Abstract
In addition to the classic methods of structural biology - X-ray crystallography and NMR, solution X-ray scattering (SAXS) is starting to play an important role in experiential structural investigation of biological macromolecules. Ease of SAXS data collection and sophistication of its data analysis tools increasingly used as black boxes can be seen as both a blessing and a curse. On one hand, a sample set aside for solution scattering will always yield experimental data, including cases when macromolecule cannot be crystallized or when it is too large for application of solution NMR. On the other hand, any sample, whether pure or contaminated, whether mono- or polydisperse, will yield scattering data and it is up to the user to ensure the absence of artifacts in them and to choose a proper structural modeling strategy. We will discuss experimental aspects of X-ray solution scattering including sample preparation, data collection, as well as the steps in data processing and preliminary analysis that need to be carried out to ensure the absence of artifacts. Our goal is to summarize everything than can possibly go wrong with SAXS data measurement so that the user can have confidence in the data before they enter structural modeling.
Keywords: solution X-ray scattering, SAXS, WAXS, sample preparation, aggregation, radiation damage, protein and RNA structure, structural biology
Introduction: Solution X-ray scattering in structural biology
Solution scattering is becoming an increasingly popular technique for structural investigation of bio-macromolecules at nearly-physiological conditions (Koch et al., 2003; Petouhkov and Svergun, 2007), particularly in cases when application of the mainstream methods of structural biology - X-ray crystallography or NMR, is difficult (Putnam et al., 2007). Such examples include systems that resist crystallization attempts and those that are too large to be studied via solution NMR. Important advantages of solution scattering include a wide range of the macromolecular sizes for which the technique is applicable - from singe kDa to hundreds of kDa, as well as a possibility to acquire meaningful interpretable data for a variety of systems that are intrinsically difficult to analyze quantitatively via crystallography or NMR such as unfolded or flexibly linked macromolecules, micelle-solubilized membrane proteins, or fibrils, to name a few. The scattering intensity data are measured as the difference between the sample containing the macromolecule of interest and the matching buffer (Figure 1). These data, collected from isotropic solutions as a function of the scattering angle, are commonly converted to be function of the scattering vector q=4π sin(θ)/λ, where 2θ is the scattering angle and λ is the wavelength of the incident radiation (typically, λ~0.6-1.5 Å for X-rays). Taken in isolation, one-dimensional solution scattering data cannot yield an unambiguously defined three-dimensional structural model. This ill-defined nature of the inverse problem (determination the structural model from the experimental scattering data) can be alleviated by imposing additional constraints on the fitted solution.
Figure 1.
Schematic representation of SAXS data collection process. Scattering data are acquired from both sample and the matching buffer, converted from the two-dimensional images to the one-dimensional scattering curves and subtracted, resulting in the scattering difference curve.
For example, a requirement that the fitted model is described by a compact shape with a smooth surface effectively restricts the solution to be no more that two-dimensional. Applications of this strategy (Chacon et al., 1998; Svergun, 1999; Walther et al., 2000; Svergun et al., 2001), so-called ab initio shape reconstructions from the scattering data, operate without any assumptions regarding the actual shape of the macromolecule and were shown to be capable of deriving low-resolution models that fit well to the atomic coordinates of the corresponding structures (Takahashi et al., 2003). The advantage of these methods is extreme ease of their usage, as they require only the measured scattering data and one additional parameter - the particle’s maximum length, dmax, readily extractable from the SAXS data. Several practical problems come at the expense of this ease. One of them is a difficulty in interpreting the derived low-resolution shapes. In many cases, the shapes determined via ab initio methods are featureless and do not allow immediate recognition of the underlying macromolecular architecture. Second, the fitted shapes are biased by the way the program code enforces particle compactness and do not have the fidelity of reconstructions from methods such as cryo-electron microscopy. This issue is not easy to alleviate by increasing the resolution range of the fitted scattering data since accurate modeling of the wide-angle scattering data requires precise definition of the atomic coordinates of the macromolecules.
Another class of techniques decreases the number of degrees of freedom in the fitted structural model in cases when it can be broken into rigidly held subunits whose coordinates are known (Petoukhov and Svergun, 2005). The accuracy of the resulting rigid-body reconstructions depend on both the accuracy and completeness of the coordinates of the individual subunits, as well as on the resolution range and the signal-to-noise ratio of the experimental scattering data. In many cases, both low-resolution ab inito shape reconstructions and rigid body fits share a common problem - so called structural degeneracy, defined as inability to obtain a unique three-dimensional structural model from fitting one-dimensional solution scattering data (Volkov and Svergun, 2003). This degeneracy can be only alleviated by imposing additional constraints on the system, which may not always be available.
SAXS data can also be fitted during high-resolution structure determination in combination with other sources of experimental restraints. Frequently, NMR data are used in these applications since global orientational restraints from NMR are ideally complementary to translational information from the solution scattering data (Mattinen et al., 2003; Grishaev and Bax, 2005; Schwieters and Clore, 2007; Grishaev et al., 2008a,b; Schwieters et al., 2010; Mittag et al., 2010; Takayama et al., 2011). Applications of this approach have been demonstrated to produce measurable increases in structural accuracy and represent some of the most accurate ways in which solution X-ray scattering data can be used for structure determination, Such applications, however, rely on having a sufficient number of restraints from a complementary technique, which should include both local restraints such as torsion angles, and global restraint such as residual dipolar couplings.
