Abstract
In this paper, we present MolFind, a highly multi-threaded pipeline type software package for use as an aid in identifying chemical structures in complex biofluids and mixtures. MolFind is specifically designed for high performance liquid chromatography/mass spectrometry (HPLC/MS) data inputs typical of metabolomics studies where structure identification is the ultimate goal. MolFind enables compound identification by matching HPLC/MS based experimental data obtained for an unknown compound with computationally derived HPLC/MS values for candidate compounds downloaded from chemical databases such as PubChem. The downloaded “bins” consist of all compounds matching the monoisotopic molecular weight of the unknown. The computational HPLC/MS values predicted include retention index (RI), ECOM50 (energy required to fragment 50% of a selected precursor ion), drift time and collision induced dissociation (CID) spectrum. RI, ECOM50, and drift time models are used for filtering compounds downloaded from PubChem. The remaining candidates are then ranked based on CID spectra matching. Current RI and ECOM50 models allow for the removal of about 28% of compounds from PubChem bins. Our estimates suggest that this could be improved to as much as 87% with additional chemical structures included in the computational models. Quantitative structure property relationship based modeling of drift times showed a better correlation with experimentally determined drift times than did Mobcal cross sectional areas. In 23/35 example cases, filtering PubChem bins with RI and ECOM50 predictive models resulted in improved ranking of the unknown compound compared to previous studies using CID spectra matching alone. In 19/35 examples, the correct candidate was ranked within the top 20 compounds in bins containing an average of 1635 compounds.
Keywords: mass spectrometry, HPLC, QSPR, metabolomics, retention index, Ecom50, CID spectrometry, ion mobility spectrometry, structure identification
Introduction
The identification of chemical structures in complex mixtures plays a vital role in many areas of chemistry and biology. A typical biological or environmental sample often consists of several hundreds to thousands of different chemical structures in varying concentrations.1–3 Thus, identification of compounds in these mixtures is difficult using classical methods which involve separation, purification and analysis by multiple analytical methods. Positive identification involves matching at least two orthogonal experimental measures against a purified standard.2 The most commonly used identification techniques include gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), and nuclear magnetic resonance (NMR). In GC-MS and LC-MS, orthogonal experimental measures may include retention index (RI), monoisotopic molecular weight (MIMW), isotope abundance pattern and mass spectrum. In metabolomics studies where sample amounts are limited and concentrations are low, MS based identification methods are generally preferred.4 LC is considered the most versatile separation method for metabolomics, as it requires less time and effort for sample preparation, while allowing for separation of a wide variety of compounds compared to GC.5
Because of the difficulties associated with purifying unknown compounds from complex mixtures2, an alternative approach involves matching experimental “features” of the unknown (RI, MIMW, CID spectrum, etc.) to predicted features of a set of candidates downloaded from a general chemical database.6–10 Candidate compounds are retrieved from a general chemical database such as PubChem11 based on the exact mass and the mass accuracy of the measurement. Experimental measurements such as RI are modeled using quantitative structure property relationships (QSPR).8,12 Collision induced dissociation (CID) spectra of unknowns have also been simulated using rule based fragmenters such as MassFrontier13 or ACD Fragmenter,14 or a combinatorial fragmenter such as MetFrag.9 Several studies have shown that it is possible to identify an unknown by using CID fragment matching alone6,9 or by combining CID fragment matching with other experimental measures such as RI.7,8,10 Although combining CID spectra matching with other experimental measures was deemed to be a viable method, there has not been software developed to achieve this seamlessly. In this study, we present a user-friendly software package that enables the rapid identification of an unknown based solely on HPLC/MS analyses. Experimental features available from a typical HPLC/MS instrument include MIMW, RI, ECOM50 (the energy in eV required to fragment 50% of a selected precursor ion)8,15, and CID spectra. In addition to these, we also include drift time as an experimental and computational variable that can be measured on ion mobility spectrometry-mass spectrometry (IMS-MS) instruments. The software package MolFind facilitates identification of an unknown by combining QSPR modeling of RI, ECOM50 and drift time, plus computationally predicted CID spectra matching. Experimental methods, general architecture of the program, QSPR modeling of drift time and benchmark calculations are presented.
