Abstract
Histidine acid phytases (HAPhy) are widely distributed enzymes among bacteria, fungi, plants, and some animal tissues. They have a significant role as an animal feed enzyme and in the solubilization of insoluble phosphates and minerals present in the form of phytic acid complex. A set of 50 reference protein sequences representing HAPhy were retrieved from NCBI protein database and characterized for various biochemical properties, multiple sequence alignment (MSA), homology search, phylogenetic analysis, motifs, and superfamily search. MSA using MEGA5 revealed the presence of conserved sequences at N-terminal “RHGXRXP” and C-terminal “HD.” Phylogenetic tree analysis indicates the presence of three clusters representing different HAPhy, that is, PhyA, PhyB, and AppA. Analysis of 10 commonly distributed motifs in the sequences indicates the presence of signature sequence for each class. Motif 1 “SPFCDLFTHEEWIQYDYLQSLGKYYGYGAGNPLGPAQGIGF” was present in 38 protein sequences representing clusters 1 (PhyA) and 2 (PhyB). Cluster 3 (AppA) contains motif 9 “KKGCPQSGQVAIIADVDERTRKTGEAFAAGLAPDCAITVHTQADTSSPDP” as a signature sequence. All sequences belong to histidine acid phosphatase family as resulted from superfamily search. No conserved sequence representing 3- or 6-phytase could be identified using multiple sequence alignment. This in silico analysis might contribute in the classification and future genetic engineering of this most diverse class of phytase.
1. Introduction
Phytate (myo-inositol 1,2,3,4,5,6-hexakisphosphate; IP6) is the major storage form of phosphorus (P), representing approximately 80% of P in soil [1], 65–80% of total P in grains [2], and up to 80% of P in manures from monogastric animals [3]. Phytate exists primarily as metal phytate complex with nutritionally important cations, that is, Ca2+, Fe2+, and Zn2+ [4].
Phytases (IP6 phosphohydrolase) are a class of phosphatases which catalyses hydrolysis of phytate to inositol phosphates, inorganic phosphorus, and myo-inositol [5], also lowers down affinity of phytate to associated minerals and proteins [6], and thus increases bioavailability of P, minerals, and proteins for growth and development of plants and animals [7–9].
Phytases are widely distributed among plants [10, 11], certain animal tissues, and microbial cells [12–15]. To date, four classes of phytases have been characterized in terrestrial organisms: histidine acid phytase (HAPhy), cysteine phytase (CPhy), purple acid phosphatase (PAP), and β-propeller phytase (BPPhy) [16, 17]. HAPhys are the most studied and diverse class of phytase. Most bacterial, fungal, and plant phytases belong to histidine acid phosphatases (EC 3.1.3.2) which are further classified as 3-phytase (EC 3.1.3.8) or 6-phytase (EC 3.1.3.26) due to their high specific activity for phytate and position specific initial hydrolysis of phytate.
Phytases have been extensively reviewed for various industrial and biotechnological applications [18–21], biochemical properties [22], and consensus phytase construct [23]. Conserved amino acid residues are reported in HAPhy sequences at N-terminal “RHGXRXP,” C-terminal “HD,” and eight cysteine residues in around sequence [16, 24, 25]. It is a well-adopted fact that all phytases have not similar and common active site; hence the initial classification system is based on catalytic mechanism [22]. Still, there is a need to devise a taxonomic system to accommodate new types of phytases with novel catalytic mechanism.
The in silico characterization of protein sequences of industrially important enzymes has been reported recently [26–28]. Biochemical features, homology search, multiple sequence alignment, phylogenetic tree construction, motif, and superfamily distribution of alkaline proteases have been analyzed using various bioinformatics tools [28]. A total of 121 protein sequences of pectate lyases were subjected to homology search, multiple sequence alignment, phylogenetic tree construction, and motif analysis [26]. Malviya et al. [27] collected forty-seven full-length amino acid sequences of PPO from bacteria, fungi, and plants and subjected them to multiple sequence alignment (MSA), domain identification, and phylogenetic tree construction.
In the present study, we performed in silico analysis of 50 HAPhy protein sequences. The biochemical features, homology search, multiple sequence alignment, phylogenetic tree construction, motif, and superfamily distribution have been analyzed using various bioinformatics tools.
2. Material and Methods
Representative genes from histidine acid phytases (E. coli AppA, GenBank accession number P07102; Aspergillus niger PhyA and PhyB, P34752 and P34754) were used as probes to BLAST microbial genome database from NCBI (http://www.ncbi.nlm.nih.gov/). The protein sequences in FASTA format from RefSeq entries, which were shown to exhibit phytase activities, were selected for further in silico study.