To summarize, the scattering data on their own can be very useful for distinguishing between candidate structural models when such models are available. A good fit between an accurate structural model and the measured SAXS data is required, but such fit is usually not sufficient to establish the model’s accuracy unambiguously. On the other hand, a poor fit necessarily implies that the model is inaccurate. Generation of the structural model based on SAXS data is a difficult problem that requires additional information, most commonly in form of structural models for the subunits, or experimental restraints from a complementary technique.
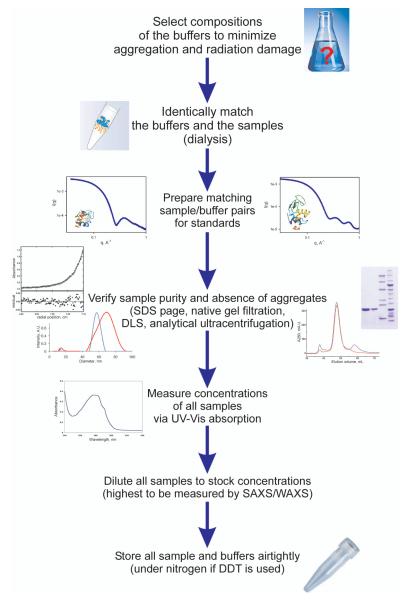
All of the above points assume that the experimental scattering data are (i) free from artifacts and (ii) accurately modeled with a correct oligomerization state (Jacques and Trewhella, 2010). The former presents the largest difficulty in dealing with the SAXS data since the artifacts are often not obvious and can be obscured by low signal/noise ratio and limited resolution range of the experimental data. The most important characteristic of high-quality scattering data is that they originate from a collection of pure, monodisperse, and non-interacting particles of interest. Both impurities and size polydispersity can render SAXS data completely uninterpretable, unless the individual concentrations of all species are accurately known. The sections below describe the procedures that can decrease the chances of encountering the artifacts in the SAXS data, and the early data analysis steps that can help to identify their presence. A schematic representation of the sample preparation procedures is shown in Figure 2.
Figure 2.
Flowchart for the SAXS sample preparation procedures.
Preparation of the matching buffer for SAXS measurements
Samples for solution scattering measurements can be often prepared at conditions that closely match the ones typically used for other experimental biophysical techniques such as crystallography or NMR. In addition to the sample containing the biomolecule of interest, a buffer is required that identically matches composition of the solvent within the sample. Such buffer cannot be simply made from the same amounts of buffer additives that were used in preparing the samples since the measured difference signal between the sample and buffer is small and can be significantly impacted even by minute imbalance of the respective solvent compositions. The most common technique to ensure the exact sample/buffer match is a multi-stage dialysis through a semi-permeable membrane with the molecular weight cutoff (MWCO) smaller than the molecular mass of the macromolecule of interest. Total dialysis time of at least 16-48 hrs is in most cases sufficient to achieve the exact sample/buffer match. The buffer needs to be stirred throughout the process and the membrane area needs to be large enough to ensure complete solvent equilibration. In most cases, microdialysis vials with volumes between 50 and 500 μL are appropriate. Buffer also needs to be degassed before dialysis with either low vacuum or sonication, to decrease the chances of bubble formation during measurements.
In cases when the samples are too unstable for a lengthy dialysis, the matching buffers can be made by centrifugation though an appropriate-MWCO filter membrane or equilibration via column filtration. Since new filter membranes contain traces of organic materials, a portion of the buffer has to be passed through them before the sample is loaded. This method should not be used unless absolutely necessary due to the generally lower quality of the resulting sample/buffer match. Prior to loading for equilibration with the buffer, the sample should be passed through a 0.2 μm membrane to remove the high-MW aggregates. This step is particularly warranted when the samples are prepared from lyophilized powders.
Buffer composition guidelines for SAXS samples
Contents of the buffer can vary significantly, but not without restrictions. In order to minimize X-ray absorption and subsequent production of the free radicals (primarily, OH), the buffers should be free from high-Z elements as much as possible. Importantly, the concentration of phosphates has to be kept to a minimum (below 20 mM). Presence of detergents, unless they are necessary to solubilize the protein, has to be controlled since large detergent assemblies formed at concentrations exceeding the critical micelle concentration produce strong scattering signals that can be noticeable on top of the signals from the macromolecules. Buffer compositions should be optimized to ensure monodispersity of samples at the scattering measurement conditions.
Reagents capable of capturing the OH free radicals formed with X-ray absorption by the buffer need be added whenever possible. Frequently, DTT at 5-10mM, or TCEP at 1-2 mM are used as additives for SAXS buffers in cases when the macromolecules are compatible with them. Since DTT is oxidizeable by air and its oxidized form absorbs at 280 nm interfering with the sample concentration measurements, both the samples and buffers containing DTT should be kept under nitrogen during dialysis and the subsequent storage up till data collection. The presence of DTT or TCEP becomes a must when the protein contains surface-exposed Cys residues. When DTT or TCEP cannot be used, organic buffering agents such as Tris or HEPES, or free radical scavengers such as glycerol, ascorbate, ethylene glycol, or sucrose can be employed.
