Abstract
We have designed a highly specific inhibitor of calpain by mimicking a natural protein-protein interaction between calpain and its endogenous inhibitor calpastatin. To enable this goal we established a new method of stabilizing an α-helix in a small peptide by screening twenty-four commercially available crosslinkers for successful cysteine alkylation in a model peptide sequence. The effects of crosslinking on the α-helicity of selected peptides were examined by CD and NMR spectroscopy, and revealed structurally rigid crosslinkers to be the best at stabilizing α-helices. We applied this strategy to the design of inhibitors of calpain that are based on calpastatin, an intrinsically unstable polypeptide that becomes structured upon binding to the enzyme. A two-turn α-helix that binds proximal to the active site cleft was stabilized, resulting in a potent and selective inhibitor for calpain. We further expanded the utility of this inhibitor by developing irreversible calpain family activity-based probes (ABPs), which retained the specificity of the stabilized helical inhibitor. We believe the inhibitor and ABPs and will be useful for future investigation of calpains, while the crosslinking technique will enable exploration of other protein-protein interactions.
Introduction
The primary goal of this work was to design and synthesize α- helical inhibitors as well as activity-based probes of human calpain, a calcium-regulated cysteine protease involved in a myriad of normal and pathological biological processes.1–12 Although there has been considerable interest in the design of α-helical peptides for the study of protein-protein/receptor-ligand interactions and drug design, to our knowledge, there has been no work to date investigating α-helices as protease inhibitors.
Inhibitor design for this class of enzyme has historically focused on the use of peptidomimetics that fit into the active site cleft in a substrate-like manner and utilize covalent, reversible or irreversible reactive groups to react with the active site cysteine.13–20 The problems with this approach are twofold: 1) the papain super-family has a highly conserved active site cleft, which complicates identification of peptidomimetic side chains that differentially bind to individual enzymes, and 2) small peptides do not bind well to calpains.
To overcome this problem we took inspiration from the recent co-crystal structure of calpain with its endogenous protein inhibitor, calpastatin and from calpain inhibitors containing constrained scaffolds or macrocycles.21–25 Calpastatin is unstructured in solution; however, upon binding to active calpain it drapes across the entire protein and undergoes structural rearrangements to form three α-helices that contact three different domains of the enzyme. One of these α-helices binds adjacent to the prime side of the active site cleft (Figure 1), forming a number of energetically favorable interactions between apolar sidechains that become buried upon complex formation. We therefore hypothesized that this α-helical motif would provide increased specificity via its unique binding mode since the helix avoids the highly conserved region of the active site while still inhibiting substrate access to the active site cleft.
Figure 1.
X-ray crystal structure of the calpain 2-calpastain complex (PDB ID: 3BOW). Key residues on the inhibitor, calpastatin, (purple) and calpain-2 (black) are labeled.
This two-turn α-helix represents a ten-residue peptide. Previous work indicated that small peptides were poor inhibitors of calpains. 26,27 We corroborated this idea by determining that the minimal calpastatin fragment peptide that formed the two-turn α-helix (IPPKYRELLA) did not inhibit calpain (Ki >100 µM). We reasoned that the entropic cost of forming an α-helix from a random coil limited the ability of small peptides to inhibit the enzyme; thus we decided to design a stabilized version of this peptide to minimize unfavorable conformational entropy.
Several strategies have previously been developed for α-helix stabilization involving main- or side-chain modifications including: disulfide bond formation,28–30 hydrogen bond surrogates,31,32 ring closing metathesis,33–36 cysteine alkylation using α-haloacetamide derivatives37 or biaryl halides,38 lactam ring formation,39–45 hydrazone linkage,46 oxime linkage,47 metal chelation,48,49 and “click” chemistry.50,51 Of the different methods used to stabilize these structures, the inclusion of a semi-rigid cross-linker52–60 has been particularly successful, and is explored herein.
Results and Discussion
1. Design of template-constrained cyclic peptides stabilizing an α-helix conformation
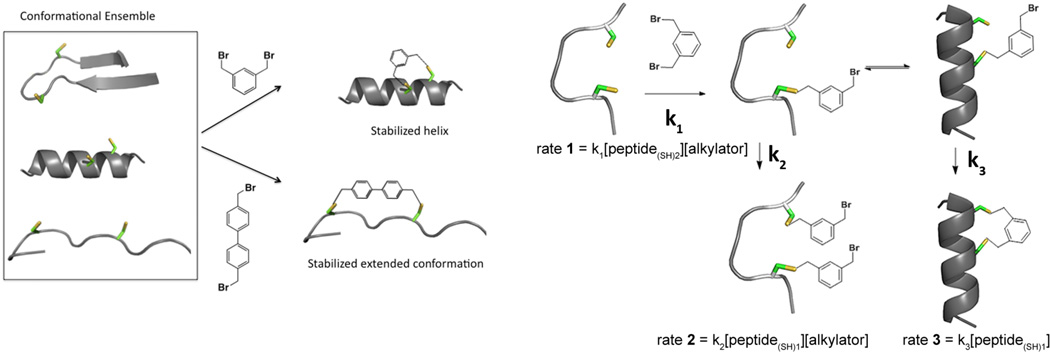
Peptides are intrinsically flexible chains, which rapidly interconvert among a large ensemble of conformations, including canonical secondary structures (helices, reversed turns, β-hairpins, etc.). Generally, only one of these conformations is required to bind a given receptor/enzyme, and very large changes in affinity (> 104) can be realized by simply restricting the structure to a single conformational state.
We were particularly interested in conformational restriction via cysteine alkylation61–64 for its chemical stability, selectivity, cost effectiveness, and ease of introduction via standard mutagenesis into recombinantly expressed peptides or proteins or by solid-phase peptide synthesis. Importantly, a number of structurally diverse thiol reactive crosslinkers are also commercially available. Thus, we envisioned that the bioactive conformation of a given peptide could be stabilized by identification of the optimal cysteine crosslinker from screening a library of crosslinkers on a peptide with two cysteines anchored in appropriate positions. We refer to α-helical peptides stabilized in this manner as template-constrained peptides.
Figure 2 (left) shows the fundamental concept of template-constrained cyclic peptides, in this case accomplished via sidechain-to-sidechain cyclizations. To do this, a pair of cysteine residues is installed at appropriate positions in order to stabilize a local conformation. Here, we placed the cysteine residues at i, i+4 positions, because this spacing brings two thioether residues into proximity when in the α-helix. In a series of parallel reactions we react the peptide with an indexed array of different crosslinking agents. Bis-alkylators with sufficient reactivity to alkylate thiols will cleanly form cyclic peptides, if the macrocycle can be formed in a low-energy conformation that matches one of the low-energy conformations of the peptide. For example, a meta-xylyl group, which matches the inter-thiol distance of the cysteine sidechains when in an α-helical conformation, should stabilize this helical structure. By contrast, the much longer distance of the 4,4’-biphenylmethyl group would not be consistent with the α-helical conformation, and would instead favor formation of a more extended conformation. Thus, depending on the template, it should be possible to stabilize any one of a number of conformations.
Figure 2.
Conformational restriction via crosslinking (left). Kinetic “selection of the fittest” reaction. Hypothetical rate constants are denoted by k1, k2, and k3 (right).
We use a kinetic “selection of the fittest” method, to screen for only those linkers that help select stable, low-energy conformations over more strained conformations. The kinetic scheme for cyclization requires two steps (Figure 2, right): The first step involves the second-order alkylation of the dithiol-peptide, which depends on the concentration of both the alkylating agent and the peptide (rate 1 = k1[peptide(SH)2][alkylator]). The rate of this reaction depends on the chemical nature of the alkylator, but to the first approximation is largely independent of the peptide structure, which is largely in a random coil in the linear form. Once mono-alkylated, the second- order process of reacting with a second equivalent of the alkylating agent (rate 2 = k2[peptide(SH)1][alkylator]) will compete with the desired first-order cyclization process (rate 3 = k3[peptide- (SH)1]). (Solvolysis reactions of the mono-alkylated product also compete with cyclization.) The cyclization reaction depends on the ability of the peptide to reach a stable, strain-free conformation as it enters the transition state for cyclization, which we presume is geometrically similar to the product for large macrocyclic rings such as those formed here. Thus, the ratio of bis-alkylated to monoalkylated compound provides a quantitative measure of the ease of cyclization that is dependent on the conformation of the cyclic form of the peptide. Bis-alkylation is dependent on the concentration of the peptide while cyclization is independent of this parameter, therefore it is possible to select for the most efficient crosslinkers by simply running the reaction at a fixed peptide concentration with increasing concentrations of bis-alkylators and examining the product distribution by mass spectrometry.
In summary, the current method of template-constrained thioether cyclization involves several steps: 1) Screening for crosslinking agents with appropriate reactivity and ability to form cyclic products under favorable conditions with nearly equimolar amounts of peptide and bis-alkylator. 2) Examining bis-alkylator “hits” with increased stringency, using higher molar concentrations of alkylators in large excess of the peptide. This step should provide template-constrained peptides with relatively strain-free conformations. 3) Testing the template-constrained peptides to determine which have been stabilized in the appropriate conformation. This can easily be accomplished by circular dichroism (CD) spectroscopy for an α-helix. 4) Finally, determining the impact of stabilizing the helix on the ability of the peptide to bind to a protein known to recognize the sequence in a helical conformation.