Physiochemical data were generated from various tools in the EXPASY proteomic server (ClustalW, ProtParam, protein calculator, Compute pI/Mw, ProtScale) [29]. The molecular weights (kDa) of the various histidine acid phytases were calculated by the addition of average isotopic masses of amino acid in the protein and deducting the average isotopic mass of one water molecule. The pI of enzyme was calculated using pK values of amino acid according to Bjellqvist et al. [30].
The evolutionary history was inferred using the Neighbor-Joining method [31]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [32] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 303 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [33]. For domain search, the Pfam site (http://www.sanger.ac.uk/resources/software/) was used. Domain analysis was done using MEME (http://meme.nbcr.net/meme/) [34]. The conserved protein motifs deduced by MEME were characterized for biological function analysis using protein BLAST, and domains were studied with InterProScan providing the best possible match based on the highest similarity score.
3. Result and Discussion
The 50 protein sequences of HAPhy were retrieved from NCBI. The accession number of retrieved sequences along with species names is listed in Table 1. The sequences were characterized for homology search, multiple sequences alignment, biochemical features, phylogenetic tree construction, motifs, and superfamily search using various bioinformatics tools. Out of 50 sequences 12 sequences belong to HAPhy gene AppA, 26 sequences to PhyA, and 12 sequences to PhyB.
Table 1.
List of retrieved protein sequences from NCBI/Entrez and their accession number.
| S. no. | Source organism | Accession number | Total sequences |
|---|---|---|---|
| 1 | Escherichia coli | P07102.2, NP_415500.1, ZP_07105303.1, YP_001462212.1 | 4 |
| 2 | Shigella boydii | YP_408643.1 | 1 |
| 3 | Shigella flexneri | YP_688514.1 | 1 |
| 4 | Shigella dysenteriae | ZP_07681338.1, YP_402619.1 | 2 |
| 5 | Escherichia albertii | ZP_02904404.1 | 1 |
| 6 | Escherichia fergusonii | YP_002384142.1 | 1 |
| 9 | Citrobacter freundii | AAR89622.1 | 1 |
| 10 | Aspergillus niger | P34752.1, XP_001401713.2, P34754.1, XP_001393206.1 | 4 |
| 11 | Aspergillus oryzae | XP_001821210.1 | 1 |
| 12 | Aspergillus awamori | P34753.1 | 1 |
| 13 | Aspergillus flavus | XP_002376973.1 | 1 |
| 14 | Aspergillus fumigates | XP_751964.2 | 1 |
| 15 | Aspergillus terreus | XP_001214511.1 | 1 |
| 16 | Neosartorya fischeri | XP_001267160.1 | 1 |
| 17 | Aspergillus nidulans | XP_659289.1 | 1 |
| 18 | Aspergillus clavatus | XP_001271757.1, XP_001271709.1 | 2 |
| 19 | Penicillium chrysogenum | XP_002561094.1 | 1 |
| 20 | Penicillium marneffei | XP_002148821.1 | 1 |
| 21 | Ajellomyces dermatitidis | XP_002629272.1 | 1 |
| 22 | Botryotinia fuckeliana | XP_001554147.1 | 1 |
| 23 | Uncinocarpus reesii | XP_002542954.1 | 1 |
| 24 | Ajellomyces capsulatus | XP_001538598.1 | 1 |
| 25 | Sclerotinia sclerotiorum | XP_001589324.1 | 1 |
| 26 | Coccidioides posadasii | XP_003065081.1 | 1 |
| 27 | Trichophyton rubrum | XP_003233631.1 | 1 |
| 28 | Arthroderma otae | XP_002849736.1 | 1 |
| 29 | Talaromyces stipitatus | XP_002483691.1 | 1 |
| 30 | Podospora anserina | XP_001906589.1 | 1 |
| 31 | Trichophyton verrucosum | XP_003021635.1 | 1 |
| 32 | Arthroderma gypseum | XP_003169494.1 | 1 |
| 33 | Penicillium marneffei | XP_002150501.1 | 1 |
| 34 | Arthroderma benhamiae | XP_003015622.1 | 1 |
| 35 | Candida albicans | XP_713416.1 | 1 |
| 36 | Candida dubliniensis | XP_002421792.1, XP_002419861.1 | 2 |
| 37 | Candida albicans | XP_713478.1 | 1 |
| 38 | Candida tropicalis | XP_002546108.1 | 1 |
| 39 | Debaryomyces hansenii | XP_458051.2 | 1 |
| 40 | Komagataella pastoris | XP_002490985.1 | 1 |
| 41 | Saccharomyces cerevisiae | NP_009650.1 | 1 |
| 42 | Coccidioides posadasii | XP_003072016.1 | 1 |
Multiple sequence alignment showed presence of conserved sites for HAPhy N-terminal “RHG/NXRXP” and C-terminal “HD” in all sequences as reported by other coworkers [25]. This is consistent with Pfam analysis of predicted active site residues, which in all sequences is shown to be N-terminal histidine residue present in conserved region and C-terminal aspartic acid. The histidine in N-terminal region seems as a nucleophile in the formation of a covalent phosphohistidine intermediate [35]. Aspartic acid at C-terminal “HD” sequence acts as a proton donor to the oxygen atom of the scissile phosphomonoester bond [36, 37]. No conserved sequence representing 3- or 6-phytase could be identified using multiple-sequence alignment.