In cases when samples are stable at higher ionic strengths, salts should be used to decrease the long-range electrostatic repulsion between the macromolecules at the measurement conditions. The presence of such repulsion manifests itself in a so-called inter-particle structure factor, in extreme cases observable as the decrease of the scattering intensity at the lowest scattering angles. Frequently, 150 mM NaCl buffer can be effective at suppressing the structure factor above 0.02-0.03A−1 for protein concentrations below 5 mg/mL. Adjusting pH of the buffer closer to the protein’s isoelectric point can also decrease long-range electrostatic repulsion but this approach should be used with caution as it can also lead to increased aggregation.
In most, cases, the exact wavelength used in the X-ray scattering measurement makes little impact on the measured data aside from the decreased X-ray absorption and radiation damage at higher energies, with an important exception of the anomalous scattering. Therefore, unless anomalous scattering data are being collected, buffers should be free from elements with K-edges below the energy of the incident X-ray radiation. For example, if 18keV radiation is to be used, such elements include Rb, Br, and Sr. In cases where the presence of any of these ions is necessary, incident X-ray energies of 8-12keV would be preferable.
Final steps in SAXS sample preparation
Following buffer equilibration, the concentrations of all stock samples used for SAXS data collections should be accurately measured, preferably via UV-Vis absorption. For samples coming from NMR studies, accurate concentrations can also be obtained from integrated intensity of well-resolved resonances in one-dimensional spectra by reference to a standard with known concentration. Sample concentration should be determined with an error not exceeding 10% for accurate determination of the molecular weight of the macromolecule via SAXS, as will be described later.
For stock solutions, concentrations not exceeding 5-10 mg/mL are suggested. Both the samples and the matching buffers should be stored in air-tight containers prior to the data collection in order to remain at the exact match. It is recommended to carry out sample preparation close to the future measurement time to minimize sample aggregation and to ensure the best sample/buffer match. Unless the sample is unstable in aqueous solution, samples should not be frozen prior to data collection. Preparation of the macromolecular stock solution for the sample has to be done keeping in mind the likely concentration range for the scattering measurements - it is best to limit the stock concentration to the highest to be measured. The preparation also has to take into account the amount of material that will be spent during the entire measurement process, considering the volumes to be loaded during each data collection. Scattering measurement typically involves collecting several concentration points (100%, 50%, 25% of the stock concentration, at the very least), possible data collection repeats, data acquisitions at a different sample-to-detector distance to expand the probed q-range, etc.
The last steps in sample preparation following the concentration measurements of all stock samples should include quality assurance procedures that help to establish sample purity and the absence of high-MW aggregates. Acceptable methods include SDS-Page, native gel filtration, dynamic light scattering, or analytical ultra-centrifugation.
In addition to the actual samples of interest, it is also recommended to prepare several standard samples using the same procedures as outlined above. The purpose of collecting SAXS data on such standards is two-fold. First, the standards allow to test proper operation of the instrument and to correct any problems that are found before samples of interest are loaded. Second, well-behaved standards with established oligomerization states and precisely measured concentrations can be used to determine the molecular weights and aggregation states for the samples of interest. For proteins, the commonly used standards include hen egg white lysozyme at pH 4.0-4.4, horse heart cytochrome C at pH 7.0, or freshly prepared bovine serum albumin.
SAXS data measurement: lab-based instruments versus synchrotron beam lines
Solution X-ray scattering data can be acquired using either lab-based instruments or synchrotron beam lines. The former operate at a fixed energy, typically Cu Kα at 8keV, while the latter allow variation of the incident X-ray energy, typically between 7 and 20 keV. Lab-based sources offer the possibility of immediate measurements once the sample and instrument are available at the expense of fairly long data collection times (typically between 30 min and several hours). Due to moderate flux from the sources of the lab-based instruments, radiation damage is typically less of a problem with them, compared to the synchrotron measurements. For the samples that are extremely radiation-sensitive, lab-based instruments might present the only chance of collecting interpretable X-ray scattering data.
Synchrotron beam lines offer a possibility to vary the energy of the incident X-ray photons, which is useful to adjust the q-range of the acquired data, and to decrease radiation damage with higher incident X-ray energies. Since, aside from the element-specific absorption edges, atomic form factors change very little with the incident radiation energy in the typical range used for the synchrotron SAXS data collections (10-20 keV), scattering data collected at multiple wavelengths within this range should be perfectly superimposable and the choice of the incident wavelength is often dictated by the desired q-range and flux/energy profile of the beam line. The incident energies at or below 6keV are not suitable due to the increased absorption by the aqueous buffers.
For both lab-based and synchrotron data collections, the space between the sample and the detector has to be kept in low vacuum (below 10−3 bar) in order to minimize air scatter which can be noticeable relative to the generally weak scattering signal from the macromolecules. Typically, an evacuated stainless steel tube is inserted between the sample and the detector for this purpose. In rare cases when vacuum is not attainable, the tube should be filled with helium since its X-ray scatter is smaller by a factor of 50 than that of the air. The placement of the tube should minimize its air gaps relative to the detector and the sample cell. The small air gap with the sample cell is more important as it decreases the exposure of the air to the direct beam. Close to the back end of the flight tube, a beam stop made from a materials with high X-ray stopping power within a wide range of energies (such as tungsten or iridium) is placed in vacuum in order to completely absorb the direct beam before it hits the detector. The beam stop, aligned with a strongly attenuated beam during the instrument setup, will frequently have a PIN diode mounded on it which reports the photon count transmitted through the sample. Since the intensity of the scattered radiation is ~105 times weaker than that of the direct beam, air scatter between the flight tube and the detector from the X-rays scattered by the sample which go around the beam stop is much less of a problem. The beam stop has to be positioned as close as possible to the detector and be as small as possible to minimize the lowest measured q. Long sample-to-detector distances (2-4 m) are used for collections of the small angle data with maximum scattering angles not exceeding ~3°, while short sample/detector distances (0.6-0.3 m) are used to collect wider-angle data corresponding to the maximum scattering angles reaching 30-45°. Beam line setup for a SAXS camera and some if its components are shown in Figure 3.