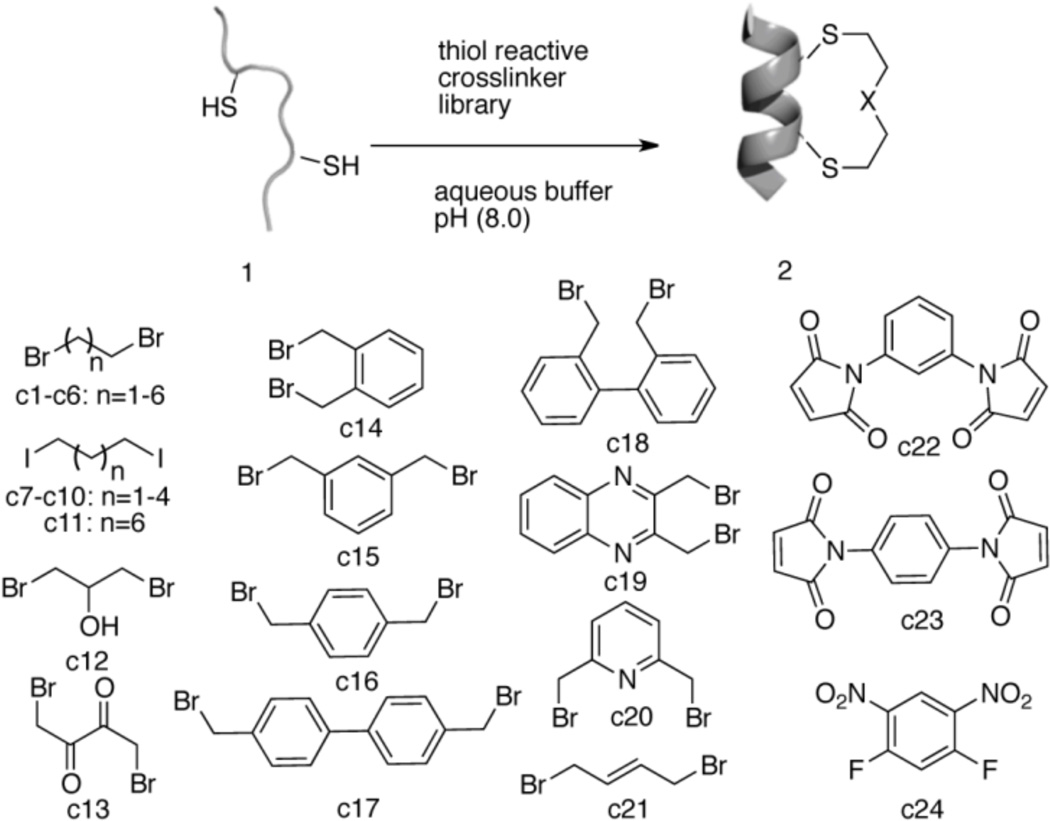
To explore template-constrained cyclization to stabilize α-helices in aqueous solution, we used the model peptide 1 (sequence: Ac-YGGEAAREACARECAARE-CONH2) which was similar to the FK-4 peptide previously described (Table S1 Supporting Information).65 The model peptide exhibited a low to moderate level of helicity without any stabilization.
We screened twenty-four crosslinkers for cys-thioether macrocyclizations. The crosslinkers included alkyl bromides c1-c6 & c12-c13, alkyl iodides c7-c11, benzyl bromides c14-c20, allyl bromide c21, maleimides c22-c23 and an electrophilic difluorobenzene c24 (Scheme 1). The initial screening reaction was performed in a 96-well plate format to identify crosslinkers that react with cysteine thiols under mild conditions (bicarbonate buffer (pH = 7.5 to 8.0)) at room temperature. The crude reaction mixture was analyzed by MALDI-TOF mass spectrometry to identify any crosslinker that was a “hit”. Additional HPLC profiling can characterize product distribution.
Scheme 1.
Helix stabilization via screening of 24 crosslinkers.
Product distribution was analyzed using MALDI-TOF and revealed that cysteine alkylation did not occur when simple alkyl halides c1-c12 were used; only intramolecular disulfide bond formation due to oxidation was observed to occur.66 Even when the leaving group was changed from bromide to the more reactive iodide c7-c11 alkylation reactions failed under these aqueous conditions. The crosslinking reaction with 1,4-dibromo 2,3-butanedione c13 produced a complex mixture of products. Crosslinking reactions with the maleimide crosslinkers c22-c23 also resulted in a mixture of epimeric products that were further complicated by hydrolysis of the imide (Figure S1 Supporting Information). Reactions using 1,5-difluoro-2,4-dinitrobenzene c24 resulted in a similar complex mixture of products. For the biaryl derivatives c17, c18, predominantly unreacted peptide was detected (MALDITOF and HPLC) accompanied by traces of the desired, cyclized product (Figures S1 & S2 Supporting Information).
The cleanest macrocyclization resulted from the reaction67,68 with benzylic/allylic halides c14-c16 & c19-c21, which provided the major peak of the cyclization product as seen by MALDI-TOF and HPLC trace analysis (Figures S1 & S2 Supporting Information). We then tested the crosslinker “hits” c14-c16 & c19-c21 under the conditions designed to increase the rate of bis-alkylation over cyclization (by increasing the concentrations of alkylating agent and peptide in solution). HPLC analysis of the “selection of the fittest” showed that the 1,3-bis(bromomethyl) benzene (α, α’-dibromo-m-xylene) crosslinker c15 and 2,6- bis(bromomethyl)pyridine crosslinker c20 gave the cleanest formation of the desired macrocycle (Figure S3 Supporting Information). By contrast, crosslinking with allyl crosslinker c21 produced multiple peaks. It is interesting that the m-xylene crosslinker c15 was most successful crosslinker out of the three α,α’-dibromoxylenes c14-c16, considering that all the three alkylating agents have relatively different reactivity profiles (ortho> meta > para). 62
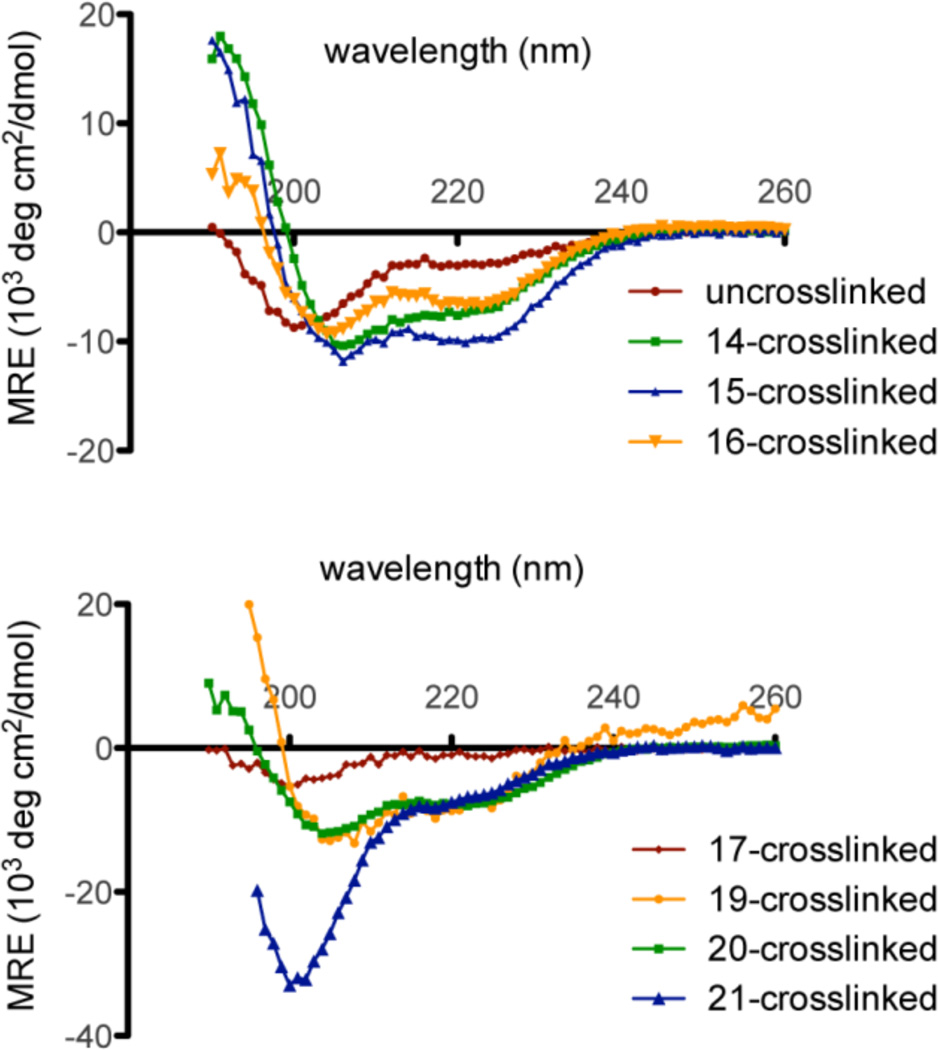
We next evaluated the CD spectra of these selected template constrained cyclic peptides to determine the effect of the template on their coil-helix equilibria (Figure 3). The determination of secondary structure was complicated somewhat by the fact that the spectra are generally interpreted using the intensity of θ222, which requires knowledge of the concentration69, generally by measuring the absorbance of an N-terminal Tyr residue. Some of our linkers contain aromatic groups that could absorb at 278 nm and complicate concentration determination. Therefore, we use dry weight to estimate the concentration, which results up to a 25% error in concentration determination (assessed by comparing gravimetric versus spectrophotometric determination of peptides containing Tyr chromophores and lacking other groups). Because θ222 is not accurately measured, we therefore interpret the data largely based on the shape of the spectra, particularly the ratio of the peak shape and relative intensities of the two exciton-coupled π- π’ bands at 190 nm and 208 nm relative to that of the n- π’ band near 222nm.70 The three xylene-based crosslinkers c14-c16 all showed an increase of the helicity in the CD spectroscopy analysis. Notably, the m-xylene based crosslinker c15 showed the most increase in helicity followed by o-xylene c14 and finally p-xylene c16.