The phylogenetic tree based on protein sequences revealed three major clusters. Cluster 1, a larger cluster containing 26 sequences under study, includes the majority of Aspergillus sp., Penicillium sp., Ajellomyces sp., Arthroderma sp., Trichophyton sp., Sclerotinia sp., Uncinocarpus sp., and Coccidioides sp. (Figure 1). Biochemical features for this cluster are listed in Table 2. The total number of amino acid residues ranged from 441 to 539 with variable molecular weights. pI values of this cluster ranged from 4.87 to 8.53. Variations among various phytase in this group in terms of other physiochemical parameters like positively charged and negatively charged residues, hydropathicity (GRAVY) are given in Table 2.
Figure 1.
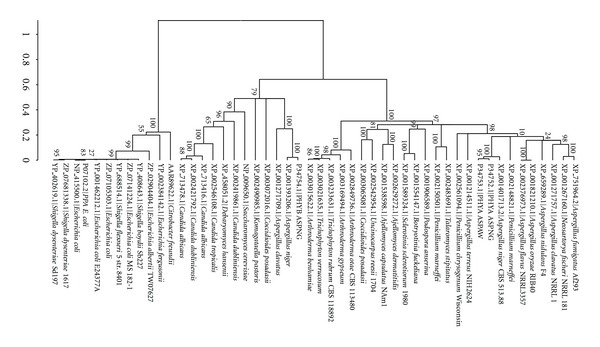
Phylogenetic tree constructed by NJ method based on HAPhy protein sequences.
Table 2.
Biochemical characteristics of HAPhy protein sequences.
| S. no. | Accession number | Source organisms | Number of amino acids | Molecular weight | Theoretical pI | Total number of negatively charged residues (Asp + Glu) | Total number of positively charged residues (Arg + Lys) | Instability index | Aliphatic index | GRAVY | Predictive active sites by Pfam |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P07102.2 | Escherichia coli | 523 | 56118.9 | 6.07 | 51 | 43 | 45.95 | 86.25 | −0.221 | 39(H), 326(D) |
| 2 | NP_415500.1 | Escherichia coli str. K-12 | 432 | 47056.8 | 6.26 | 40 | 37 | 38.08 | 93.08 | −0.157 | 39(H), 326(D) |
| 3 | ZP_07105303.1 | Escherichia coli MS 119-7 | 442 | 48081 | 6.09 | 41 | 37 | 39.71 | 92.08 | −0.147 | 49(H), 336(D) |
| 4 | YP_001462212.1 | Escherichia coli E24377A | 432 | 47029.8 | 6.09 | 40 | 36 | 38.56 | 93.31 | −0.138 | 39(H), 326(D) |
| 5 | ZP_07141224.1 | Escherichia coli MS 182-1 | 442 | 48081 | 6.26 | 41 | 38 | 39.52 | 92.08 | −0.148 | 49(H), 336(D) |
| 6 | YP_408643.1 | Shigella boydii Sb227 | 432 | 47063.8 | 6.09 | 40 | 36 | 37.99 | 92.41 | −0.141 | 39(H), 326(D) |
| 7 | YP_688514.1 | Shigella flexneri | 432 | 47105.9 | 5.94 | 40 | 35 | 38.2 | 92.87 | −0.131 | 39(H), 326(D) |
| 8 | ZP_07681338.1 | Shigella dysenteriae 1617 | 434 | 47354.1 | 5.55 | 42 | 34 | 38.57 | 93.99 | −0.142 | 41(H), 328(D) |
| 9 | YP_402619.1 | Shigella dysenteriae Sd197 | 434 | 47328 | 5.55 | 42 | 34 | 38.57 | 93.09 | −0.152 | 41(H), 328(D) |
| 10 | ZP_02904404.1 | Escherichia albertii | 439 | 48000.6 | 9.35 | 33 | 44 | 37.78 | 96.88 | −0.094 | 46(H), 333(D) |
| 11 | YP_002384142.1 | Escherichia fergusonii | 428 | 46608.5 | 8.37 | 32 | 35 | 40.79 | 93.48 | −0.132 | 39(H), 322(D) |
| 12 | AAR89622.1 | Citrobacter freundii | 433 | 48506.5 | 6.29 | 49 | 47 | 35.09 | 86 | −0.322 | 39(H), 325(D) |
| 13 | P34752.1 | Aspergillus niger | 467 | 51086 | 4.94 | 51 | 34 | 44.72 | 76.62 | −0.211 | 82(H), 382(D) |
| 14 | P34753.1 | Aspergillus awamori | 467 | 51074.9 | 4.89 | 52 | 33 | 42.73 | 76.85 | −0.221 | 82(H), 382(D) |
| 15 | XP_001401713.2 | Aspergillus niger | 497 | 54579.1 | 5.25 | 53 | 38 | 44.74 | 77.69 | −0.225 | 112(H), 392(D) |
| 16 | XP_001821210.1 | Aspergillus oryzae | 466 | 51257.1 | 4.87 | 57 | 39 | 34.89 | 70.49 | −0.316 | 81(H), 361(D) |
| 17 | XP_002376973.1 | Aspergillus flavus | 496 | 54729.2 | 5.11 | 60 | 46 | 35.72 | 72.7 | −0.31 | 111(H), 391(D) |