Figure 3.
A typical synchrotron SAXS setup. Station 12-IDC, Advanced Photon Source, Argonne National Laboratory, Argonne, IL.
SAXS data measurement on a lab-based instrument
Data collection on a lab-based instrument includes fewer adjustables than when using a synchrotron due to the fixed X-ray energy and small chances of radiation damage. In cases when the detector can be moved relative to the sample, an appropriate set of the sample/detector distances has to be decided upon based on the needed q-range. These distances should be calibrated using a strong scatterer such as silver behenate. Sample and buffer data collections should be done with the same exposures, long enough to obtain the needed data uncertainty. When CCD detector is used, dark current measurement should be done with the same exposure as for the sample and buffer, and subtracted from both data sets. In a case when line-shaped beam and Kratky camera setup is used, beam profile should be measured in both dimensions and used for data desmearing. GNOM software can be used for this purpose (Svergun, 1992) with the trapezoidal parameterization of the beam profile. Typical range of concentrations for the biological macromolecules with the lab-based measurements is 2-10 mg/mL.
Synchrotron SAXS data measurement: q-range selections(s)
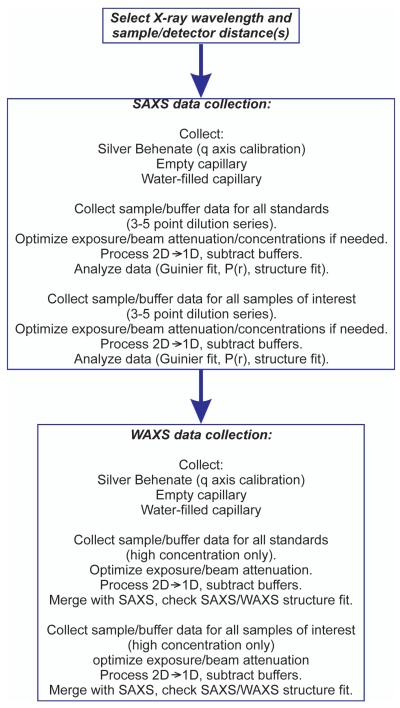
Setup and planning of the synchrotron SAXS data collection starts from selection of the sample/detector distance(s) and the incident X-ray energy in order to cover the desired q-range. For most macromolecular samples, minimum q of 0.006-0.01 Å−1 is needed for reliable determination of the particle size and evaluation of particle aggregation and/or structure factor effects. Maximum q to be measured would largely depend on the size of the macromolecule with smaller particles requiring larger qmax. Typically, qmax should be at least 0.20-0.25Å−1 for particles above 100kDa and at least 0.5 Å−1 for smaller macromolecules. Protein SAXS data at q above 1 Å−1 are often very similar and rarely used for structural analysis. On the other hand, oligonucleotides often exhibit distinct features within a wide angular range due to higher regularity of the RNA/DNA structure at small length scales. Data that include the water scattering peak at ~2 Å−1 can be very helpful to detect sample/buffer mismatches. For this purpose, separate wide-angle x-ray scattering (WAXS) data collections with a short sample/detector distance are recommended at synchrotron beam lines. For example, a 50 cm sample-to-detector distance and incident photon energy of 18 keV can result in measurable q range as wide as ~0.1 to ~2.2Å−1. The SAXS and WAXS data collections should have a minimum overlap region spanning ~0.1Å−1 for accurate merging of the two data sets. In addition to expansion of the measured q-range, WAXS data collection frequently brings a benefit of higher signal/noise data within the region overlapping with the SAXS acquisition. The sequence of steps for collection of the scattering data is outlined in Figure 4.
Figure 4.
Flowchart for the collection of the experimental scattering data.
Measurement of the synchrotron SAXS data: preliminary steps
The main advantage of using a synchrotron is the extremely high photon flux attainable with modern undulator beam lines. Reaching ~1012-1014 photons/sec, which brings a real possibility of radiation damage, high beam intensities allow data collections that are fast (0.1-10.0 sec exposure time) and can be done at very low sample concentrations (as low as ~0.1 mg/mL for proteins), which are extremely useful for suppressing macromolecular aggregation.
Preparations for synchrotron SAXS measurements include calibration of the q axis with a strong scatterer standard such as silver behenate with 0.1076 Å−1 spacing between the successive powder diffraction peaks. Preliminary measurements with a water-filled capillary should include estimation of the maximum exposure that keeps the measured data within the range of the linear response of the detector. For SAXS measurements, maximum exposure time is limited by the intensities the lowest measurable q values, and for WAXS by the scattering intensity near the 2.0 Å−1 water peak. The estimated exposure times may be subsequently decreased depending on the resistance of the samples to radiation damage.