Figure 3.
CD spectra of the model peptide and the crosslinked peptides in phosphate buffer [50mM, pH=7.0, 25 °C].
Interestingly, the CD spectrum of the crosslinked peptides by crosslinkers c17 and c21 showed some structural differences from those seen using the xylene crosslinkers. As expected, the 4,4’- biphenyl (c17) crosslinked peptide showed little helicity, likely due to destabilization of the α-helix and stabilization of an extended conformation of the peptide because the end-to-end length of the biphenyl template is much longer than the typical α-helix pitch. Likewise, peptide crosslinked with the butenyl derivative c21 showed a CD spectrum with a deep minimum near 200 nm, similar to that of the random coil (Figure 3). It would be interesting to test whether this peptide, after the reduction of the double bond, could stabilize a 310 helix as shown in the Grubbs’s work35. This crosslinker could be an alternative to ring closing metathesis (RCM) stapling and subsequent double bond reduction strategy.
Heterocyclic templates were also capable of stabilizing the α- helix. 2,3-quinoxaline c19 and 2,6-pyridine c20 crosslinked peptides showed CD spectra similar to those of the o-xylene c14 and m-xylene c15 crosslinked peptides (Figure 3).
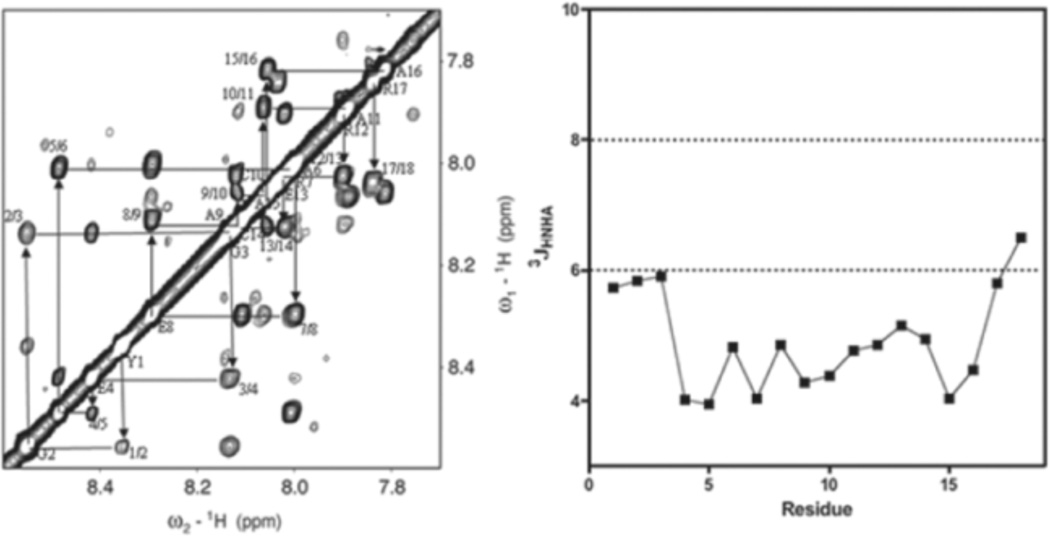
NMR spectroscopy experiments demonstrate that the cyclic template restraint strongly stabilized the helical conformation within the macrocyclic ring, and that the helix extended towards the C-terminus of the peptide (Figure 4). Typical stepwise NH(i)/NH(i + 1) NOE connections were observed from the first residue to the last residue, which are indicative of a helical conformation. Closer inspection showed that the cross-peak intensity became stronger after the residue 6, suggesting that the crosslinked region in the helix was more organized than frayed region of the N-terminus, which included two glycines. Furthermore, 3JNH-HA coupling was evaluated by the INFIT (inverse Fourier transformation of in-phase multiplets) procedure.71 The J coupling constant is a good indicator of secondary structure. It is generally averaged to ~7 Hz if the residue is in a random coil or in equilibrium between different structures. It is less than 6 Hz if it is in α-helical structure and is larger than 8 Hz if the secondary structure is a β-sheet. Our J coupling constant was mostly below 6 Hz suggesting an α-helical structure. In addition, the chemical shift index of α-H strongly demonstrated helix formation even in the fraying N-terminus. Secondary chemical shifts which were calculated by subtracting the experimental values from the intrinsic values and clearly showed the effect of the crosslinker. The most dramatic changes were observed on Cys10, Ala11, Arg12 and Cys14, influenced in part by the anisotropy effect from the benzene ring in the crosslinker (Figure S4 Supporting Information).
Figure 4.
NMR of m-xylyl c15-constrained cyclic peptide (left). NOE sequential walk of backbone amide region of NOESY (250 ms) for the peptide. The cross peaks are labeled as NH(i)/NH(i + 1) 3JNH-HA coupling as function of residue (right). The small 3JNH-HA(< 6Hz) and strong sequential NH-NH NOEs denote helix formation in the peptide.
2. Application of i, i+4 m-xylene crosslinker-based stabilization for calpain inhibitor design
Turning back to calpain inhibitor design we chose to use the calpastatin fragment IPPKYRELLA (previously shown to be inactive against calpain) as the backbone since this sequence, in the context of full-length calpastatin, forms a two-turn helix in the prime side of the active site of calpain-1 as shown in figure 1. Three different sets of double cysteine mutants, 3a-c, along with their m-xylene crosslinked partners, 3a-c, were synthesized (Figure 5, Table S3 Supporting Information). Cysteine locations were chosen by both visual inspection and virtual alanine scanning mutagenesis (Table S2 Supporting Information) so as not to disturb key interactions at the protein-helix interface, which includes Pro51 (inhibitor) ring stacking against Trp288 (calpain) and Tyr54 (inhibitor) H-bonding to His169 (calpain) as shown in Figure 1.
Figure 5.
Sequence of double cysteine mutants (3a, 3b, and 3c) and their crosslinked counterparts (3a, 3b, and 3c) (left). A helical wheel representation to indicate the crosslinked regions (right).72
 denotes the m-xylyl c15 crosslinking between the cysteines.
denotes the m-xylyl c15 crosslinking between the cysteines.
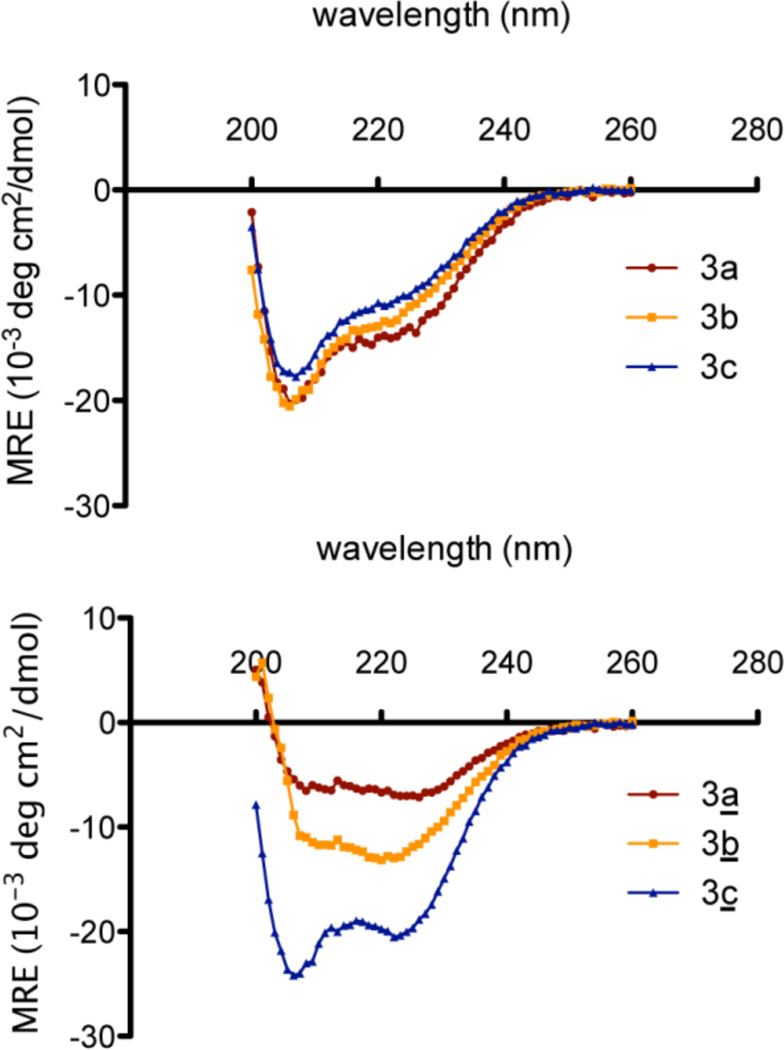
Next, the difference in structural changes as a result of cysteine crosslinking was examined via CD spectroscopy (Figure 6).69,73 The helical content of the uncrosslinked peptides was low in the absence of added trifluoroethanol (TFE), so the experiments were conducted in the presence of 40% TFE.74 CD analysis revealed a clear trend whereby all unlinked peptides showed little secondary structure, while the crosslinked peptides demonstrated varying degrees of α-helicity. Peptide 3c showed the greatest helicity after crosslinking, followed by 3b, while 3a showed negligible helicity after crosslinking. The lack of increased helicity for 3a may be due to the fact that it lacks the proline that is frequently found as an helix initiator of an α-helix.75 A possible salt bridge between the glutamic acid and lysine may also be enhancing helical content in 3c.76–78 Thus, we believe that the primary sequence of the peptide as well as the crosslinker can influence the final helical content of the product peptide.