| 18 | XP_751964.2 | Aspergillus fumigates | 498 | 54538.8 | 8.53 | 48 | 53 | 29.31 | 77.59 | −0.197 | 114(H), 393(D) |
| 19 | XP_001214511.1 | Aspergillus terreus | 466 | 51088.1 | 5.12 | 51 | 33 | 35.3 | 72.94 | −0.226 | 82(H), 382(D) |
| 20 | XP_001267160.1 | Neosartorya fischeri | 464 | 50787.1 | 6.17 | 47 | 43 | 31.33 | 73.17 | −0.206 | 80(H), 359(D) |
| 21 | XP_659289.1 | Aspergillus nidulans | 463 | 51816.2 | 5.35 | 52 | 39 | 32.09 | 72.48 | −0.287 | 80(H), 358(D) |
| 22 | XP_001271757.1 | Aspergillus clavatus | 465 | 51531.3 | 7.14 | 52 | 52 | 29.94 | 72.6 | −0.332 | 81(H), 360(D) |
| 23 | XP_002561094.1 | Penicillium chrysogenum | 483 | 53668.6 | 7.11 | 51 | 51 | 46.27 | 69.65 | −0.393 | 96(H), 378(D) |
| 24 | XP_002148821.1 | Penicillium marneffei | 465 | 50878 | 5.19 | 48 | 37 | 32.37 | 73.46 | −0.172 | 80(H), 360(D) |
| 25 | XP_002629272.1 | Ajellomyces dermatitidis | 528 | 58565.2 | 6.21 | 55 | 49 | 44 | 80.49 | −0.167 | 140(H), 420(D) |
| 26 | XP_001554147.1 | Botryotinia fuckeliana | 529 | 57902.2 | 5.08 | 55 | 42 | 40.6 | 69.38 | −0.329 | 140(H), 423(D) |
| 27 | XP_002542954.1 | Uncinocarpus reesii | 501 | 56117.4 | 8.51 | 52 | 57 | 33.68 | 70.08 | −0.448 | 108(H), 388(D) |
| 28 | XP_001538598.1 | Ajellomyces capsulatus | 441 | 49342.6 | 5.88 | 46 | 35 | 44.48 | 79.59 | −0.238 | 53(H), 333(D) |
| 29 | XP_001589324.1 | Sclerotinia sclerotiorum | 465 | 50709.3 | 4.88 | 45 | 29 | 38.75 | 73.25 | −0.188 | 76(H), 359(D) |
| 30 | XP_003065081.1 | Coccidioides posadasii | 539 | 60239.3 | 7.93 | 61 | 63 | 29.66 | 75.25 | −0.368 | 145(H), 425(D) |
| 31 | XP_003233631.1 | Trichophyton rubrum | 474 | 52142.7 | 6.23 | 54 | 50 | 41.86 | 69.22 | −0.331 | 87(H), 362(D) |
| 32 | XP_002849736.1 | Arthroderma otae | 466 | 51429.8 | 5.58 | 53 | 45 | 40.37 | 67.04 | −0.33 | 85(H), 360(D) |
| 33 | XP_002483691.1 | Talaromyces stipitatus | 523 | 58182.2 | 4.9 | 65 | 42 | 42.55 | 78.7 | −0.26 | 129(H), 409(D) |
| 34 | XP_001906589.1 | Podospora anserina | 514 | 57367.6 | 5.55 | 63 | 53 | 36.91 | 77.8 | −0.396 | 125(H), 406(D) |
| 35 | XP_003021635.1 | Trichophyton verrucosum | 456 | 50332.6 | 6.27 | 53 | 49 | 41.1 | 66.36 | −0.372 | 69(H), 344(D) |
| 36 | XP_003169494.1 | Arthroderma gypseum | 473 | 52338.9 | 6.33 | 54 | 51 | 38.25 | 67.06 | −0.353 | 86(H), 361(D) |
| 37 | XP_002150501.1 | Penicillium marneffei | 510 | 57067.8 | 5.07 | 64 | 45 | 40.04 | 74.18 | −0.345 | 116(H), 396(D) |
| 38 | XP_003015622.1 | Arthroderma benhamiae | 456 | 50490.7 | 6.14 | 54 | 49 | 43.2 | 65.72 | −0.4 | 69(H), 344(D) |
| 39 | P34754.1 | Aspergillus niger | 479 | 52611.5 | 4.65 | 48 | 28 | 34.04 | 71.96 | −0.279 | 82(H), 382(D) |
| 40 | XP_001393206.1 | Aspergillus niger | 479 | 52486.2 | 4.62 | 49 | 27 | 33.71 | 71.17 | −0.289 | 82(H), 382(D) |
| 41 | XP_001271709.1 | Aspergillus clavatus | 460 | 50746.9 | 4.61 | 54 | 31 | 39.64 | 79.96 | −0.17 | 69(H), 329(D) |
| 42 | XP_713416.1 | Candida albicans | 461 | 51283.1 | 5.8 | 48 | 43 | 29.87 | 74.27 | −0.44 | 73(H), 335(D) |
| 43 | XP_002421792.1 | Candida dubliniensis | 462 | 51275.9 | 5.44 | 48 | 41 | 28.02 | 72.62 | −0.411 | 73(H), 335(D) |