Scattering data collection should include, at a very minimum, measurement of the scattering from the cell filled with the buffer and the sample, with the empty cell and the cell filled with pure water measurements recommended as well. Both sample and buffer data collections should be done using the same cell, identically positioned and thoroughly cleaned between the measurements to remove any macromolecular deposits from the cell walls. For SAXS measurements on aqueous samples, cell path length of 1 mm is close to optimum. For cleaning the flow-through cell, a sequence of flushes with water, 2% bleach, isopropanol, and water works well. In most cases, the cell does not need to be cleaned with this procedure after the buffer data collection and can be just flushed with the buffer. When a static cell is used, it should also be thoroughly cleaned and dried prior to loading the sample/buffer for each measurement. The cell needs to be kept at a constant temperature during both sample and buffer measurements for which Peltier elements are well suited.
It is recommended to start with buffer measurement and follow it with the sample using the same exposure times. When acquiring a concentration series on the same sample, data subtractions are most accurate if separate buffers are acquired before each sample dilution, as opposed to using a single buffer for all sample concentrations. Flow-though setup is recommended for synchrotron data collections in order to decrease radiation damage. Measurements at several exposures/beam attenuations are also recommended to detect the presence of radiation damage. Data collections are frequently done in 10-40 sequential frames that should be analyzed for any systematic time-dependent changes between frames associated with the radiation damage. If a flow-through setup is used, it is important that a plug of the matching buffer is loaded immediately preceding the sample load for the measurement. In this case, mixing of the plug material and the sample during pump operation will, at worst, dilute the sample, preserving the sample/buffer match.
Bubbles that are formed within, or pass though the irradiated volume resulting from either exposure to X-rays or pump operation can be a serious problem since they increase the transmitted photon counts and decrease the scattered intensities causing substantial systematic errors in data subtraction. They are more likely to be happen during the sample measurement, more frequent for proteins rather than RNA/DNA, and are especially easily formed when detergents are present in the buffer. Bubbles due to X-ray absorption can be minimized by decreasing exposure times or attenuating incident beam intensity and by flowing the sample during the measurement. Bubbles caused by flow-through operation can be minimized by decreasing the flow rate and using the tubing with the internal diameter that matches the cell. Degassing the samples/buffers and thoroughly cleaning both the cell and the tubing that leads to it after each sample measurement also helps to decrease bubble formation.
SAXS measurement should include a series of concentrations in order to evaluate oligomerization and inter-particle repulsion (structure factor) effects. At least 3-5 concentration points should be acquired with dilutions by a factor of 2. For proteins with MW above 100 kDa, highest measured concentration of 1-2 mg/mL is recommended, while for smaller macromolecules the highest concentration can be in the range between 5 and 10 mg/mL. For RNA or DNA samples, increased electrostatic repulsion between oligonucleotides due to their higher surface charge density results in a larger magnitude of the structure factor compared to the proteins at the same concentration and buffer ionic strength. Therefore, it is important that the lowest measured concentration for an RNA/DNA sample falls down to at least 0.5-1.0 mg/mL for a buffer containing 150 mM salt. At these conditions, RNA/DNA structure factors are often minimal above q=0.02-0.03 Å−1. Buffers with lower ionic strengths would require sample measurements at even lower concentrations. Since the effects of the structure factor rarely extend into the WAXS region, wide-angle data can be collected at a single concentration only, typically that of the stock solution.
SAXS data processing: low-angle data analysis via Guinier approximation
Successful data collection during a time-limited frame of a synchrotron session requires that data are processed and undergo a preliminary analysis as soon as possible after the sample/buffer pair is acquired so that repeat collections or modifications to the data collection procedures can be done in a timely manner. The conversion of the two-dimensional scattering intensity image to the one-dimensional scattering profile usually involves standard scripts with little freedom for user modifications. Such procedures include detector masking, subtraction of the detector dark current, scaling of each detector pixel for its previously measured sensitivity and the solid angle as seen from the sample, removal of abnormally intense pixels, and azimuthal integration to yield the 1D scattering intensity curve, followed by conversion of the scattering angle to the q axis. Collection of the scattering data should be accompanied by measurements of both the incident and transmitted beam intensities for each acquired time frame. These measurements are necessary since subtraction of the buffer from the sample data requires normalization of both data sets for the incident photon counts and transmissions. The subtracted scattering data are calculated with buffer scaled by α, the volume fraction of the solvent in the sample:
| Equation 1 |
The factor α can be approximately calculated as 1-cmg/mL*7.4 10−4 for proteins and 1-cmg/mL*5.4 10−4 for RNA/DNA, I° are the incident photon counts and T stand for transmission factors. Sample/buffer subtraction that uses the empty capillary scattering is more accurate and should be used when very precise data are acquired within a wide angular range:
| Equation 2 |
Needless to say, precise measurements of all incident and transmitted photon counts, as well as sample concentrations are crucial for accurate data subtraction. Negative intensities in the final subtracted data can be linked to either sample/buffer mismatch, or inaccuracies of the measured incident/transmitted photon counts, or the presence of bubbles within the irradiated volume during the sample measurement.