Figure 6.
CD spectra of uncrosslinked peptides 3a-c (top) and crosslinked peptides 3a-c (bottom), [~125 µM peptide, 50 mM Tris (pH 7.5), 40% TFE)]. Crosslinked peptide 3c demonstrates the greatest helical content. (See Figure S5 & S6 Supporting Information for CD analysis without 40% TFE.)
The inhibitors, both crosslinked and uncrosslinked, were tested for their ability to inhibit calpain-1 (Table 1, Figure S7 & S9 Supporting Information). No appreciable inhibition (Ki>100 µM) of calpain-1 was observed for the uncrosslinked peptides 3a-c. These results corroborate previous reports stating that the minimum length of a standard calpastatin derived peptide needed to achieve reasonable calpain inhibition is 27 amino acids long.79 However, the crosslinked peptide, 3c, which is only 10 amino acids long, showed good inhibition of calpain-1 in the low micromolar range (Table 1, Figure S9 Supporting Information). Furthermore, a trend relating higher helical content (Figure 6) positively correlated with better inhibition of calpain-1 (Table 1). This trend is likely directly related to helical content stabilized by the crosslinker c15, although it is also possible that the crosslinker itself could contribute to enzyme recognition of the inhibitor.
Table 1.
Ki against calpain-1.80 The calpain assay was done as described in Materials and Methods.
| Peptide | 3a | 3b | 3c | 3a | 3b | 3c |
|---|---|---|---|---|---|---|
| Calpain-1 (µM) |
>100 | >100 | >100 | >100 | 95.6 ± 25.5 | 10.2 ± 2.9 |
Kinetic studies were then performed to understand the mechanism of 3c inhibition of calpain-1; standard Michaelis- Menten and Lineweaver-Burke analysis showed that 3c behaved as a competitive inhibitor (Figure 7, Figure S10 & Table S4 Supporting Information). These results are consistent with the idea that 3c binds to the α-helix binding site in the primed side of the active site of calpain and physically blocks substrate binding, and subsequently proteolysis, as predicted from the initial co-crystal data (Figure 1).
Figure 7.
Lineweaver-Burke analysis shows that calpain inhibitor 3c to be a competitive inhibitor. Lineweaver-Burke plot was constructed from standard Michaelis-Menten kinetics.
There has been considerable difficulty in achieving good selectivity within the papain superfamily of enzymes as these enzymes contain highly conserved active sites.21,81 To determine whether the helical inhibitor 3c was specific for calpain we tested it against a set of canonical papain family cysteine proteases including: papain, cathepsin B and cathepsin L (Table 2, Figure S11 Supporting Information). Significantly, no inhibition (Ki > 100 µM) was observed using the crosslinked peptide 3c against papain or cathepsin B. The inhibitor was about four fold more potent against calpain over cathepsin L (Ki 39.9 ± 1.09 µM). These results indicate that this α-helical motif may represent a uniquely selective binding element for inhibition of calpains and further validates our structure-based approach. Furthermore, structure activity relationship studies of these helical inhibitors may result in a more potent and specific inhibitors of calpain and also shed some light on to how the calpastatin helix interacts with human calpains.
Table 2.
The Ki of crosslinked inhibitor 3c against other papain family proteases.
| Enzyme | Calpain-1 | Papain | Cathepsin B | Cathepsin L |
|---|---|---|---|---|
| 3c (µM) | 10.2 ± 2.9 | >100 | >100 | 39.2 ± 1.1 |
The crosslinking reaction was performed with the crosslinker c15 and the three peptides in aqueous buffer system. However, in instances where there are multiple cysteines, we believe that solid-phase cysteine crosslinking could be useful for selective crosslinking. To this end, we tested the on-resin crosslinking the peptide 3c. Fmoc-Cys(Mmt)-OH was used instead of Fmoc-Cys(Trt)-OH and selective deprotection of specific cysteine side chains was achieved by 1% TFA/DCM treatment while the peptide was still resin bound.82,83 (See the Materials and Methods). The same kinetic results were achieved with on resin crosslinked inhibitor.
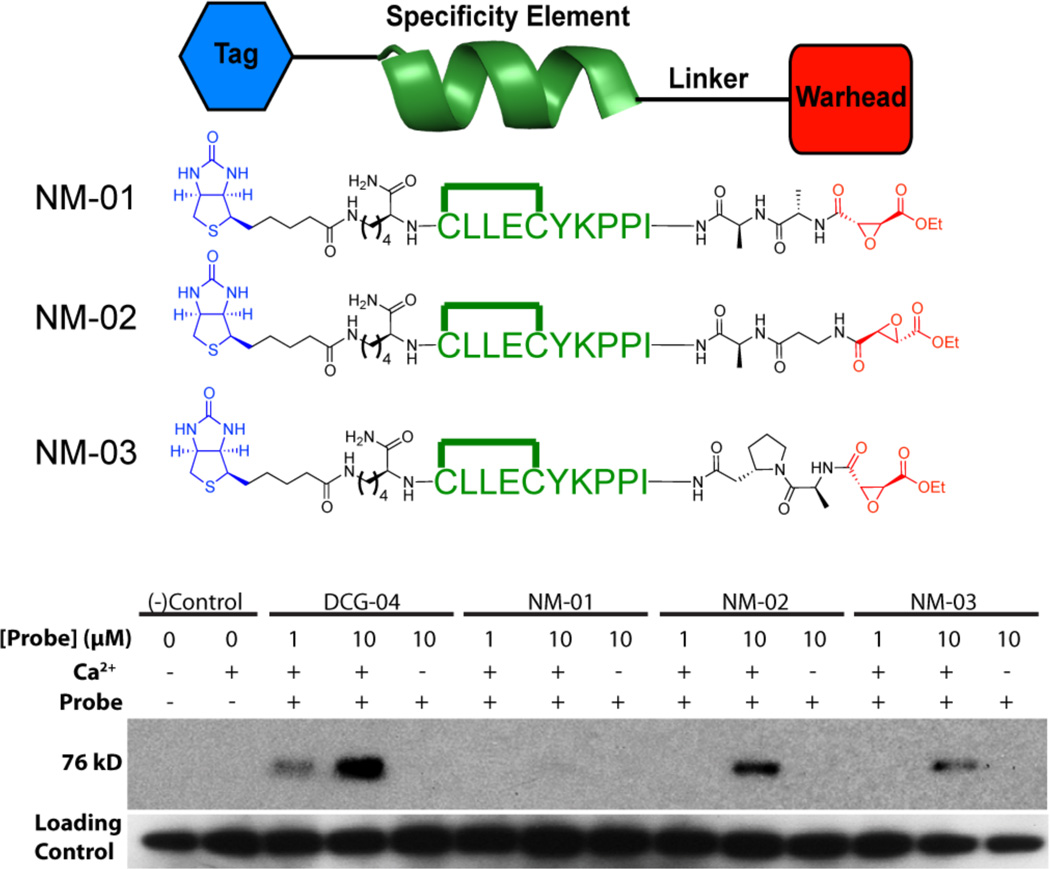
Based on our initial success with a stabilized, α-helical-based inhibitor of calpain we next endeavored to develop an activitybased probe (ABP) specific for calpains. ABPs are complementary chemical tools to traditional genomic and proteomic techniques; ABPs are used for identification of enzymatic targets and to evaluate dynamics of enzyme activity regardless of levels of expression.84- 89 This is important because in many cases translation and transcription do not correlate with enzyme activity90; this is especially true for calpains as their proteolytic activity is finely regulated post-translationally by intracellular calcium levels. Basic ABP design includes a mechanism based inhibitor, a specificity element, and a tag (Figure 8, top). In this case, the crosslinked peptide 3c was used for the specificity element and the succinyl epoxide functions as the warhead group that reacts with the cysteine thiol. This warhead has been established to react in a mechanism dependent manner only with active papain family proteases91. Three dipeptide linkers (NM-01, 02 and 03) of different lengths and rigidities were chosen via visual inspection in PyMOL92 based on the crystallographic structure of calpastatin-bound calpain 2 (PDB code 3BOW).21 Lastly, we chose to use either biotin or fluorescein isothiocyanate (FITC) as a tag.