Materials and Methods
Detailed information describing the measurements of retention indices, ECOM50 values and drift times are presented in the supporting information.
Software Architecture
MolFind is a highly multithreaded desktop application written in Java. Much of Molfind’s underlying cheminformatics functionality is based on the open source chemo and bioinformatics library Chemistry Development Kit (CDK).16,17 In addition to CDK, MolFind uses several open source and commercial programs such as MetFrag9, Mobcal18,19, JPMML20, MolConn21, and ChemAxon’s conformer plugin.22 MolFind is available as a free download from http://metabolomics.pharm.uconn.edu and will require Java 1.6 or a later version to operate. Third party libraries used in MolFind allow free academic usage of the program. Molconn based calculations are done on a remote server with the permission of Hall Associates Consulting. Users are not required to purchase a license for running Molconn based calculations.
A schematic of the MolFind workflow is shown in Fig 1. Upon user input of an exact mass and a mass accuracy in parts per million (ppm), MolFind retrieves from the PubChem database all compounds that correspond to the mass window. The initial set of structures is then filtered according to experimentally measured RI, ECOM50 and drift time values. QSPR models are used for predicting RI, ECOM50, and drift times for candidate structures. A progressively smaller set of compounds is passed into the next filter based on a 99.8% confidence interval for each QSPR model. The set of candidate compounds remaining after the last filter, along with the experimental CID spectrum of the test compound are then passed into the combinatorial fragmenter MetFrag for ranking of the remaining unknowns.
Fig 1.
Schematic of MolFind Workflow
Candidate Structures
Candidate compounds are downloaded from the National Center for Biotechnology Information’s (NCBI) PubChem database which is the largest freely accessible compound database currently available. As of March 5, 2012, PubChem contained approximately 32 million unique compounds. Alternatively, a compound database downloaded from a different compound repository such as KEGG23 or HMDB24 or a compound database generated with a combinatorial structure generator such as Molgen25 can be supplied as an SD file or compressed SD file.
Pre-Processing Filters
Candidates downloaded from PubChem contain salts, disconnected structures, heavy isotopes and compounds with an overall charge. User selectable pre-processing filters are applied to curate the initial set of downloaded structures. The pre-processing filters include a filter to remove disconnected structures, a filter to remove compounds containing heavy isotopes, a stereochemistry filter and a set of filters to select different types of elements and charged compounds. The stereochemistry filter allows removal of structures with the same connectivity but different stereochemistry. All stereoisomers except the first are eliminated from the candidate list. The PubChem CID numbers of the eliminated stereoisomers are stored as a property field inside the molecular record of the first stereoisomer. An elemental filters option restricts the candidate structures to a particular set of elements. For example C, H, N, O, P and S for biological applications.
QSPR Modeling of Retention Index and ECOM50
Both HPLC RI and ECOM50 values are modeled using Molconn topological molecular descriptors. Implementation details of these models are discussed elsewhere.8 MolFind communicates with Molconn by passing an SD file and retrieving the predicted RI and ECOM50 data as a text file. Errors associated with our current RI and ECOM50 models are ±114 RI units and ±2.1 eV respectively.8 These errors are based on three times the standard deviation of each model, and are equivalent to a 99.8% confidence interval.
Drift Time Models
The dataset for building drift time models was developed using 62 organic compounds with monoisotopic molecular weights (MIMWs) ranging from 114.0252 to 609.2951 and experimentally determined effective drift times of 2.30 to 9.72 milliseconds. Four different models were built for predicting drift time from structure. Initially, two-dimensional structures for the compounds were downloaded from NCBI’s PubChem database as an SD file. Following this, ChemAxon’s conformer plugin22 was used to generate three-dimensional coordinates, which were produced by exploiting conformational analysis and molecular mechanics (MMFF94) based optimization of lowest energy conformers. Finally, a modified version of Mobcal, optimized for room temperature He and N2-based trajectory method (TM) calculations was used for average cross sectional area calculations.18,19,26
Poly-Alanine Power-Law Model
A model was built using helium drift tube-derived average cross sectional areas of poly-alanine standards. Seven poly-alanine standards (3-Ala to 9-Ala) were run at six different time points during the drift time measurements of 62 compounds. The average effective drift times of the poly-alanine standards were used in the model (Table S2 in supporting information). Helium drift tube derived cross sectional areas were converted into mass and charge independent values by multiplying by the square root of the reduced mass and dividing by the charge. The relationship between mass and charge independent average cross sectional area (Ω) and effective drift time (t) is given by an empirically derived power-law function of the form Ω = A (t)B where, A and B are constants derived by fitting data to a power-law function.