SAXS data analysis
The intensity of radiation that is elastically and coherently scattered by an ensemble of randomly oriented identical particles can be generally expressed as
| Equation 3 |
Here, N is the number particles in the irradiated volume, V is the particle volume and δρ is the difference between the average electron density of the particle and that of the solvent. The form factor F(q) reflects the shape of a single particle and the associated distribution of the inter-atomic distances within the particle, P(r):
| Equation 4 |
The structure factor S(q), equal to 1 independently of q for completely uncorrelated particles, is otherwise linked to g(r) - probability distribution function for the inter-particle distances in solution:
| Equation 5 |
Importantly, separabilty of the form and structure factors can only be guaranteed for centro-symmetric interaction potentials, and therefore does not apply to the concentrated solutions of strongly anisometric particles.
Experimental SAXS data are typically visualized as either log I(q) vs. q, or log I(q) vs. log q. A flat plateau at the lowest angles for the latter plot is a visual characteristic of the data free from the effects of aggregation and inter-particle repulsion. The first step in data analysis should be a linear fit of the lowest-q data in so-called Guinier coordinates (ln I(q) vs q2). In this regime, the intensity scattered by a monodisperse system of non-interacting particles is described by a simple formula
| Equation 6 |
The slope of such Guinier fit can be used to extract the gyration radius Rgyr of the scattering particle (r.m.s. distance from all its points to the center). The zero angle scattering intensity, unobservable directly due to the beam stop shadow, is related via I(0)~V2*(δρ)2*c to the volume of the scattering particle V, its electron density contrast relative to the solvent δρ, and its concentration, c. Since the highest scattered intensities come at the lowest angles, Rgyr and I(0) are the most precise parameters that can be obtained from SAXS data, assuming that aggregation and inter-particle repulsion effects are negligible. The limited range of applicability of the Guinier relationship (qmaxRgyr <1.3 for globular and qmaxRgyr < 1.0 for elongated shapes) requires interactive adjustments of the minimum and maximum fitted q while inspecting the fit residual for any systematic deviations, most readily accomplished using PRIMUS software (Konarev et al., 2003). Absence of aggregation and inter-particle repulsion in the sample manifest themselves in a linear Guinier plots with no bias in the fit residual and Rgyr and I(0)/c values that are independent of the sample concentration. In cases where the concentration dependence of these parameters is detected, the scattering data have to be either extrapolated to zero concentration using programs such as PRIMUS, or the structure and form factors have to be fitted simultaneously using an approach such as GIFT (Fritz et al., 2000).
For a monodisperse sample with negligible structure factor in the fitted Guinier range, I(0) can be used to obtain the molecular mass of the scattering particle and, correspondingly, its oligomerization state. This analysis can be performed based on the measured scattering intensities of the standard samples since the I(0) values, after normalization by the sample concentration in mg/mL, are proportional to the proteins’ molecular masses (Mylonas aand Svergun, 2007). Such MW estimates are most accurate for folded proteins, which are characterized by an approximately constant electron density. On the other hand, the concentrations of intrinsically unfolded proteins are less accurate when determined via SAXS due to their data’s narrower regions of linearity in the Guinier coordinates and the difference between their contrasts relative to the solvent vs. that of the folded proteins. Due to the contrast dependence of the I(0) values, it is best if the nature of the standard sample (protein vs. RNA vs. DNA) matches the sample of interest. For proteins, the accuracy of the molecular weight obtained via this method has been estimated to be ~10% (Mylonas aand Svergun, 2007). In case where the sample of interest is RNA or DNA and standards are proteins, the standards’ I(0) values normalized by their concentrations in mg/mL should be scaled up by a factor of ~3.1 due to the electron density difference between oligonucleotides and proteins. Since the scattering intensity is also impacted by a higher electron density of the solvent surrounding oligonucleotides relative to the proteins, the error of such MW estimates is higher than the ~10% uncertainty for the proteins. Alternatively, molecular weight of the scattering particle can be determined by placing the data on the absolute scale using the pure water and empty capillary measurements (Orthaber et al., 2000)
Polydisperse samples can fall into two classes (i) a very large aggregate, and (ii) a continuum of oligomerization states, in addition to the species of interest. The former case is easier to deal with since the effect of very large particles is primarily limited to the lowest scattering angles and can be either removed by truncating the lowest-q data, or the aggregates themselves can be removed by filtering through a high-MWCO membrane or native gel filtration. This case is typically characterized by a linear Guinier plot with an abrupt upward turn at the lowest q. The case of the oligomerization equilibrium can only be resolved by shifting it towards the monomeric species with sample dilution or change in the buffer composition (ionic strength, pH, etc.) and is generally more difficult to address. This situation manifests itself in a Guinier plot that exhibits upward curvature throughout the entire low-q range, visually resembling that of the intrinsically unfolded proteins. Examples of several types of Guinier plots corresponding to common artifacts are shown in Figure 5.
Figure 5.
Analysis of the low-q SAXS data via Guinier fits. Quality of the fit is more obvious from the difference between the measured and fitted data, shown at the bottom of the panels. (A) Normal fit, monodisperse system. (B) Large aggregate in addition to the species of interest. The parameters of the Guinier fit within the truncated range and the resulting Rgyr and I(0) values are normal. (C) Polydisperse system. Note the upward curvature in the fit discrepancy throughout the entire low-q range. (D) Intrinsically unfolded protein. Note a similar upward curvature in the fit discrepancy throughout the entire low-q range. (E) The effect of a subtle inter-particle repulsion, only noticeable with the concentration series. All data were normalized to their concentrations. Note the absence of any systematic trends in the fit residuals. The presence of the structure factor is only noticeable from concentration dependence of the Rgyr and I(0)/c values. (F) The effect of a substantial inter-particle repulsion. Note the downward curvature in the fit discrepancy throughout the entire low-q range.