Figure 8.
Design of a calpain specific ABP (top). ABPs contain a mechanism based inhibitor, specificity element, and tag. Only the chemical structures ABPs containing a biotin tag are shown here.  denotes the m-xylyl c15 crosslinking between the cysteines. ABP binding to calpain-1 (bottom). The linker length and rigidity between the crosslinked peptide and succinyl epoxide was evaluated via reaction with calpain-1 in vitro. A five-carbon backbone, flexible linker appears optimal. Loading control lanes beneath the panel show Western blot analysis using anti-calpain-1.
denotes the m-xylyl c15 crosslinking between the cysteines. ABP binding to calpain-1 (bottom). The linker length and rigidity between the crosslinked peptide and succinyl epoxide was evaluated via reaction with calpain-1 in vitro. A five-carbon backbone, flexible linker appears optimal. Loading control lanes beneath the panel show Western blot analysis using anti-calpain-1.
We used three different amino acid sequences as linkers: alanine-alanine, β-alanine-alanine, and alanine-β-homoproline, (NM-01, NM-02, and NM-03, respectively) (Table S5 Supporting Information). NM-01 is the shortest linker by one carbon but has similar flexibility as NM-02. NM-02 and NM-03 should cover a similar distance between the helix and succinyl epoxide, however the β-homoproline provides more rigidity than the β-alanine.
To evaluate the best linker, we initially tested biotinylated versions of either NM-01, -02, or -03 on purified, activated calpain-1 at two concentrations, 1 and 10 µM, and on unactivated calpain at 10 µM (Figure 8, bottom). Each ABP was added to purified calpain (pH 7.0), followed by the addition of calcium to activate the enzyme. The probe was allowed to react for 20 min. at room temperature. No calcium addition was used as a control to demonstrate that labeling only occurred with active calpain, and DCG-04, a pan-papain family cysteine protease ABP91, was used as a positive control as it is known to label calpains. Samples were analyzed by SDS PAGE electrophoresis; proteins were transferred to PVDF membrane and analyzed by western blot for biotin using streptavidin-HRP. Our results show that two ABPs, NM-02 and NM-03, labeled calpain in an activity dependent manner, which indicated that an extra carbon in the amino acid backbone of the linker was necessary for the epoxide to react with the active site cysteine (Figure 8). The intensity of the bands in the blot suggested that the use of the linker β-alanine-alanine resulted in the most potent probe (NM-02) (Figure 8, bottom). The ABP with the alanine-β-homoproline linker (NM-03) also bound to calpain but the rigidity in the linker induced by the pyrrolidine ring in homoproline may have contributed to less labeling. These results further support our hypothesis that the helix is binding at the active site as measurements of the probe visualized in PyMOL92 show that a β-alanine-alanine linker would position the epoxide at the correct distance from the active site cysteine.
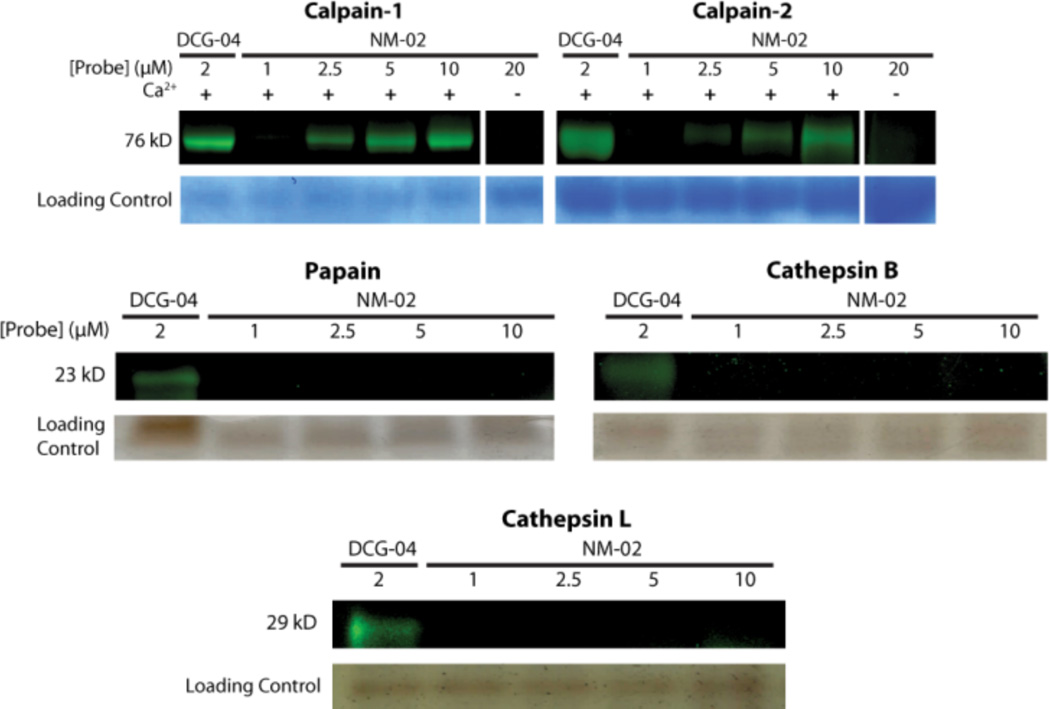
The presence of the succinyl epoxide warhead could reduce the specificity of the inhibitor due to its reactivity against most papain family active site cysteines. However, based on the previous kinetic studies, we reasoned that if the crosslinked peptide bound to the enzyme followed by a covalent reaction between the warhead and the active site cysteine, the ABPs had a high probability of being specific for calpain despite the addition of this reactive warhead. To investigate the specificity of NM-02, we tested a FITC tagged NM-02 against calpain-1 and calpain-2, and a panel of papain family proteases including papain, cathepsin B, and cathepsin L (Figure 9). FITC-NM-02 was added in increasing concentrations to either papain, cathepsin B, or cathepsin L and allowed to react for 20 min. at room temperature. Labeled enzymes were analyzed by SDS-PAGE and were visualized using a flatbed fluorescent scanner (Typhoon). We found that even at 10 µM, NM-02 did not bind to any of the other papain family cysteine proteases, which was in good agreement with the Ki (Table 2) determined in the binding studies of the crosslinked peptide 3c. This further suggests that NM-02 is specific for calpain at concentrations that would be appropriate for protease labeling experiments.
Figure 9.
FITC-NM-02 as a calpain specific ABP. We tested FITCNM- 02 (probe) in vitro against purified calpain-1, calpain-2, papain, cathepsin B, and cathepsin L. Only active calpain-1 and -2 are labeled and both are increasingly labeled with increased amounts of probe. Papain, Cathepsin B, and Cathepsin L are not labeled by NM-02. Loading control lanes beneath each panel show colloidal blue staining or silver staining of the respective gel.
Conclusions
In summary, we have demonstrated a simple screening of inexpensive, commercially available crosslinkers on an i, i+4 double cysteine mutant peptide to identify the best crosslinker to stabilize an α-helix. We identified five crosslinkers that increase α-helical character. Out of these five crosslinkers, dibromo-m-xylene c15, reacted in a simple, one-pot reaction, both in solution and on solid-phase, with the cysteine side chain and best increased the helicity of the peptide.
We have also applied this helix stabilization method to mimic a protein-protein interaction between a protease and its endogenous protein inhibitor to create, to our knowledge, the first active site directed, α-helical inhibitor of a protease. Importantly, we demonstrate that this inhibitor is shows good potency and high specificity for calpains over other highly similar cysteine proteases.
Lastly, we show that we can use the α-helical inhibitor as a scaffold to create an activity-based probe for examination of calpain activity. We determined that a β-amino acid is needed in the linker to bridge the gap between the helix and the active site cysteine. Furthermore it appeared that the ABP, NM-02, retained specificity for calpains over closely related cathepsin proteases. Given this specificity, we hope that these inhibitors and probes will allow for future studies of calpain function in multiple biological systems. We believe that the methodology used to stabilize this α-helical inhibitor will be another useful technique for α-helix stabilization for use in multiple biological applications.
Materials and Methods
Crosslinker Screen
To each well of a black round-bottomed 96- well plate (polypropylene) 90 µL of the stock solution, a peptide solution (0.114 mM) in NH4HCO3 buffer (12mL, 50 mM, pH=8.0), treated with TCEP (1M solution in the same NH4HCO3 buffer, 1.1 eq.) at room temperature (rt) for 1 h was added. Then 10 µL of the freshly prepared alkylating agent solution (1.5 mM in anhydrous DMF, 1.5 eq.) was applied to the well at rt and stirred for 2 h under protection from light. MALDI spectra were taken to monitor reaction progress and more alkylating agent was added if needed. The reaction was quenched by addition of 5% HCl which resulted in acidic conditions (pH=3-4). If necessary, 100 µL of ether was added to dissolve the excess reagent and organic byproducts into the organic layer. The ether layer could be removed by pipetting. MALDI spectra were taken from the sample in the remaining aqueous solution mixture.
