The opioid receptor family is comprised of three members, the μ, δ and κ opioid receptors, that respond to classical opioid alkaloids such as morphine and heroin as well as endogenous peptide ligands like endorphins. They belong to the G-protein-coupled receptor (GPCR) superfamily, and are excellent therapeutic targets for pain control. The δ opioid receptor (δ-OR) plays a role in analgesia, as well as other neurological functions that remain poorly understood1. The structures of μ-OR and κ-OR have recently been solved2,3. Here we report the crystal structure of the δ-OR, bound to the subtype-selective antagonist naltrindole. Together with the structures of μ-OR and κ-OR, the δ-OR structure provides insights into conserved elements of opioid ligand recognition while also revealing structural features associated with ligand subtype selectivity. The binding pocket of opioid receptors can be divided into two distinct regions. While the lower part of this pocket is highly conserved among opioid receptors, the upper part contains divergent residues that confer subtype selectivity. This provides a structural explanation and validation for the “message-address” model of opioid receptor pharmacology4,5, in which distinct “message” (efficacy) and “address” (selectivity) determinants are contained within a single ligand. Comparison of the “address” region of the δ-OR with other GPCRs reveals this structural organization may be a more general phenomenon, extending to other GPCR families as well.
Opioid receptors play an important role in the central nervous system, regulating pain perception, hedonic homeostasis, mood, and well-being1. Thus, they have long been the focus of physiological and pharmacological studies, as well as important therapeutic targets. The opioid receptors are G-protein coupled-receptors (GPCRs), and were classified based on their pharmacology and their tissue distribution into three subclasses: the μ (for morphine), the δ (for vas deferens) and the κ (for ketocyclazocine) receptors6. The sequence identity within the transmembrane domains (TMs) between μ-OR/δ-OR, μ-OR/κ-OR and δ-OR/κ-OR is 76%, 73% and 74%, respectively7. Another receptor identified by cloning, the nociceptin/orphanin FQ receptor, was classified in this family due to a high sequence identity (67% in the TM)7. However, morphinans and most other classical opioid ligands have little affinity for the nociceptin receptor8. The μ, δ, and the κ opioid receptors are activated by endogenous peptides: the endorphins, enkephalins, and dynorphins8. They are also the targets of chemically diverse small molecules presenting a variety of efficacy and selectivity profiles8. In an effort to better understand the structural basis for opioid receptor pharmacology and function, we used the in meso crystallization method to solve a 3.4 Å structure of a Mus musculus δ-OR T4 lysozyme fusion protein (Supplementary Fig. 1) bound to naltrindole (NTI), a non-covalent δ-OR selective morphinan antagonist9.
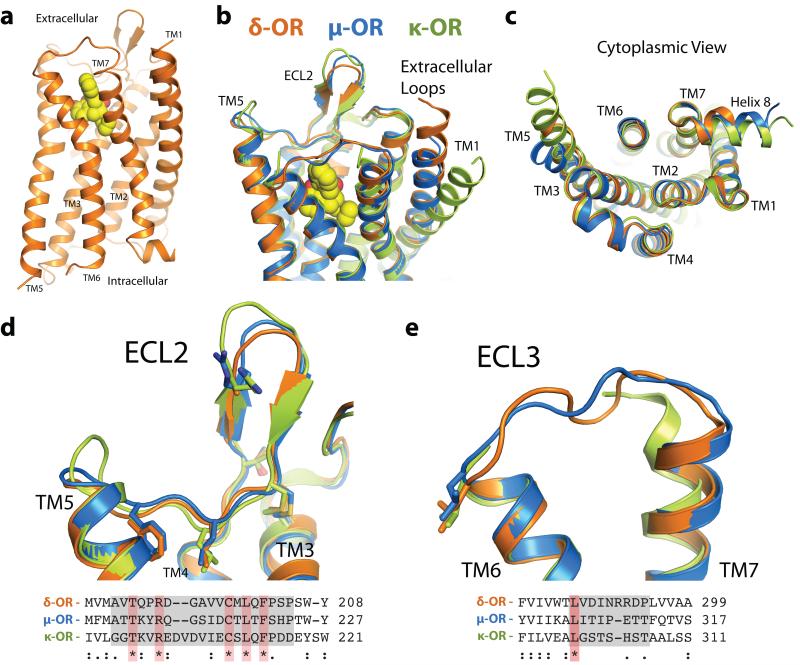