Mobcal Power-Law Model
A five-fold cross validation model was built using Mobcal calculated cross sectional areas. A modified version of Mobcal parameterized for N2 based cross sectional area calculations was used to calculate the cross sectional areas for the 62 compounds in the data set. The dataset was split into 5 folds using the classification and regression training (caret) library27 of R statistical software package.28 A power function of the form Ω = A (t)B was fitted to each training data set using the non linear least squares method of R. Drift times of test folds were predicted using power-law equations of corresponding training folds.
Random Forest Models
A random forest is an ensemble learning technique that builds multiple decision trees with random sampling of both data and variables.29 In the context of regression, the final prediction given by a random forest is the average prediction of all trees in the ensemble. Decision trees were built with two-thirds of the data while one-third of the data was left out. The data portions left out of the tree building process are called the out of bag samples and are used for a type of in parallel cross validation that provides a measure of model quality analogous to an explicit cross validation. In this study, several random forest models were built with 2D and 3D molecular descriptors. Optimized 3D structures of compounds were used in molecular descriptor calculations. The CDK descriptor engine was used to calculate 295 electronic, topological, geometrical, constitutional and hybrid molecular descriptors. Missing values, descriptors with all zero or constant values, molecular weight descriptors and LogP descriptors were removed using the Rattle library30 of R leaving 223 descriptors. Nitrogen based Mobcal trajectory method (TM) cross sectional area was added to the pool of molecular descriptors making the final descriptor count 224. The Boruta library31,32 of R was used for descriptor selection. 100 Boruta runs were carried out in force selection mode. Only those descriptors that were confirmed to be irrelevant after 100 runs were removed from the descriptor pool. The final descriptor pool comprised 41 molecular descriptors (Table S3 in supporting information). Two sets of random forest cross validation models were built; one with and one without Mobcal cross sectional area as a descriptor. The same set of training and testing folds used in the Mobcal power-law model were used in the random forest cross validation.
Compound Identification
In addition to RI, ECOM50 and drift time, an unknown can be identified by matching the experimental CID spectrum of the test compound with the computationally predicted CID spectra of all compounds in the bin. Each compound in the bin is given a rank based on either the number of peaks matching (“Peaks Rank”) or the score (“Score Rank”), which is calculated based on the number of peaks matching, the intensity of the peaks and bond dissociation energies. Details explaining the MetFrag peaks matching and score are discussed elsewhere.9 Benchmark calculations were done with 35 compounds selected from a previous study published by Hill et al.6 In the aforementioned study, unknowns were identified by matching experimental CID spectra with MassFrontier (version 4.0) predicted fragments. Compound bins used in this study (henceforth called “old bins”) correspond to a snapshot of the PubChem database on February 6, 2006. Old bins were filtered using RI and ECOM50 models. Compounds were ranked before and after applying RI and ECOM50 filters using MassFrontier (version 4.0) and peaks matching. The experimental CID spectrum of lowest energy that resulted in <22% precursor ion remaining was used for MassFrontier ranking.6 Filtered bins were also ranked with MetFrag using 22% precursor ion spectra, composite spectra and intensity corrected composite spectra. Composite spectra were constructed from 5 CID spectrums (10-50 eV in 10 eV intervals) by averaging peak masses that fell within 10 ppm from any given peak. Peak intensity was taken as the sum of intensities of individual peaks that made up an average peak. Peak intensities were corrected to account for the effect of RF voltage on ion transfer efficiency as described.33 MetFrag based CID matching was repeated using current PubChem bins (as of March 5, 2012). A random forest based drift time model (without the Mobcal cross sectional area descriptor) was constructed by removing from the drift time model the five worst ranked compounds based on MetFrag score rank with intensity corrected composite spectra. PubChem bins of the five worst ranked compounds were then additionally filtered using drift time model.