“Selection of the fitness” Screen
Screens were performed in 1.5 mL microcentrifuge tubes. 1 mL of the stock peptide solution (1mM) in NH4HCO3 buffer (50mM, pH=8.0) was pre-treated with TCEP as described above and incubated for 1 h. Then, 100 µL of the concentrated alkylating agent solution (250mM or saturated solution in anhydrous DMF) was added and shaken for 2 h under protection from light. The reaction was quenched by the addition of 5% HCl which resulted in acidic conditions (pH=3–4) and purified by Reverse Phase HPLC.
Crosslinking with the unpurified peptide
The lyophilized crude peptide solution (app. 3–5mg/mL) in NH4HCO3 buffer (100mM, pH=8.0) was treated with TCEP (1.5 eq.) and stirred for 1 h. The alkylating agent in DMF (app. 3eq) was added to the solution and shaken for the 2 h. The reaction was quenched by adjusting the pH of the mixture to slightly acidic conditions through the addition of 0.5 N HCl or TFA. The crude mixture was either purified by HPLC or lyophilized for the next step.
Preparation of crosslinked peptides 3c from model peptide 3c by solid-phase peptide crosslinking
The uncrosslinked peptide 3c was similarly prepared on the CLEAR™ Rink Amide MBHA resin using the standard Fmoc peptide synthesis protocol (See Supporting Information). Fmoc-Cys(Mmt)-OH was used for cysteine for ease of deprotection. After the final coupling and cooling down to room temperature, the resin was washed with NMP(x3) and DMF(x3) followed by DCM(x3). The resin was then treated with 1% TFA solution in DCM for 10 min then washed with dichloromethane. This process was repeated until the solution lost its yellow color, which indicated the complete removal of Mmt protecting group. Then, the resin was washed with hexane and dried. After re-swelling in DMF, a solution of α,α’-dibromo-m-xylene (2 eq.) in DMF and DIPEA (4eq) was added. Alternatively, the resin was re-swollen in NH4HCO3 buffer (pH=8.0, 100 mM) for 1 h, a solution of α,α’-dibromo-m-xylene (5 eq.) in a minimal volume of DMF was added. The solution was stirred for 3 h at room temperature. The solvent was then removed and the resin was washed thoroughly with DMF. The Fmoc group on N-terminus was removed by treatment with 20% piperidine in DMF and acetylated by Ac2O and DIPEA. The cleavage/deprotection was done using TFA/thioanisole/EDT/anisole (90/5/3/2). The crude mixture was purified by reverse phase HPLC.
CD spectroscopy
Peptide solutions were prepared at ~50 µM in 50 mM phosphate buffer (pH 7.0) without TFE. The molar concentration of the peptide determined was by the weight (after lyophilization of the HPLC fractions) with consideration for molecular weight increase due to the presence of TFA salt for basic residues (Lys, Arg) as well as hydration (average 10%). Concentrations of the uncrosslinked peptides were determined by absorbance of Tyr residue at 280 nm with an extinction coefficient of 1280 M−1 cm−1.93 Circular dichroism studies were conducted at 25°C on a JASCO J-810 spectropolarimeter equipped with a Peltier temperature control unit.
NMR spectroscopy
The peptide sample was prepared with peptide concentrations of 2 mM in 0.6 mL of 9:1 v/v water/D2O mixture in 50mM sodium phosphate, pH 5.5. All spectra were recorded at 10 °C on a Bruker Avance III 500 MHz spectrometer equipped with a cryogenic probe. All 2D homonuclear spectra were recorded with standard pulse sequences.94 Spectra were processed and analyzed using the programs nmrPipe95 and XEASY,96 respectively. (See Supporting Information.)
Protease Activity Assays
Peptides were evaluated for ability to bind and subsequently inhibit the cysteine proteases using standard proteolytic fluorescence activity assays. Inhibition was assayed using a standard donor-quencher strategy using a previously published peptide substrates.14,97,98
Enzyme concentration for Calpain-1 was 25 nM. Enzyme concentration for papain was 25 nM. Enzyme concentrations for cathepsin B and cathepsin L was 3 nM. Calpain and papain buffer contained 10 mM dithioreitol (DTT), 100 mM KCl, 2 mM EGTA, 50 mM Tris-HCl (pH 7.5), and 0.015% Brij-35. Substrate concentration for calpain and papain was 0.25 µM H-Glu(Edans)-Pro-Leu-Phe- Ala-Glu-Arg-Lys(Dabcyl)-OH (Km calculation in Supporting Information, Figures S8 & S10).14,97,98 Cathepsin buffer contained 10 mM DTT, 500 mM sodium acetate (pH 5.5), and 4 mM EGTA.14,97,98 Substrate concentration for the cathepsins was 0.25 µM Z-FR-Amc. Calpain was activated by the injection of CaCl2 to a final concentration of 5 mM. Papain and cathepsin assays were activated by the addition of the substrate via a multichannel pipette. Varying concentrations of inhibitor, 1–100 µM, were used for each assay. All assays were done at a total well volume of 100 µL in 96-well plate, and each well contained a separate inhibitor concentration. Fluorescence was read in a Berthold Tri-Star fluorimeter. The excitation wavelength was 380 nm and the emission wavelength was 500 nm for H-Glu(Edans)-Pro-Leu-Phe-Ala-Glu-Arg-Lys(Dabcyl)-OH. The excitation wavelength 351 nm and emission wavelength was 430 nm for Z-FR-Amc.
Kinetic analysis of Calpain-1 by 3c
To identify inhibition type we used standard Michaelis-Menten treatment. Initial velocities (obtained from the linear segment of the progress curves) were plotted against substrate concentration.99 Due to the linearity of the first segment of the progress curve we believe that autoproteolysis during the first 500 seconds was not substantial enough to prevent the use of simple Michaelis-Menten kinetics, i.e. loss of enzyme did not change the velocity enough to cause it to deviate from linearity and incorporation of this additional complex would severely complicate the kinetics. Velocities were determined in RFU/sec then converted to µM/sec using the conversion factor 1386 RFU/µM. The conversion factor was obtained by the total hydrolysis of the substrate H-Glu(Edans)-Pro-Leu-Phe-Ala-Glu-Arg-Lys(Dabcyl)- OH in a known concentration by papain. To avoid weighting errors we used the values of Kmapp and Vmaxapp determined directly from the non-linear least-squares best fits of the untransformed data and put these values into the reciprocal equation:
.99.99 We then plotted the resulting reciprocal velocities against the respective reciprocal substrate concentrations.
Determination of IC50 against Enzymes
IC50 curves were generated identifying the initial rate of the enzyme at each inhibitor concentration from the respective progress curves. The conversion factor (1386 RFU/µM) was obtained by the total hydrolysis of the substrate H-E(Edans)-PLFAER-K(Dabcyl)-OH in a known concentration by papain. Initial velocities were converted from RFU/sec to µM/sec. Fractional activity was calculated by dividing the initial velocity at each inhibitor concentration by the initial velocity of the uninhibited enzyme. Data obtained up to 500 seconds was used for the initial rate calculation. The initial rate was then plotted against the log of the inhibitor concentration, and IC50 was calculated by GraphPad Prism.
Activity Based Probe Linker Experiments
Experimental conditions included 10 mM dithioreitol (DTT), 1.5 µg calpain, 100 mM KCl, 2 mM EGTA, 50 mM Tris-HCl (pH 7.5), 0.015% Brij-35, and either 1 µM or 10 µM of biotinylated probe (DCG-04, NM-01, NM-02, NM-03). Calpain was activated by the addition of calcium (3.33 µM of 50 mM CaCl2) to a final concentration of 8.3 mM in tubes containing either 1 µM or 10 µM ABP. For the negative control, water, instead of CaCl2, was added to the calpain solution containing 10 µM probe. Probes were allowed to bind to the calpain for 20 minutes at room temperature. The reaction was stopped by the addition of 10 µL NuPage® LDS Running Buffer (Life Technologies, Grand Island, NY). 10 µL of each labeled enzyme was loaded on a 10% Bis-Tris NuPAGE® gel (Life Technologies, Grand Island, NY) and separated via gel electrophoresis for 1.5 h, 140 V. The bands were then transferred to a PVDF membrane at 30 V for 70 min. The membrane was blocked and blotted using the Vectastain® Elite® ABC kit (Vector Laboratories, Burlingame, CA). Kodak film was exposed to the membrane and developed.