| 44 | XP_713478.1 | Candida albicans | 462 | 51305 | 5.57 | 48 | 42 | 27.7 | 71.99 | −0.426 | 73(H), 335(D) |
| 45 | XP_002546108.1 | Candida tropicalis | 465 | 52540.6 | 4.41 | 67 | 35 | 33.65 | 67.74 | −0.543 | 73(H), 337(D) |
| 46 | XP_458051.2 | Debaryomyces hansenii | 464 | 51835.7 | 5.13 | 52 | 37 | 39.65 | 73.1 | −0.404 | 73(H), 337(D) |
| 47 | XP_002419861.1 | Candida dubliniensis | 457 | 52259.6 | 5.2 | 57 | 46 | 36.41 | 70.55 | −0.493 | 73(H), 332(D) |
| 48 | XP_002490985.1 | Komagataella pastoris | 468 | 52690.7 | 4.41 | 68 | 33 | 37.25 | 84.19 | −0.27 | 84(H), 346(D) |
| 49 | NP_009650.1 | Saccharomyces cerevisiae | 467 | 52776.5 | 4.43 | 67 | 36 | 30.82 | 71.46 | −0.373 | 75(H), 338(D) |
| 50 | XP_003072016.1 | Coccidioides posadasii | 403 | 45575.2 | 5.82 | 51 | 42 | 31.24 | 70.97 | −0.473 | 4(H), 269(D) |
Aliphatic index analysis reveals uniformity in this group of phytases within the range of 75 ± 5 except for some sequences of Arthroderma sp. (XP_002849736.1, XP_003169494.1, XP_003015622.1) and Trichophyton sp. (XP_003021635.1). Aliphatic index of protein measures the relative volume occupied by aliphatic side chains of the amino acids: alanine, valine, leucine, and isoleucine. Globular proteins with high aliphatic index have high thermostability, and an increase in aliphatic index increases protein thermostability [38, 39].
Cluster 2 includes 12 protein sequences and represents PhyB gene sequences including the majority of Candida sp., S. cerevisiae, C. posadasii, and D. hansenii. Total number of sequences in this group is in the range of 457 to 479, and the pI values range from 4.41 to 5.82. It has less variation in its pI as compared to cluster 1 sequences (PhyA). Aliphatic index of this cluster sequences is uniform in the range of 75 ± 5 except for Candida tropicalis (XP_002546108.1) with a value of 67.74 and Komagataella pastoris (XP_002490985.1) with a value of 84.19.
Cluster 3 represents protein sequences from phytase gene AppA, also abbreviated as PhyC [22], which includes E. coli (in majority) along with various Shigella sp. and Citrobacter freundii. Various biophysical parameters for this group of sequences reveal amino acid residues ranging from 428 to 523, while pI value of the majority of sequences is in range of 5.5 to 6.5 except for E. albertii (9.35) and E. fergusonii (8.37). Aliphatic index of this group of sequences reveals highest thermostability among all three clusters. Predominantly positively charged amino acids are present in all three clusters.
The instability index is used to measure in vivo half-life of a protein [40]. The proteins which have been reported as in vivo half-life of less than 5 hours showed instability index greater than 40, whereas those having more than 16 hours half-life [41] have an instability index of less than 40. Instability index of HAP sequences under the study is found higher than 40 (Table 2) for 15 sequences including fully characterized E. coli and A. niger phytases, indicating an in vivo half-life of less than 5 hours. Superfam tool on ExPASy server for superfamily analysis of phytase sequences reveals the identity of all sequences to histidine acid phosphatase family belonging to phosphoglycerate mutase-like superfamily [42] (Table 3).