Projected rank improvement using more accurate ECOM50 and RI models
Using MolFind’s model accuracy window options, test calculations were done to evaluate the level of filtering possible with improved RI and ECOM50 models. PubChem bins were filtered using 16 combinations of RI and ECOM50 error windows. Predicted ECOM50 and RI values were used in the filtering to ensure that the “unknown” was retained in the filtered bin. The RI window was varied from ±100 to ±40 in 20 RI unit intervals and the ECOM50 window was varied from ±2.0 to ±0.5 in 0.5 ECOM50 unit intervals. Upper limits for the RI and ECOM50 windows were selected based on the errors of our current QSPR models. The lower limits were set well above three times the standard deviation of experimental RI and ECOM50 replicate values (data in Tables S4 and S5 of the supporting information). The percentage of compounds that remained for each combination of RI and ECOM50 window was calculated by averaging all bins.
Results and Discussion
Ranking of unknowns
The effect of filtering PubChem bins with predicted RI and ECOM50 was evaluated using 40 old PubChem bins (February 6, 2006) previously ranked with CID spectra matching alone. These bins contained 253 compounds on average after applying pre filters. Applying RI and ECOM50 filters resulted in the removal of 29% of compounds from the pre filtered bins. In five cases, the “unknown” was lost in the filtering due to limitations in the QSPR models. Thus, only 35 bins were considered in calculating average rankings. When using MassFrontier ranking, the average rank of an unknown improved from 19 to 13 (Table 1).
Table 1.
Average rankings of test compounds (n = 35 bins).
| MEDIAN | AVERAGE | STD | |
|---|---|---|---|
| Old Bins | |||
|
| |||
| No of compounds after pre filters | 253 | ||
| No of compounds after RI filter | 195 | ||
| No of compounds after RI and ECOM50 filters | 180 | ||
| MassFrontier rank after pre filters (22% prec spectra) | 6 | 19 | 36 |
| MassFrontier rank after RI and ECOM50 filters (22% prec spectra) |
5 | 13 | 21 |
| MetFrag peaks rank after RI and ECOM50 filters (22% prec spectra) |
6 | 18 | 23 |
| MetFrag score rank after RI and ECOM50 filters (22% prec spectra) |
6 | 17 | 31 |
| MetFrag peaks rank after RI and ECOM50 filters (composite spectra) |
7 | 18 | 32.1 |
| MetFrag score rank after RI and ECOM50 filters (composite spectra) |
5 | 15 | 28.6 |
| MetFrag score rank after RI and ECOM50 filters (intensity corrected composite spectra) |
4 | 12 | 15.9 |
|
| |||
| New Bins | |||
|
| |||
| No of compounds after pre filters | 1635 | ||
| No of compounds after RI filter | 1296 | ||
| No of compounds after RI and ECOM50 filters | 1184 | ||
| MetFrag peaks rank after pre filters (22% prec spectra) |
28.5 | 142 | 228 |
| MetFrag peaks rank after RI and ECOM50 filters (22% prec spectra) |
22 | 102 | 157 |
| MetFrag score rank after RI and ECOM50 filters (intensity corrected composite spectra) |
18 | 56 | 91 |
Table 1 summarizes data in Table S6 and S7 of supplementary information. An overview of the effect of different fragmentation algorithms, CID spectra and bin sizes on rankings is given.
In 8 bins, the unknown compound was ranked first before applying RI and ECOM50 filters and was ranked within the top 10 for 20 of the 35 bins. With the application of RI and ECOM50 filters, the unknown was ranked within the top 10 for 23 bins, and in 9 cases was found at the top of the list. Experimental CID spectrum of lowest energy that resulted in <22% precursor ion remaining was used in MassFrontier rankings. Filtered bins were also ranked in various ways with the open source combinatorial fragmenter MetFrag by employing several types of CID spectra. MetFrag ranking with peak intensity corrected composite spectra was found to be slightly better than other methods based on average, standard deviation and median of rankings (Table 1). The detailed rankings of unknowns in old bins are listed in Table S6 of supporting information.
