xing S, Shan L, O’Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009;119:3473–3486. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Gilmour DS, Henderson AJ. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem. 2007;282:16981–16988. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- Zhou M, Huang K, Jung KJ, Cho WK, Klase Z, Kashanchi F, Pise-Masison CA, Brady JN. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J Virol. 2009;83:1036–1044. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.