The presence of aggregation can be further ruled out by the analysis of the distribution of the inter-atomic distances, P(r), within the scattering particle, which can be obtained via regularized Fourier transform of the scattering data using programs such as GNOM (Svergun, 1992) or GIFT (Fritz et al., 2000). Such programs typically have one adjustable parameter - maximum particle dimension dmax, which needs to be iteratively optimized. From the Nyquist-Shannon sampling theorem, such optimization requires that the smallest fitted q is below π/dmax. The bounds on the dmax can be obtained by overestimating it, in which case the long-distance tail of the P(r) distribution can turn negative close to dmax, or underestimating it, in which case P(r) falls to zero very abruptly towards dmax. A properly defined dmax will correspond to P(r) distribution that is positive everywhere, exhibits no oscillations, provides a good fit to the data, and smoothly decays to zero while approaching dmax (Figure 6). Detergent micelles are an exception to the last of these rules, since their P(r) curves frequently have negative regions at intermediate distances due to a core/headgroups contrast sign difference. In the cases when the approximate maximum particle dimension is known, the agreement between dmax determined from the SAXS data and the value measured from the structural coordinates confirms that the oligomerization state in solution matches the assumed state of the macromolecule. A point to note is that, since dmax extracted from the experimental SAXS data is also impacted by the layer of solvent surrounding the macromolecule, it can be 5-10 Å larger that the value determined from the macromolecular coordinates. In practice, the uncertainty of the dmax value determined via P(r) fits can be assumed to be no less than ~3-5 Å, making it a fairly “soft” parameter. Distributions of the inter-atomic distances obtained via regularized Fourier transform methods can also be used to extract I(0) and Rgyr values as the zeroth and first moments of P(r), respectively. Such estimates tend to be less affected by the aggregation or long-range inter-particle repulsion but can be more susceptible to the errors in data subtraction or a sample/buffer mismatch. SAXS data of good quality should exhibit a close agreement (within their reported uncertainties) between the I(0) and Rgyr values extracted from Guinier and P(r) analyses.
Figure 6.
Regularized Fourier transforms of the SAXS data using the program GNOM (Svergun, 1992) to obtain the P(r) distribution and the associated maximum dimension of the particle, dmax. The red curve corresponds to the correct dmax=46Å, the pink to the underestimated dmax=38Å, and the green to the overestimated dmax=60Å. Note the negative tail of the P(r) distribution for dmax=60Å and the abrupt drop to zero for dmax=38Å. Panel (A) shows the fits of the SAXS data with the Fourier transforms of the three P(r) distributions shown in panel (B). Insets show the expanded view of the high-q data fits in panel (A) and the long-distance tails of the P(r) distributions in panel (B).
When macromolecular coordinates are known, even if only approximately, the presence of aggregation, radiation damage, and inter-particle structure factor can be sometimes detected from the fit of the experimental data to the structural coordinates. Such fits can be done using a variety of methods (CRYSOL, Svergun et al., 1995; Fast-SAXS, Yang et al., 2009; FoXS, Shneiderman-Duhovny et al. 2010, AXES, Grishaev et al., 2010; AquaSAXS, Poitevin et al., 2011) with CRYSOL being the most popular. Systematic discrepancy between the fitted and measured data at the lowest q values can serve as an indicator of either inter-particle repulsion (in which case the measured Rgyr is lower than model Rgyr) or aggregation or radiation damage (when measured Rgyr is higher that model Rgyr). This analysis can be used when the structural model is complete and has an accurate Rgyr.
Flexibility of the macromolecular architecture in solution leads to representation of the scattering particle by an ensemble of distinct models from which the observed SAXS data are averaged. By itself, flexibility does not cause any problems with data collection or lead to artifacts. However, its presence should be recognized for proper modeling of the scattering particle. In particular, SAXS data collected from flexible/unfolded systems should not be fitted via tools that use a single structural model such as most of the ab initio or rigid body modeling programs. For flexible macromolecules, models can be fitted to the scattering curves using programs specifically designed to deal with the ensemble-averaged data such as EOM (Bernado et al., 2007) or BILBOMD (Pelikan et al., 2009). The most frequently used method to detect flexibility from SAXS data is via Kratky plot (q2 I(q) vs. q). This plot is based on change in the appearance of an unfolded macromolecule from a Gausssian coil (I(q)~q−2) to a thin rod (I(q)~q−1), as q is increased. A random polymer would thus have a Kratky plot that shifts from a plateau at intermediate q values to a straight line at higher q. In reality, unless the protein is unfolded (in which case the disorder is also often obvious from the upwards curved Guinier plot), recognition of structural disorder/flexibility from Kratky plot can be challenging as there are no clear criteria for distinguishing between a limited degree of disorder and a completely rigid system (Figure 7). For a multi-domain system, when NMR data are available, a better indicator of flexibility is the difference between the rotational diffusion tensors of the individual domains fitted to the NMR relaxation data, or the difference between the molecular alignment tensors of the individual domains fitted to the residual dipolar couplings data.
Figure 7.
Kratky plot used to detect structural flexibility from SAXS data. While the appearance of an unfolded protein is clearly distinct, the differences between flexibly linked and completely rigid proteins can be very minor. Application of this analysis requires a wide angular range and high signal/noise of the scattering data.