At the time the old bins were downloaded, PubChem contained about 6 million compounds. Currently (as of March 5, 2012), PubChem contains about 32 million compounds, which is roughly 5 times the size of PubChem in 2006. On average, the current 35 PubChem bins contained 1635 compounds after applying pre filters: a 6.5 fold increase compared to old PubChem bins. Filtering current PubChem bins with RI and ECOM50 filters resulted in a 28% reduction in bin size, compared to 29% with old bins. Although RI and ECOM50 filters performed similarly with old and new bins, the large number of compounds that remain in current filtered bins leads to substantial degradation of MetFrag ranking. We selected MetFrag peak ranking for comparing average ranks before and after applying RI and ECOM50 filters, as MetFrag score ranking is not directly comparable between bins of different size (due to normalization of the MetFrag score based on maximum values for a given bin). The average MetFrag peak rank (with CID spectrum of lowest energy that resulted in <22% precursor ion remaining) degraded from 18 with old bins to 102 with new bins. However, the significance of RI and ECOM50 filters is clearly seen as MetFrag peak rank improved from 142 to 102 with RI and ECOM50 filters. The detailed rankings of unknowns in new bins are found in Table S7 of supporting information.
Projected rank improvement upon improving ECOM50 and RI filters
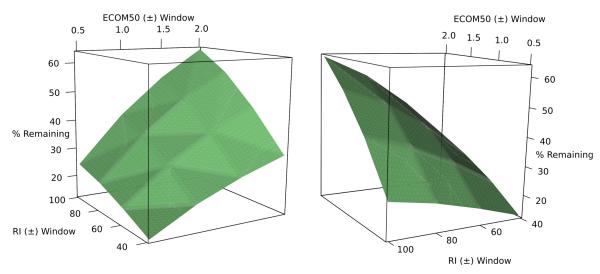
Identifying the correct compound using CID spectrum matching becomes more and more challenging as the number of candidates in a bin grows. Currently the RI and ECOM50 models allow for the removal of 28% of candidates (on average) from any given pre-filtered PubChem bin. It is important to note that our current RI and ECOM50 models are far from optimal. The ECOM50 model for example was built with 54 training compounds and should be regarded as an initial model. The more robust RI model (built with 400 diverse training compounds) is also limited by the lack of certain functional groups. Both the RI and ECOM50 models could be improved by including a much large number of diverse chemical structures. Thus, a set of test calculations was carried out to understand the level of filtering possible with improved RI and ECOM50 models. As shown in Fig 2, improving the RI model to a window of ±40 RI units and the ECOM50 model to a window of ±0.5 ECOM50 units would remove 87.2% of compounds from the pre-filtered PubChem bins. An average MetFrag score ranking (with intensity corrected composite spectra) of 15.5 would also be expected with improved RI and ECOM50 windows (Table S8 in supporting information).
Fig 2.
Average percentage of compounds remaining in bins using various RI and ECOM50 windows (left and right: two views of the 3D surface plot)
Drift Time Models
Ion mobility spectrometry coupled to mass spectrometry (IMS-MS) allows separation of ions with identical m/z values based on their physical shapes.34,35 Modern IMS-MS instruments are capable of separating closely related compounds such as enantiomers,36 chiral stereoisomers,37–39 diastereomers,26 isomeric drug metabolites40 and protein conformers.41,42 In a conventional drift tube based IMS-MS instrument, ions with identical m/z values are separated by the time (drift time) an ion takes to traverse a gas-filled cell under the influence of a uniform electric field. In a conventional drift tube, the drift time is proportional to the average collisional cross sectional area (Ω) of the gas phase ion. Average collisional cross sectional area can be theoretically calculated with the 3D structure of a gas phase ion. It is often used to correlate the structure with the experimentally observed drift time. In travelling wave (T-Wave) IMS-MS instruments, the drift time cannot be directly correlated with the cross sectional area as the electric field used is neither constant nor uniform. However, T-Wave instruments can be calibrated using a set of standards (a series of poly alanine standards are commonly used for calibration) with known cross sectional areas. Several studies26,39,40 using closely related small molecules with identical m/z values have shown that cross sectional areas calculated with T-wave calibration curves are in quantitative agreement with cross sectional areas calculated with the open source program Mobcal.