The δ-OR structure presents the typical GPCR seven-pass transmembrane helix fold (Fig. 1a), and shows striking conservation of backbone structure with other opioid receptors (Fig. 1b,c), even in regions with low sequence conservation (Fig. 1d,e). The ligand naltrindole sits in an exposed binding pocket, similar in shape to that observed for μ-OR and κ-OR2,3. The CXCR4 receptor also has a solvent-exposed binding pocket, suggesting that this may be a feature common to some GPCRs activated by peptides. The β-hairpin in extracellular loop (ECL) 2 (Fig. 1d) is observed in all three opioid receptors, despite the low sequence identity in this domain. ECL3, which is also poorly conserved among the three opioid receptors, was unresolved in the κ-OR structure and has high temperature factors in both μ-OR and δ-OR (Fig. 1e), suggesting a relatively flexible link between TMs 6 and 7. Of note, the κ-OR structure shows a clear difference in the position of the extracellular half of TM1, with an outward displacement of approximately 10 Å (Fig. 1b) compared to the μ-OR and δ-OR. In this respect, the μ-OR and δ-OR resemble each other and the CXCR4 chemokine receptor more closely than the κ-OR. However, this structural difference may simply reflect differences in crystallization conditions or crystal packing influences, as is seen in the turkey β1AR structure (PDB 2VT4) where two different TM1 conformations are observed in the same crystal10.
Figure 1. Overall structure of the δ-opioid receptor.
(a) The δ-OR, orange, exhibits a typical seven-pass transmembrane architecture common to other GPCRs. (b-c) This fold is highly conserved among all three classical members of the opioid receptor family. (d) The opioid family possesses a conserved beta strand fold in the second extracellular loop (ECL2), creating a wide, open binding pocket. Despite the structural similarity, only five residues in this region are absolutely conserved. Conserved residues are highlighted red in sequence alignment and shown as sticks (e) ECL3, an important selectivity determinant for ligand binding, shows modest structural variability in the μ-OR and δ-OR. In the κ-OR receptor structure this region could not be resolved due to poor electron density. A single leucine residue is conserved among in ECL3 among opioid subtypes.
δ- and μ-ORs have been observed to form homo-oligomers in trasfected cells, and functional studies suggest that they form pharmacologically distinct hetero-oligomers when they are co-expressed11. It is therefore interesting that in the μ-OR crystal lattice two parallel dimeric interfaces were observed2 (Supplementary Fig. 2). The most extensive interface involves TM5 and TM6 of adjacent protomers. The other interface, which is also observed in the κ-OR, involves TM1, TM2 and Helix 8. In addition to this common interface, an antiparallel interaction is also observed in the κ-OR crystal lattice. In contrast, the δ-OR crystallizes with only an anti-parallel arrangement of receptor molecules (Supplementary Fig 2). However, inferences regarding the physiologic relevance of oligomeric interfaces observed in GPCR crystal structures should be made with caution. Purified, detergent solubilized δ-OR and μ-OR are monomeric prior to crystallization and the association into either parallel or antiparallel dimers occurs during crystallogenesis. The differences between the μ-OR and δ-OR dimeric interfaces most likely reflect differences in the most energetically favorable interactions under the crystallography conditions and may be the consequence of one or more of the following: different crystallization conditions, a different T4 lysozyme arrangement, sequence differences in the protein, and subtle differences in the structures stabilized by the different ligands.