Table 3.
Distribution of superfamily among HAPhy protein sequences determined using superfam server.
| Family | Superfamily | Accession number (range of amino acids residues) |
|---|---|---|
| Histidine acid phosphatase | Phosphoglycerate mutase-like | XP_001401713.2 (61–496), P07102.2 (26–429), NP_415500.1 (26–429), ZP_07105303.1 (36–439), YP_001462212.1 (26–429), ZP_07141224.1 (36–439), YP_408643.1 (26–429), YP_688514.1 (26–429), ZP_07681338.1 (28–431), YP_402619.1 (28–431), ZP_02904404.1 (33–436) YP_002384142.1 (27–424), AAR89622.1 (27–427), P34752.1 (30–466), P34753.1 (31–466), XP_001821210.1 (29–465), XP_002376973.1 (59–495), XP_751964.2 (62–497), XP_001214511.1 (32–466), XP_001267160.1 (28–463), XP_659289.1 (28–461), XP_001271757.1 (30–464), XP_002561094.1 (45–482), XP_002148821.1 (28–464), XP_002629272.1 (89–527), XP_001554147.1 (90–527), XP_002542954.1 (57–495), XP_001538598.1 (6–440), XP_001589324.1 (26–463), XP_003065081.1 (97–532), XP_003233631.1 (36–469), XP_002849736.1 (35–466), XP_002483691.1 (87–517), XP_001906589.1 (76–512), XP_003021635.1 (23–451), XP_003169494.1 (35–468), XP_002150501.1 (73–505), XP_003015622.1 (23–451), P34754.1 (35–470), XP_001393206.1 (35–470), XP_001271709.1 (23–452), XP_713416.1 (28–455), XP_002421792.1 (28–455), XP_713478.1 (28–455), XP_002546108.1 (28–457), XP_458051.2 (28–451), XP_002419861.1 (28–445), XP_002490985.1 (42–464), NP_009650.1 (34–460), XP_003072016.1 (1–393) |
Histidine acid phytase from all three clusters shares a large α/β and a small α-domain [22]. MEME analysis results in frequently observed 10 motifs (Table 4). A set of 41 amino acid residues “SPFCDLFTHEEWIQYDYLQSLGKYYGYGAGNPLGPAQGIGF” representing motif 1 were conserved and uniformly observed in 38 phytase protein sequences from clusters 1 and 2, that is, PhyA and PhyB, revealing their identity with HP_HAP like, histidine acid phosphatase superfamily. Other motifs are associated with HAP superfamily (Table 2). Cluster 3, representing AppA, does not have motif 1 in its sequences, but it does contain a 50 amino acid residues long unique motif 9 “KKGCPQSGQVAI IADVDERTRKTGEAFAAGLAPDCAITVHTQADTSSPDP.” Motif 5 “YAFLKTYNYSL GADDLTPFGEQQLVDSGIKFYQRYESLAKDIVPFIRASG” is present in all protein sequences representing PhyA cluster 1. PhyB protein sequences also contain a unique 41 amino acid residues long motif 8 “ETSPENSEGPYAGTTNALRHGAAFRARY GSLYDENSTLPVF.”
Table 4.
Distribution of commonly observed motifs in different HAPhy protein sequences along with their functional domains.
| Motifs number | Motif present in number of sequence | Motif width | Sequence | Domain |
|---|---|---|---|---|
| 1 | 38 | 41 | SPFCDLFTHEEWIQYDYLQSLGKYYGYGAGNPLGPAQGIGF | HP_HAP_like, histidine phosphatases superfamily |
| 2 | 49 | 29 | VPPGCKITFVQVLSRHGARYPTKSKSKMY | Histidine phosphatase superfamily |
| 3 | 47 | 30 | VRVLVNDRVVPLHGCLVDPLGRCKLDDFVA | Local conserved domain |
| 4 | 49 | 29 | TLYADFSHDNDMTSIFTALGLYNGTEPLS | Histidine phosphatase superfamily |
| 5 | 26 | 50 | YAFLKTYNYSLGADDLTPFGEQQLVDSGIKFYQRYESLAKDIVPFIRASG | Histidine phosphatase superfamily |
| 6 | 49 | 29 | RLNKALPGVNLTSADVVSLMDMCSFETVA | Histidine phosphatase superfamily |
| 7 | 48 | 21 | GYSAAWTVPFGARAYFEKMQC | Histidine phosphatase superfamily |
| 8 | 11 | 50 | TEIFLLQQAQGMPEPGWGRITDSHQWNTLLSLHNAQFYLLQRTPEVARSR | Local conserved domain |
| 9 | 12 | 50 | KKGCPQSGQVAIIADVDERTRKTGEAFAAGLAPDCAITVHTQADTSSPDP | Histidine phosphatase superfamily |
| 10 | 9 | 50 | TPHPPQKQAYGVTLPTSVLFIAGHDTNLANLGGALELNWTLPGQPDNTPP | Histidine phosphatase superfamily |
4. Conclusion
Phylogenetic clustering and variation among biochemical features of different phytases might contribute in further classification of highly diverse HAPhys and their selection for various application purposes. Conserved sequences in motifs may be utilized for designing specific degenerate primers for identification and isolation of type and class of phytase (HAPhy) as numerous phytases are being isolated to fulfill the need of efficient phytase for feed application in various systems. Variation in biochemical features may be a key source of information for the screening of novel phytases and comparison with other classes of phytases. Functional attributes are needed to verify experimentally for conserved motifs found. This in silico analysis might be used for future genetic engineering of industrially important phytase.