As a first step, we attempted to build a calibration curve for predicting drift times based on drift tube derived cross sectional areas of a series of polyalanine (PolyA) standards. As shown in Fig 3 (bottom right), a quantitative agreement between the predicted and the experimental drift times is reached only for those compounds with smaller masses. Predicted drift times of larger compounds (presumably with more flexible or more extended structures) were found to deviate substantially from the experimental values. In the second approach, a power function of the form Ω = A (t)B (where Ω = Mobcal cross sectional area, t = experimental drift time) was fitted to each training data set by splitting the dataset into five cross validation folds. The cross-validated power-law model showed a better agreement with the actual values than the standard PolyA curve (MSE of 0.228 compared to 0.585). Mobcal cross sectional area was calculated with the lowest energy conformer (minimized with MMFF94 force field) obtained from a conformational analysis. When an ensemble of minimum energy structures was used (all structures that were within 2 kcal/mol of the lowest energy conformer) instead of a single structure or quantum mechanically optimized structures (using B3LYP/6-31g*), the predicted drift times did not show any improvement. Our results are in agreement with the recent study by Zakharova et al.43 where they assessed the accuracy of Mobcal calculations for moderately flexible molecules. According to that study, a quantitative agreement would require quantum mechanics based re-optimization of the structures obtained from a lengthy molecular dynamics (MD) simulation. Furthermore, the outcome was found to be highly dependent on the parameters used in the MD simulation making such an approach not amenable to automation.
Fig 3.
Correlation between experimental and predicted drift time for different drift time models
In the current study, we propose a different approach to predict drift times using QSPR models. In this approach, molecular descriptors are used to: 1) compensate for inaccuracies associated with the 3D geometry optimization and, 2) include characteristics of flexible or extended molecules. Two sets of random forest based cross validation models were built; one with and one without Mobcal cross sectional area included as a descriptor. Several molecular descriptors showed a direct correlation with measured drift time. The best-correlated descriptors (ones listed at the top of the heat map in Fig S1 of supporting information) include chi path descriptors (VP.0, VP.1, and SP.1), pseudo molecular volume descriptor (VABC), Eccentric connectivity index (ECCEN), Wiener path number (WPATH) and nitrogen based Mobcal cross sectional area (MobCSA). Interestingly, a number of molecular descriptors showed a better or as good a correlation as the widely used Mobcal cross sectional area. The removal of Mobcal cross sectional area as a descriptor resulted in only a minor degradation of the predictive accuracy of the model (MSE of 0.125 vs. 0.129). Eliminating Mobcal cross sectional area calculations resulted in a significant decrease in computational time. This is especially true for nitrogen based cross sectional area calculations where the calculation time is substantially increased due to terms such as ion-quadrupole interactions. The utility of using experimental drift time as a potential filter was assessed via a model built by leaving out five compounds from the original 62 compound model. One out of bag error (±0.35 msec), roughly equivalent to an 80% confidence interval, was used as the window for filtering compounds. As might be expected, the drift time model had a minimum effect on small molecular weight bins with presumably more rigid molecules. For example, in the case of CID bin 2206, the drift time model was unable to remove any compounds. In the cases of CID bins 2136, 2151 and 4893, the drift time model was only able to improve the ranking by one or two. However, the drift time model was found to be quite useful for large molecular weight bins with more flexible molecules. For example, in the case of the CID 4544 bin, applying the drift time filter resulted in removal of 67% of compounds.