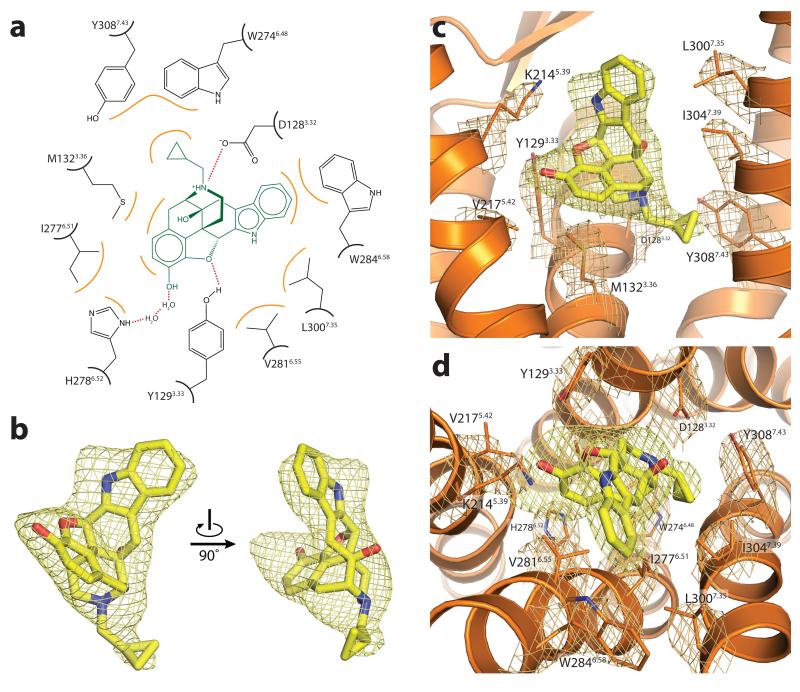
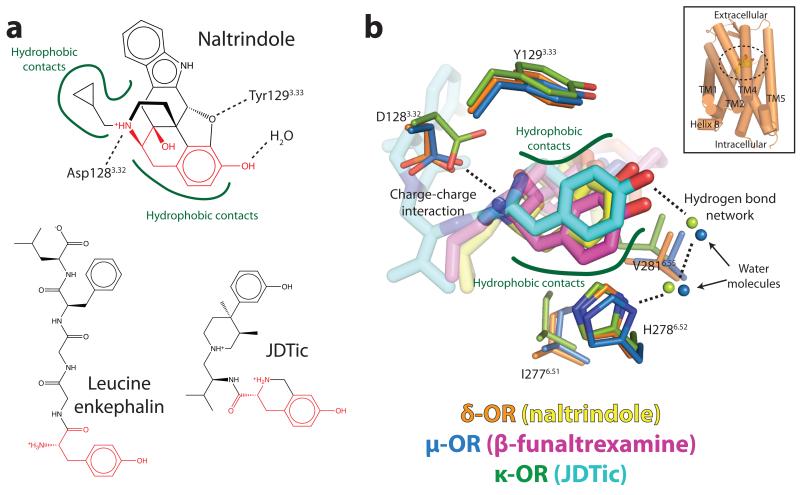
Opioid receptors bind exceptionally well to highly diverse ligands, including morphinans, a wide variety of other small molecules, and peptides of varying length. Details of naltrindole binding to the δ-OR are presented in Fig. 2 and Supplementary Fig. 3). Despite their chemical diversity, many opioid ligands display conserved features, most notably a phenolic hydroxyl separated by six carbons from a positive charge, which mimics the amino-terminal tyrosine of all endogenous opioid peptides (Fig. 3). The morphinan ligand naltrindole used in crystallization of the δ-OR is non-covalent9, and is therefore not subject to possible distortions in its binding mode due to a covalent tether like that in the structure of the μ-OR. Indeed, the position of naltrindole in the binding pocket is shifted slightly relative to the position of the covalent morphinan ligand β-FNA12 bound the μ-OR (Fig. 4), although all major interactions are present in both structures. As anticipated from the μ-OR structure, the leucine in the position 3007.35 is in contact with the indole group of naltrindole (Fig. 2). This residue is responsible for naltrindole selectivity13, since W3187.35 in μ-OR and Y3127.35 in κ-OR are sterically incompatible with naltrindole binding. As with μ-OR and κ-OR, electron density near the phenolic hydroxyl of naltrindole suggests the presence of water molecules (Fig. 3b and Supplementary Fig. 4). The resolution of the δ-OR structure, however, is not sufficient to place solvent molecules with confidence. Nonetheless, this feature suggests that a two-water hydrogen bond network linking H2786.52 and the ligand phenolic hydroxyl is likely a conserved feature of opioid ligand recognition, and certainly this appears to be the case in μ-OR and κ-OR.
Figure 2. Ligand binding site of the δ-OR.
Naltrindole binds in a deep but open binding site within the δ-OR. Selected contacts are highlighted at in (a), and a ligand Fo-Fc omit map within 2 Å radius of naltrindole is shown in (b) at a 3σ contour. The complete binding site is shown in (c) and (d). 2Fo-Fc electron density maps within a 2 Å radius of binding site amino acid side chains are shown in orange at a 1.5σ contour. The ligand omit density is shown as in (b).
Figure 3. A conserved opioid ligand recognition mode.