References
- 1.Turner BL, Papházy MJ, Haygarth PM, McKelvie ID. Inositol phosphates in the environment. Philosophical Transactions of the Royal Society B. 2002;357(1420):449–469. doi: 10.1098/rstb.2001.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lott JNA, Ockenden I, Raboy V, Batten GD. Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Science Research. 2000;10(1):11–33. [Google Scholar]
- 3.Barnett GM. Phosphorus forms in animal manure. Bioresource Technology. 1994;49(2):139–147. [Google Scholar]
- 4.Asada K, Tanaka K, Kasai Z. Formation of phytic acid in cereal grains. Annals of the New York Academy of Sciences. 1969;165(2):801–814. [PubMed] [Google Scholar]
- 5.Wyss M, Brugger R, Kronenberger A, et al. Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Applied and Environmental Microbiology. 1999;65(2):367–373. doi: 10.1128/aem.65.2.367-373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell DB, Vogel K, Weimann BJ, Pasamontes L, Van Loon APGM. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila . Microbiology. 1997;143(1):245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- 7.Augspurger NR, Webel DM, Lei XG, Baker DH. Efficacy of an E. coli phytase expressed in yeast for releasing phytate-bound phosphorus in young chicks and pigs. Journal of Animal Science. 2003;81(2):474–483. doi: 10.2527/2003.812474x. [DOI] [PubMed] [Google Scholar]
- 8.Olukosi OA, Cowieson AJ, Adeola O. Age-related influence of a cocktail of xylanase, amylase, and protease or phytase individually or in combination in broilers. Poultry Science. 2007;86(1):77–86. doi: 10.1093/ps/86.1.77. [DOI] [PubMed] [Google Scholar]
- 9.Rutherfurd SM, Chung TK, Moughan PJ. The effect of microbial phytase on ileal phosphorus and amino acid digestibility in the broiler chicken. British Poultry Science. 2002;43(4):598–606. doi: 10.1080/0007166022000004516. [DOI] [PubMed] [Google Scholar]
- 10.Gibson DM, Ullah AHJ. Purification and characterization of phytase from cotyledons of germinating soybean seeds. Archives of Biochemistry and Biophysics. 1988;260(2):503–513. doi: 10.1016/0003-9861(88)90475-4. [DOI] [PubMed] [Google Scholar]
- 11.Hegeman CE, Grabau EA. A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiology. 2001;126(4):1598–1608. doi: 10.1104/pp.126.4.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greiner R, Alminger ML, Carlsson NG. Stereospecificity of myo-inositol hexakisphosphate dephosphorylation by a phytate-degrading enzyme of baker’s yeast. Journal of Agricultural and Food Chemistry. 2001;49(5):2228–2233. doi: 10.1021/jf0100090. [DOI] [PubMed] [Google Scholar]
- 13.Kim YO, Lee JK, Kim HK, Yu JH, Oh TK. Cloning of the thermostable phytase gene (phy) from Bacillus sp. DS11 and its overexpression in Escherichia coli . FEMS Microbiology Letters. 1998;162(1):185–191. doi: 10.1111/j.1574-6968.1998.tb12997.x. [DOI] [PubMed] [Google Scholar]
- 14.Tseng YH, Fang TJ, Tseng SM. Isolation and characterization of a novel phytase from Penicillium simplicissimum . Folia Microbiologica. 2000;45(2):121–127. doi: 10.1007/BF02817409. [DOI] [PubMed] [Google Scholar]
- 15.Ullah AH, Gibson DM. Extracellular phytase (E.C. 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Preparative Biochemistry. 1987;17(1):63–91. doi: 10.1080/00327488708062477. [DOI] [PubMed] [Google Scholar]
- 16.Mullaney EJ, Ullah AHJ. The term phytase comprises several different classes of enzymes. Biochemical and Biophysical Research Communications. 2003;312(1):179–184. doi: 10.1016/j.bbrc.2003.09.176. [DOI] [PubMed] [Google Scholar]
- 17.Chu HM, Guo RT, Lin TW, et al. Structures of Selenomonas ruminantium phytase in complex with persulfated phytate: DSP phytase fold and mechanism for sequential substrate hydrolysis. Structure. 2004;12(11):2015–2024. doi: 10.1016/j.str.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Afinah S, Yazid AM, Anis Shobirin MH, Shuhaimi M. Phytase: application in food industry. International Food Research Journal. 2010;17(1):13–21. [Google Scholar]