Conclusions
We developed a user-friendly software package for enabling the rapid identification of chemical structures in complex mixtures based solely on high performance liquid chromatography/mass spectrometry. The stand-alone java based software package MolFind facilitates identification of an unknown by matching experimentally measured RI, ECOM50, drift times and CID spectra with computational predictions. We show that filtering a bin of candidate compounds based on multiple QSPR filters leads to improved ranking of the unknown compound. The molecular descriptor based drift time model developed in this study outperformed the widely used Mobcal method. As RI, ECOM50, drift time and CID spectra computational predictions are improved by broadening the chemical space included in each model, this approach should dramatically enhance our ability to identify the structures of unknown compounds in complex mixtures.
Supplementary Material
Table 2.
Leave 5 out drift time model results
| CID | Experimental Drift Time (msec) |
Predicted Drift Time (msec) |
# cpds After Pre Filters |
# cpds After RI and ECOM50 |
# cpds After RI, ECOM50 and Drift Filter |
MetFrag peaks rank after RI and ECOM50 |
MetFrag peaks rank after RI, ECOM50 and Drift Time Filter |
|---|---|---|---|---|---|---|---|
| 2136 | 4.1 | 4.2 | 1334 | 927 | 923 | 35 | 34 |
| 2151 | 3.4 | 3.67 | 1447 | 1182 | 1165 | 205 | 204 |
| 2206 | 3.19 | 3.29 | 1490 | 1423 | 1423 | 605 | 605 |
| 4544 | 6.32 | 6.65 | 1662 | 1439 | 538 | 1057 | 349 |
| 4893 | 6.26 | 6.46 | 1223 | 1019 | 926 | 23 | 19 |
Acknowledgements
The authors would like to thank Ritvik Dubey for performing some of the mass spectral analysis used in this study. We wish to thank Egon Willighagen and Rajarshi Guha from the CDK project for technical help. This research was funded by NIH grant 1R01GM087714.
References
- (1).Mihaleva VV, Verhoeven H. a, Vos R. C. H. de, Hall RD, Ham R. C. H. J. van. Bioinformatics (Oxford, England) 2009;25:787–794. doi: 10.1093/bioinformatics/btp056. [DOI] [PubMed] [Google Scholar]
- (2).Wishart DS. Bioanalysis. 2011;3:1769–1782. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- (3).Zwiener C, Frimmel FH. Anal. Bioanal. Chem. 2004;378:862–874. doi: 10.1007/s00216-003-2412-1. [DOI] [PubMed] [Google Scholar]
- (4).Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner H-P. Angew. Chem. Int. Ed. Engl. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- (5).Moco S, Vervoort J, Bino RJ, Vos R. C. H. De, Bino R. Trends Anal. Chem. 2007;26:855–866. [Google Scholar]
- (6).Hill DW, Kertesz TM, Fontaine D, Friedman R, Grant DF. Anal. Chem. 2008;80:5574–5582. doi: 10.1021/ac800548g. [DOI] [PubMed] [Google Scholar]
- (7).Kertesz TM, Hill DW, Albaugh DR, Hall LH, Hall LM, Grant DF. Bioanalysis. 2009;1:1627–1643. doi: 10.4155/bio.09.145. [DOI] [PubMed] [Google Scholar]
- (8).Hall LM, Hall LH, Kertesz TM, Hill DW, Sharp TR, Oblak EZ, Dong YW, Wishart DS, Chen M-H, Grant DF. J. Chem. Inf. Model. 2012;52:1222–1237. doi: 10.1021/ci300092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wolf S, Schmidt S, Müller-Hannemann M, Neumann S. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schymanski EL, Gallampois CMJ, Krauss M, Meringer M, Neumann S, Schulze T, Wolf S, Brack W. Anal. Chem. 2012;84:3287–3295. doi: 10.1021/ac203471y. [DOI] [PubMed] [Google Scholar]
- (11).Bolton EE, Wang Y, Thiessen PA, Bryant SH. Annual Reports in Computational Chemistry. Vol. 4. American Chemical Society; Washington, DC: 2008. PubChem: Integrated Platform of Small Molecules and Biological Activities; pp. 217–241. [Google Scholar]
- (12).Albaugh DR, Hall LM, Hill DW, Kertesz TM, Parham M, Hall LH, Grant DF. J. Chem. Inf. Model. 2009;49:788–799. doi: 10.1021/ci9000162. [DOI] [PubMed] [Google Scholar]
- (13).Mass Frontier 4.0. http://www.highchem.com.