ABP Labeling Experiments
Buffer conditions for calpain and papain experiments were 10 µM dithioreitol (DTT), 100 mM KCl, 2 mM EGTA, 50 mM Tris-HCl (pH 7.5), and 0.015% Brij-35. 1.5 µg calpain-1 or 6 µg calpain-2 (calpain-2 was not as active) was used. (For labeling experiments greater concentrations of enzyme were used for ease of visualization of the enzyme on stained gels.) Buffer conditions for cathepsin experiments were 10 µM DTT, 500 mM sodium acetate (pH 5.5), and 4 mM EGTA. 1.5 µg of each cathepsin was labeled.14,97,98 Probes were allowed to bind for 20 min. at room temperature. Labeled enzymes were separated via gel electrophoresis on 10% (calpain, papain) or 12% (cathepsins) Bis-Tris NuPAGE® gels for 1 hr, 140 V. A Typhoon Fluorescent Imager was used for FITC visualization of the probe bound enzyme. Following fluorescent scanning the gels were colloidal blue stained (calpain-1 and calpain-2) or silver stained (papain, cathepsin B, and cathepsin L) to demonstrate that the same amount of enzyme had been used in all lanes. (See Supporting Information).
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from the Penn Genome Frontiers Institute (D.C.G), and National Institutes of Health 1R01AI09727301A1(D.C.G.), and Chemistry-Biology Interface Training Grant 5T32GM071339 (N.M.). This work was also supported by the National Institutes of Health GM54616 (W.F.D) and by a grant to P.L.D. from the Canadian Institutes for Health Research. K.E.L. was supported by a R.J. Wilson Fellowship. We would like to thank Prof. Walter Englander for the use of his NMR.
ABBREVIATIONS
- CD
circular dichroism
- MALDI
matrix-assisted laser desorption/ionization
- TOF
time-of-flight
- HPLC
high performance liquid chromatography
- NOESY
nuclear Overhauser effect spectroscopy
- Fmoc
fluorenyl methyloxycarbonyl
- Mmt
monomethoxytrityl
- Trt
trityl
- TCEP
tris(2-carboxyethyl) phosphine
- DMF
N,N-dimethylformamide
- NMP
N-methyl-2-pyrrolidinone
- DIPEA
N,N-diisopropylethylamine
- TFA
trifluoroacetic acid
- Edans
5-((2-aminoethyl)amino)naphthalene-1-sulfonic acid
- Dabcyl
4-(((4-dimetylamino)phenyl)azo) benzoic acid
- EGTA
ethylene glycol tetraacetic acid
- PVDF
polyvinylidene difluoride
- Z
carboxybenzyl
- Amc
7-amino-4-methyl coumarin
- RFU
relative fluorescence units
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Procedures for peptide and probe synthesis, crosslinking procedure for screening, MALDI-TOF analysis, scheme for competition between inter- and intramolecular reactions, analytical HPLC traces, CD spectra, alanine scanning mutagenesis, activity assay progress curve example, Km determination for calpain substrate, Michaelis-Menten plots and values, IC50 curves, and procedures for protease labeling experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zatz M, Starling A. N Engl J Med. 2005;352:2413. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 2.Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. J Biol Chem. 2005;280:6447. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- 3.Janossy J, Ubezio P, Apati A, Magocsi M, Tompa P, Friedrich P. Biochem Pharmacol. 2004;67:1513. doi: 10.1016/j.bcp.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Sreenan SK, Zhou YP, Otani K, Hansen PA, Currie KP, Pan CY, Lee JP, Ostrega DM, Pugh W, Horikawa Y, Cox NJ, Hanis CL, Burant CF, Fox AP, Bell GI, Polonsky KS. Diabetes. 2001;50:2013. doi: 10.2337/diabetes.50.9.2013. [DOI] [PubMed] [Google Scholar]
- 5.Franco SJ, Huttenlocher A. J Cell Sci. 2005;118:3829. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 6.Kulkarni S, Reddy KB, Esteva FJ, Moore HC, Budd GT, Tubbs RR. Oncogene. 2010;29:1339. doi: 10.1038/onc.2009.422. [DOI] [PubMed] [Google Scholar]
- 7.Shintani-Ishida K, Yoshida K. Biochim Biophys Acta. 2011;1812:743. doi: 10.1016/j.bbadis.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Liang B, Duan BY, Zhou XP, Gong JX, Luo ZG. J Biol Chem. 2010;285:27737. doi: 10.1074/jbc.M110.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portbury AL, Willis MS, Patterson C. J Biol Chem. 2011;286:9929. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H, Wei M, Wang Q, Li J, Wang H, Liu W, Lacefield JC, Greer PA, Karmazyn M, Fan G, Peng T. J Biol Chem. 2012;287:27480. doi: 10.1074/jbc.M112.358929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho WC, Pikor L, Gao Y, Elliott BE, Greer PA. J Biol Chem. 2012;287:15458. doi: 10.1074/jbc.M112.349308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Y, Wu C, De Veyra T, Greer PA. J Biol Chem. 2006;281:17689. doi: 10.1074/jbc.M601978200. [DOI] [PubMed] [Google Scholar]
- 13.Cuerrier D, Moldoveanu T, Campbell RL, Kelly J, Yoruk B, Verhelst SH, Greenbaum D, Bogyo M, Davies PL. J Biol Chem. 2007;282:9600. doi: 10.1074/jbc.M610372200. [DOI] [PubMed] [Google Scholar]
- 14.Gil-Parrado S, Assfalg-Machleidt I, Fiorino F, Deluca D, Pfeiler D, Schaschke N, Moroder L, Machleidt W. Biol Chem. 2003;384:395. doi: 10.1515/BC.2003.045. [DOI] [PubMed] [Google Scholar]
- 15.Ma H, Yang HQ, Takano E, Hatanaka M, Maki M. J Biol Chem. 1994;269:24430. [PubMed] [Google Scholar]
- 16.Sasaki T, Kikuchi T, Fukui I, Murachi T. J Biochem. 1986;99:173. doi: 10.1093/oxfordjournals.jbchem.a135457. [DOI] [PubMed] [Google Scholar]
- 17.Qian J, Cuerrier D, Davies PL, Li Z, Powers JC, Campbell RL. J Med Chem. 2008;51:5264. doi: 10.1021/jm800045t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donkor IO. Expert Opin Ther Pat. 2011;21:601. doi: 10.1517/13543776.2011.568480. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Ortega-Vilain AC, Patil GS, Chu DL, Foreman JE, Eveleth DD, Powers JC. J Med Chem. 1996;39:4089. doi: 10.1021/jm950541c. [DOI] [PubMed] [Google Scholar]
- 20.Ovat A, Li ZZ, Hampton CY, Asress SA, Fernandez FM, Glass JD, Powers JC. J Med Chem. 2010;53:6326. doi: 10.1021/jm901221v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna RA, Campbell RL, Davies PL. Nature. 2008;456:409. doi: 10.1038/nature07451. [DOI] [PubMed] [Google Scholar]
- 22.Moldoveanu T, Gehring K, Green DR. Nature. 2008;456:404. doi: 10.1038/nature07353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donkor IO, Korukonda R. Bioorg Med Chem Lett. 2008;18:4806. doi: 10.1016/j.bmcl.2008.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietsch M, Chua KC, Abell AD. Curr Top Med Chem. 2010;10:270. doi: 10.2174/156802610790725489. [DOI] [PubMed] [Google Scholar]
- 25.Abell AD, Jones MA, Coxon JM, Morton JD, Aitken SG, McNabb SB, Lee HY, Mehrtens JM, Alexander NA, Stuart BG, Neffe AT, Bickerstaffe R. Angew Chem Int Ed Engl. 2009;48:1455. doi: 10.1002/anie.200805014. [DOI] [PubMed] [Google Scholar]
- 26.Wendt A, Thompson VF, Goll DE. Biol Chem. 2004;385:465. doi: 10.1515/BC.2004.054. [DOI] [PubMed] [Google Scholar]
- 27.Goll DE, Thompson VF, Li H, Wei W, Cong J. Physiol Rev. 2003;83:731. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 28.Jackson DY, King DS, Chmielewski J, Singh S, Schultz PG. Journal of the American Chemical Society. 1991;113:9391. [Google Scholar]
- 29.Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF. P Natl Acad Sci USA. 2003;100:11273. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida AM, Li R, Gellman SH. J Am Chem Soc. 2012;134:75. doi: 10.1021/ja208856c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Liao W, Arora PS. Angew Chem Int Ed Engl. 2005;44:6525. doi: 10.1002/anie.200501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimartino G, Wang D, Chapman RN, Arora PS. Org Lett. 2005;7:2389. doi: 10.1021/ol0506516. [DOI] [PubMed] [Google Scholar]
- 33.Schafmeister CE, Po J, Verdine GL. Journal of the American Chemical Society. 2000;122:5891. [Google Scholar]
- 34.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackwell HE, Grubbs RH. Angew Chem Int Edit. 1998;37:3281. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, Chen K, Kulp JL, Arora PS. Journal of the American Chemical Society. 2006;128:9248. doi: 10.1021/ja062710w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolley GA. Accounts of chemical research. 2005;38:486. doi: 10.1021/ar040091v. [DOI] [PubMed] [Google Scholar]