(a) Opioid receptors bind a wide variety of ligands, including morphinans like naltrindole, other small molecules such as JDTic, and peptides like enkephalins. Most opioid ligands, including these, contain a “tyrosine” pharmacophore (red) with a phenolic hydroxyl in close proximity to a positive charge. Conserved recognition features for morphinan ligands observed in the μ-OR and δ-OR are highlighted on naltrindole (top). (b) The tyrosine pharmacophores of naltrindole, β-funaltrexamine, and JDTic are shown in their three dimensional context as observed in the crystal structures of δ-OR, μ-OR, and κ-OR. Specific conserved interactions in all three receptors are highlighted.
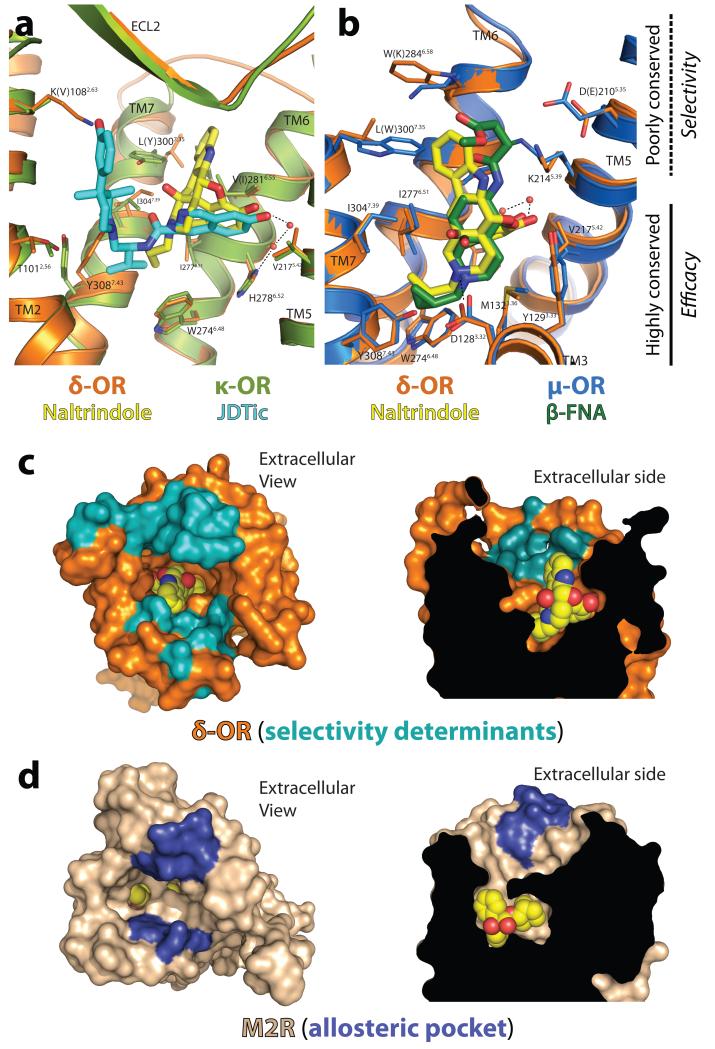
Figure 4. The message-address hypothesis is reflected in opioid receptor structure.
Comparison of the δ-OR with κ-OR (a) and with μ-OR (b) reveals high conservation in both sequence and structure in the base of the ligand binding pocket, while extracellular regions are more divergent. Residue numbers and labels are those for δ-OR, with κ-OR residues in parentheses in (a) and μ-OR residues in parentheses in (b) where sequence differs from δ-OR. These regions interact with ligand moieties that can target binding to a particular opioid subtype. (c) Residues previously characterized as important for opioid subtype selectivity18-20 are clustered around the upper part of the binding pocket, delineating an “address” region of the receptor. (d) This region is structurally analogous to the allosteric site in muscarinic receptors (blue) suggesting that the high selectivity of muscarinic ligands occupying this space is also a manifestation of the message-address features structurally encoded within GPCRs.