- (14).ACD/MS Fragmenter. http://www.acdlabs.com/products/adh/ms/ms_frag/
- (15).Kertesz TM, Hall LH, Hill DW, Grant DF. J. Am. Soc. Mass Spectrom. 2009;20:1759–1767. doi: 10.1016/j.jasms.2009.06.002. [DOI] [PubMed] [Google Scholar]
- (16).Steinbeck C, Han Y, Kuhn S, Horlacher O, Luttmann E, Willighagen E. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- (18).Shvartsburg A. Chem. Phys. Lett. 1996;261:86–91. [Google Scholar]
- (19).Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- (20).JPMML. http://www.jpmml.org.
- (21).winMolconn 1.1.1.4. Hall Associates Consulting; Quincy, MA: 2008. [Google Scholar]
- (22).ChemAxon, 5.4.1.1. http://www.chemaxon.com.
- (23).Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly M, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Benecke C, Grüner T, Kerber A, Laue R, Wieland T. Fresenius. J. Anal. Chem. 1997;359:23–32. [Google Scholar]
- (26).Campuzano I, Bush MF, Robinson CV, Beaumont C, Richardson K, Kim H, Kim HI. Anal. Chem. 2012;84:1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- (27).Kuhn M. J. Stat. Softw. 2008;28:1–26. [Google Scholar]
- (28).R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- (29).Breiman L. Machine Learning. 2001;45:5–32. [Google Scholar]
- (30).Williams G. The R Journal. 2009;1:45–55. [Google Scholar]
- (31).Kursa MB, Jankowski A, Rudnicki WR. Fundamenta Informaticae. 2010;101:271–285. [Google Scholar]
- (32).Kursa MB, Rudnicki WR. J. Stat. Softw. 2010;36:1–13. [Google Scholar]
- (33).Hill DW, Baveghems CL, Albaugh DR, Kormos TM, Lai S, Ng HK, Grant DF. Rapid Commun. Mass Spectrom. 2012;26:2303–2310. doi: 10.1002/rcm.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. J. Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- (35).Creaser CS, Griffiths JR, Bramwell CJ, Noreen S, Hill CA, Thomas CLP. The Analyst. 2004;129:984–994. [Google Scholar]
- (36).Dwivedi P, Wu C, Matz LM, Clowers BH, Siems WF, Hill HH. Anal. Chem. 2006;78:8200–8206. doi: 10.1021/ac0608772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Borsdorf H, Nazarov EG, Miller RA. Anal. Chim. Acta. 2006;575:76–88. doi: 10.1016/j.aca.2006.05.066. [DOI] [PubMed] [Google Scholar]
- (38).McCooeye M, Ding L, Gardner GJ, Fraser CA, Lam J, Sturgeon RE, Mester Z. Anal. Chem. 2003;75:2538–2542. doi: 10.1021/ac0342020. [DOI] [PubMed] [Google Scholar]
- (39).Williams JP, Bugarcic T, Habtemariam A, Giles K, Campuzano I, Rodger PM, Sadler PJ. J. Am. Soc. Mass Spectrom. 2009;20:1119–1122. doi: 10.1016/j.jasms.2009.02.016. [DOI] [PubMed] [Google Scholar]
- (40).Dear GJ, Munoz-Muriedas J, Beaumont C, Roberts A, Kirk J, Williams JP, Campuzano I. Rapid Commun. Mass Spectrom. 2010;24:3157–3162. doi: 10.1002/rcm.4742. [DOI] [PubMed] [Google Scholar]
- (41).Smith DP, Giles K, Bateman RH, Radford SE, Ashcroft AE. J. Am. Soc. Mass Spectrom. 2007;18:2180–90. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Valentine SJ, Clemmer DE. J. Am. Chem. Soc. 1997;119:3558–3566. [Google Scholar]
- (43).Zakharova NL, Crawford CL, Hauck BC, Quinton JK, Seims WF, Hill HH, Clark AE. J. Am. Soc. Mass Spectrom. 2012;23:792–805. doi: 10.1007/s13361-012-0339-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.