- 38.Muppidi A, Wang Z, Li X, Chen J, Lin Q. Chemical communications. 2011;47:9396. doi: 10.1039/c1cc13320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houston ME, Jr, Gannon CL, Kay CM, Hodges RS. Journal of peptide science : an official publication of the European Peptide Society. 1995;1:274. doi: 10.1002/psc.310010408. [DOI] [PubMed] [Google Scholar]
- 40.Phelan JC, Skelton NJ, Braisted AC, McDowell RS. Journal of the American Chemical Society. 1997;119:455. [Google Scholar]
- 41.Osapay G, Taylor JW. Journal of the American Chemical Society. 1992;114:6966. [Google Scholar]
- 42.Felix AM, Heimer EP, Wang CT, Lambros TJ, Fournier A, Mowles TF, Maines S, Campbell RM, Wegrzynski BB, Toome V, Fry D, Madison VS. International journal of peptide and protein research. 1988;32:441. doi: 10.1111/j.1399-3011.1988.tb01375.x. [DOI] [PubMed] [Google Scholar]
- 43.Fujimoto K, Kajino M, Inouye M. Chem-Eur J. 2008;14:857. doi: 10.1002/chem.200700843. [DOI] [PubMed] [Google Scholar]
- 44.Geistlinger TR, Guy RK. J Am Chem Soc. 2001;123:1525. doi: 10.1021/ja005549c. [DOI] [PubMed] [Google Scholar]
- 45.Geistlinger TR, Guy RK. J Am Chem Soc. 2003;125:6852. doi: 10.1021/ja0348391. [DOI] [PubMed] [Google Scholar]
- 46.Cabezas E, Satterthwait AC. Journal of the American Chemical Society. 1999;121:3862. [Google Scholar]
- 47.Haney CM, Loch MT, Horne WS. Chemical communications. 2011;47:10915. doi: 10.1039/c1cc12010g. [DOI] [PubMed] [Google Scholar]
- 48.Ghadiri MR, Choi C. Journal of the American Chemical Society. 1990;112:1630. [Google Scholar]
- 49.Ruan FQ, Chen YQ, Hopkins PB. Journal of the American Chemical Society. 1990;112:9403. [Google Scholar]
- 50.Kawamoto SA, Coleska A, Ran X, Yi H, Yang CY, Wang S. Journal of medicinal chemistry. 2012;55:1137. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holland-Nell K, Meldal M. Angew Chem Int Edit. 2011;50:5204. doi: 10.1002/anie.201005846. [DOI] [PubMed] [Google Scholar]
- 52.Freidinger RM. J Med Chem. 2003;46:5553. doi: 10.1021/jm030484k. [DOI] [PubMed] [Google Scholar]
- 53.Hruby VJ. Life Sci. 1982;31:189. doi: 10.1016/0024-3205(82)90578-1. [DOI] [PubMed] [Google Scholar]
- 54.Satterthwait AC, Arrhenius T, Hagopian RA, Zavala F, Nussenzweig V, Lerner RA. Vaccine. 1988;6:99. doi: 10.1016/s0264-410x(88)80007-0. [DOI] [PubMed] [Google Scholar]
- 55.Oneil KT, Hoess RH, Jackson SA, Ramachandran NS, Mousa SA, Degrado WF. Proteins. 1992;14:509. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 56.Bach AC, Eyermann CJ, Gross JD, Bower MJ, Harlow RL, Weber PC, Degrado WF. Journal of the American Chemical Society. 1994;116:3207. [Google Scholar]
- 57.Cheng RP, Suich DJ, Cheng H, Roder H, DeGrado WF. J Am Chem Soc. 2001;123:12710. doi: 10.1021/ja0116932. [DOI] [PubMed] [Google Scholar]
- 58.Suich DJ, Mousa SA, Singh G, Liapakis G, Reisine T, DeGrado WF. Bioorgan Med Chem. 2000;8:2229. doi: 10.1016/s0968-0896(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 59.Cheng RP, Scialdone MA, DeGrado WF. Abstr Pap Am Chem S. 1999;218:U138. [Google Scholar]
- 60.Nestor JJ. Curr Med Chem. 2009;16:4399. doi: 10.2174/092986709789712907. [DOI] [PubMed] [Google Scholar]
- 61.Szewczuk Z, Rebholz KL, Rich DH. International journal of peptide and protein research. 1992;40:233. doi: 10.1111/j.1399-3011.1992.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 62.Timmerman P, Beld J, Puijk WC, Meloen RH. Chembiochem. 2005;6:821. doi: 10.1002/cbic.200400374. [DOI] [PubMed] [Google Scholar]
- 63.Lindman S, Lindeberg G, Gogoll A, Nyberg F, Karlen A, Hallberg A. Bioorgan Med Chem. 2001;9:763. doi: 10.1016/s0968-0896(00)00294-7. [DOI] [PubMed] [Google Scholar]
- 64.Walker MA, Johnson T. Tetrahedron Lett. 2001;42:5801. [Google Scholar]
- 65.Blanco-Lomas M, Samanta S, Campos PJ, Woolley GA, Sampedro D. Journal of the American Chemical Society. 2012 doi: 10.1021/ja301868p. [DOI] [PubMed] [Google Scholar]
- 66.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, Marmorstein R, Cole RN, Khochbin S, Cole PA. Journal of the American Chemical Society. 2010;132:9986. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dewkar GK, Carneiro PB, Hartman MCT. Organic Letters. 2009;11:4708. doi: 10.1021/ol901662c. [DOI] [PubMed] [Google Scholar]
- 68.Smeenk LEJ, Dailly N, Hiemstra H, van Maarseveen JH, Timmerman P. Organic Letters. 2012;14:1194. doi: 10.1021/ol203259a. [DOI] [PubMed] [Google Scholar]
- 69.Johnson WC. Proteins. 1990;7:205. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 70.Woody RW, Koslowski A. Biophys Chem. 2002;101:535. doi: 10.1016/s0301-4622(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 71.Szyperski T, Guntert P, Otting G, Wuthrich K. J Magn Reson. 1992;99:552. [Google Scholar]
- 72.Armstrong DaZ. Raphael; v 1.4 ed. 2009 [Google Scholar]
- 73.Greenfield NJ. Anal Biochem. 1996;235:1. doi: 10.1006/abio.1996.0084. [DOI] [PubMed] [Google Scholar]
- 74.Filippi B, Borin G, Moretto V, Marchiori F. Biopolymers. 1978;17:2545. [Google Scholar]
- 75.MacArthur MW, Thornton JM. J Mol Biol. 1991;218:397. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 76.Marqusee S, Baldwin RL. Proc Natl Acad Sci U S A. 1987;84:8898. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradley EK, Thomason JF, Cohen FE, Kosen PA, Kuntz ID. J Mol Biol. 1990;215:607. doi: 10.1016/S0022-2836(05)80172-X. [DOI] [PubMed] [Google Scholar]
- 78.Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Nature. 1987;326:563. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- 79.Betts R, Weinsheimer S, Blouse GE, Anagli J. J Biol Chem. 2003;278:7800. doi: 10.1074/jbc.M208350200. [DOI] [PubMed] [Google Scholar]
- 80.Cheng Y, Prusoff WH. Biochem Pharmacol. 1973;22:3099. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 81.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Biochim Biophys Acta. 2012;1824:68. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barlos K, Gatos D, Hatzi O, Koch N, Koutsogianni S. International journal of peptide and protein research. 1996;47:148. doi: 10.1111/j.1399-3011.1996.tb01338.x. [DOI] [PubMed] [Google Scholar]
- 83.Benito JM, Meldal M. Qsar Comb Sci. 2004;23:117. [Google Scholar]
- 84.Adam GC, Cravatt BF, Sorensen EJ. Chem Biol. 2001;8:81. doi: 10.1016/s1074-5521(00)90060-7. [DOI] [PubMed] [Google Scholar]
- 85.Cravatt BF, Sorensen EJ. Curr Opin Chem Biol. 2000;4:663. doi: 10.1016/s1367-5931(00)00147-2. [DOI] [PubMed] [Google Scholar]
- 86.Kidd D, Liu Y, Cravatt BF. Biochemistry. 2001;40:4005. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 87.Puri AW, Broz P, Shen A, Monack DM, Bogyo M. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Albrow VE, Ponder EL, Fasci D, Bekes M, Deu E, Salvesen GS, Bogyo M. Chem Biol. 2011;18:722. doi: 10.1016/j.chembiol.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan F, Verhelst SH, Blum G, Coussens LM, Bogyo M. J Am Chem Soc. 2006;128:5616. doi: 10.1021/ja060835v. [DOI] [PubMed] [Google Scholar]
- 90.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol Cell Biol. 1999;19:1720. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Chemistry & Biology. 2000;7:569. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 92.The PyMOL Molecular Graphics System. Version 1.3 ed. Schrödinger, LLC; [Google Scholar]
- 93.Edelhoch H. Biochemistry-Us. 1967;6:1948. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 94.Wüthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. [Google Scholar]
- 95.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 96.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. Journal of Biomolecular Nmr. 1995;6:1. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 97.Kelly JC, Cuerrier D, Graham LA, Campbell RL, Davies PL. Bba-Proteins Proteom. 2009;1794:1505. doi: 10.1016/j.bbapap.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 98.Pfizer J, Assfalg-Machleidt I, Machleidt W, Schaschke N. Biol Chem. 2008;389:83. doi: 10.1515/BC.2008.004. [DOI] [PubMed] [Google Scholar]
- 99.Copeland RA. Enzymes : a practical introduction to structure, mechanism, and data analysis. New York: VCH Publishers; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.