Summary
Increasing application of X-rat solution scattering to study the structure of biological macromolecules leads to a situation when such data are collected by users with relatively little experimental experience in this technique. However, with improvements in instrumentation and data analysis tools, even a novice user should be able to collect high-quality SAXS data as long as proper procedures are followed. In practice, these procedures require that data aquisition and preliminary analysis include steps to ensure the absence of common artifacts including sample/buffer mismatch, sample impurities, aggregation/oligomerization, inter-particle repulsion, and radiation damage. When present, these factors can significantly affect the parameters extracted from SAXS data and, therefore, introduce substantial artifacts in the models derived from their analysis. Many of these problems can be overcome with proper preparation and thorough preliminary characterization of the samples. Usage of secondary standards is strongly recommended as they allow to determine the molecular weight of the scattering particle. When collecting scattering data, care needs to be taken to ensure the absence of radiation damage. SAXS data should be always acquired in concentration series in order to account for the effects of the inter-particle repulsion. Data should be immediately analyzed using Guinier fit, P(r) distribution via regularized Fourier transform, and fits to the atomic coordinates, when available. Many of the issues discovered during such analysis are immediately addressable with acquisitions at lower flux intensities and/or concentrations. System flexibility should be recognized for proper modeling of the scattering particle. When these guidelines are followed, SAXS data can be an extremely valuable source of structural information for a wide variety of biomolecular systems.
Acknowledgements
The author would like to thank Drs. Jill Trewhella, Ad Bax, Soenke Seifert and Xiaobing Zuo for training and helpful discussions. This work was supported by the Intramural Research Program of the NIDDK, NIH, and by the Intramural Antiviral Target Program of the Office of the Director, NIH.
Literature cited
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural chracterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Chacon P, Moran F, Diaz JF, Pantos E, Andreu JM. Low-resolution structures of proteins in solution retrieved from x-ray scattering with a genetic algorithm. Biophys. J. 1998;74:2760–2775. doi: 10.1016/S0006-3495(98)77984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G, Bergmann A, Glatter O. Evaluation of small-angle scattering data of charged particles using the generalized indirect Fourier transformation technique. J. Chem. Phys. 2000;113:9733–9740. [Google Scholar]
- Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle x-ray scattering and NMR data. J. Am. Chem. Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. J. Biomol. NMR. 2008a;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Ying J, Canny M, Pardi A, Bax A. Solution structure of tRNA(Val) from refinement of homology model against residual dipolar couplings and SAXS data. J. Biomol. NMR. 2008b;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishaev A, Guo L, Irving T, Bax A. Improved fitting of solution x-ray scattering data to macromolecular structural ensembles by explicit water modeling. J. Am. Chem. Soc. 2010;132:15484–15486. doi: 10.1021/ja106173n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. Small-angle scattering for structural biology - expanding the frontier while avoiding the pitfalls. Prot. Sci. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MHJ, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Quart. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Mattinen ML, Paakkonen K, Ikonen T, Craven J, Drakenberg T, Serimaa R, Waltho J, Annilla A. Quaternary structure built from subunits combining NMR and small-angle x-ray scattering data. Biophys. J. 2003;83:1177–1183. doi: 10.1016/S0006-3495(02)75241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay J. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Crystallogr. 2007;40:s245–s249. [Google Scholar]
- Orthaber D, Bergmann A, Glatter O. SAXS experimenta on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 2000;33:218–225. [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins identified through small-angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Poitevin F, Orland H, Doniach S, Koehl P, Delarue M. AquaSAXS: a web server for computation and fitting of SAXS profiles with non-uniformly hydrated atomic models. Nucl. Acids Res. 2011;39:W184–W189. doi: 10.1093/nar/gkr430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura G, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quart. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Schneiderman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucl. Acids Res. 2010;28:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Clore GM. A physical picture of atomic motions within the Dickerson DNA dodecamer in solution derived frosm joint ensemble refinement against NMR and large-angle x-ray scattering data. Biochemistry. 2007;46:1152–1166. doi: 10.1021/bi061943x. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Suh J-Y, Grishaev A, Ghirlando R, Takayama Y, Clore GM. Solution structure of the 128 kDa Enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide angle x-ray scattering. J. Am. Chem. Soc. 2010;132:13026–13045. doi: 10.1021/ja105485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL- a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 2001;80:2001. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nishikawa Y, Fujisawa T. Evaluation of three algorithms for ab initio determination of three-dimensional shape from one-dimensional solution scattering profiles. J. Appl. Crystallogr. 2003;36:549–552. [Google Scholar]
- Takayama Y, Schwieters CD, Grishaev A, Ghirlando R, Clore GM. Combined use of residual dipolar couplings and solution x-ray scattering to rapidly probe rigid-body conformational transitions in a non-phosphorylatable active-site Mutant of the 128 kDa Enzyme I dimer. J. Am. Chem. Soc. 2011;133:424–427. doi: 10.1021/ja109866w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D, Cohen FE, Doniach S. Reconstruction of low-resolution three-dimensional density maps from one-dimensional small-angle X-ray scattering data for biomolecules. J. Appl. Crystallogr. 2000;33:350–363. [Google Scholar]
- Yang S, Park S, Makowski L, Roux B. A rapid coarse residue-based computational method for X-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes. Biophys. J. 2009;96:4449–4463. doi: 10.1016/j.bpj.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]