- 19.Konietzny U, Greiner R. Bacterial phytase: potential application, in vivo function and regulation of its synthesis. Brazilian Journal of Microbiology. 2004;35(1-2):11–18. [Google Scholar]
- 20.Cao L, Wang W, Yang C, et al. Application of microbial phytase in fish feed. Enzyme and Microbial Technology. 2007;40(4):497–507. [Google Scholar]
- 21.Lei XG, Porres JM. Phytase enzymology, applications, and biotechnology. Biotechnology Letters. 2003;25(21):1787–1794. doi: 10.1023/a:1026224101580. [DOI] [PubMed] [Google Scholar]
- 22.Oh BC, Choi WC, Park S, Kim YO, Oh TK. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Applied Microbiology and Biotechnology. 2004;63(4):362–372. doi: 10.1007/s00253-003-1345-0. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann M, Loch C, Middendorf A, et al. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Engineering. 2002;15(5):403–411. doi: 10.1093/protein/15.5.403. [DOI] [PubMed] [Google Scholar]
- 24.Van Etten RL, Davidson R, Stevis PE, MacArthur H, Moore DL. Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. Journal of Biological Chemistry. 1991;266(4):2313–2319. [PubMed] [Google Scholar]
- 25.Mullaney EJ, Ullah AHJ. Conservation of cysteine residues in fungal histidine acid phytases. Biochemical and Biophysical Research Communications. 2005;328(2):404–408. doi: 10.1016/j.bbrc.2004.12.181. [DOI] [PubMed] [Google Scholar]
- 26.Dubey AK, Yadav S, Kumar M, Singh VK, Sarangi BK, Yadav D. In silico characterization of pectate lyase protein sequences from different source organisms. Enzyme Research. 2010;2010:14 pages. doi: 10.4061/2010/950230.950230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malviya N, Srivastava M, Diwakar SK, Mishra SK. Insights to sequence information of polyphenol oxidase enzyme from different source organisms. Applied Biochemistry and Biotechnology. 2011;165:397–405. doi: 10.1007/s12010-011-9259-2. [DOI] [PubMed] [Google Scholar]
- 28.Morya VK, Yadav S, Kim EK, Yadav D. In silico characterization of alkaline proteases from different species of Aspergillus . Applied Biochemistry and Biotechnology. 2012;166:243–257. doi: 10.1007/s12010-011-9420-y. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Bjellqvist B, Hughes GJ, Pasquali C, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14(10):1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Zuckerkandl E, Pauling L. Evolving Genes and Proteins. 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology; 1994; Menlo Park, Calif, USA. AAAI Press; pp. 28–36. [PubMed] [Google Scholar]
- 35.Mullaney EJ, Ullah AHJ. Inositol Phosphates: Linking Agriculture and the Environment. Oxfordshire, UK: CAB International; 2007. Phytases: attributes, catalytic mechanisms and applications; pp. 97–110. [Google Scholar]
- 36.Lindqvist Y, Schneider G, Vihko P. Crystal structures of rat acid phosphatase complexed with the transition-state analogs vanadate and molybdate. Implications for the reaction mechanism. European Journal of Biochemistry. 1994;221(1):139–142. doi: 10.1111/j.1432-1033.1994.tb18722.x. [DOI] [PubMed] [Google Scholar]
- 37.Porvari KS, Herrala AM, Kurkela RM, et al. Site-directed mutagenesis of prostatic acid phosphatase. Catalytically important aspartic acid 258, substrate specificity, and oligomerization. Journal of Biological Chemistry. 1994;269(36):22642–22646. [PubMed] [Google Scholar]
- 38.Ikai A. Thermostability and aliphatic index of globular proteins. Journal of Biochemistry. 1980;88(6):1895–1898. [PubMed] [Google Scholar]
- 39.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Research. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guruprasad K, Reddy BVB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering. 1990;4(2):155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 41.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 42.Gough J. The SUPERFAMILY database in structural genomics. Acta Crystallographica Section D. 2002;58(11):1897–1900. doi: 10.1107/s0907444902015160. [DOI] [PubMed] [Google Scholar]