With the structure of the δ-OR, all classical opioid receptors have now been crystallized and solved in inactive conformations. A closely related receptor, that for the nociceptin/orphanin FQ peptide, is often classified within the opioid receptor family. However, this receptor has low or negligible affinity for most opioid alkaloids8 despite high sequence conservation within the TM domains. Examination of the morphinan binding site of δ-OR reveals that only a few of the critical interacting residues differ between δ-OR and the nociceptin receptor (Supplementary Fig. 5). However, mutation of certain residues in the nociceptin receptor to their δ-OR counterparts can create a high affinity alkaloid binding site14. These mutations change smaller amino acid side chains in the nociceptin receptor to larger residues in the corresponding positions of the δ-OR, so it is likely that the binding pocket of the nociception receptor is somewhat enlarged relative to that of the δ-OR. The loss of tightly fitting hydrophobic interactions with the morphinan ring of opioid alkaloids may therefore account, at least in part, for the much higher affinity of most morphinans for δ-OR, μ-OR, and κ-OR than for the nociceptin receptor.
Opioid pharmacology has long been described in terms of the “message-address” concept4,5, in which the ligand can be viewed as composed of two distinct modules carrying information about efficacy (message) and selectivity (address). The structure of the δ-OR and other opioid receptors reveals this pharmacological phenomenon to be a direct consequence of opioid receptor structure. The lower portion of the binding pocket is well-conserved in both sequence and structure. In the δ-OR, this portion of the binding pocket recognizes the core morphinan group, which entails the “message” of the ligand (Fig. 4a,b). In contrast, the upper binding pocket is divergent among subtypes, and is rich in selectivity determinants (Fig. 4a,b). The indole “address” of naltrindole extends into this region, conferring its δ-OR selectivity (Fig. 4a,b). Similarly, the 3-hydroxyphenyl group of JDTic extends into the address region, and would clash with δ-OR residue K1082.63. This feature may account for the selectivity of JDTic for κ-OR and μ-OR (Fig. 4a), although it is likely that other factors contribute as well.
Development of highly subtype-selective ligands has proven to be possible for the classical opioid receptors15. However, for another GPCR family, the muscarinic acetylcholine receptors, this has proven significantly more challenging. Comparison of the message and address regions of the δ-OR with the M2 muscarinic receptor16 (Fig. 4c,d) reveals that the address region corresponds to the allosteric site of these receptors. This region is separated by a layer of tyrosines from the highly conserved orthosteric site, which matches the message region of opioid receptors. The physical separation of the two regions may therefore explain the relatively greater challenges associated with development of selective muscarinic ligands compared to opioid receptors, as well as the promising results of selective “dualsteric” or “bitopic” ligands that occupy both orthosteric and allosteric sites simultaneously17. The distinct message and address regions of δ-OR and other opioid receptors then appear to be a more general feature of GPCRs, which may have implications for development of ligands even for distantly related GPCR families.
Together with μ-OR and κ-OR, the structure of δ-OR completes the initial structural characterization of the opioid receptors, offering the first experimental views of the atomic details of ligand recognition and selectivity in this important GPCR family. However, such antagonist-bound structures are only the first step toward a complete understanding of opioid receptor function. Given the importance of opioid agonists in clinical medicine, active state structures, as well as signaling complexes, will be required to fully leverage structural methods towards the development of a new generation of opioid drugs.
Methods summary
The δ-opioid receptor-T4 lysozyme fusion protein was expressed in Sf9 insect cells and purified by nickel affinity chromatography followed by FLAG antibody affinity chromatography and size exclusion chromatography. It was crystallized using the lipidic mesophase technique, and diffraction data were collected at GM/CA-CAT beamline 23ID-B at the Advanced Photon Source at Argonne National Laboratory. The structure was solved by molecular replacement using merged data from 20 crystals. Full details are provided in the online methods.
Supplementary Material
Acknowledgements
We thank Roger Sunahara for helpful suggestions on opioid receptor pharmacology and biochemistry. We acknowledge support from INSERM (S.G.), from the Stanford Medical Scientist Training Program (A.M.), from the National Science Foundation (A.C.K.), from the National Institutes of Health Grants NS028471 (B.K.K) and DA031418 (B.K.K), and from the Mathers Foundation (B.K.K. and W.I.W.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions A.M., A.C.K., and S.G. designed experiments, performed research and analyzed data. T.S.K. and F.S.T expressed and purified receptor. W.I.W supervised diffraction data analysis and model refinement. A.M., A.C.K., S.G., and B.K.K prepared the manuscript. S.G. and B.K.K supervised the research.
Author Information Coordinates and structure factors for δ-OR-T4L are deposited in the Protein Data Bank (accession code 4EJ4).
Reprints and permission information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
REFERENCES
- 1.Pradhan AA, Befort K, Nozaki C, Gaveriaux-Ruff C, Kieffer BL. The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol Sci. 2011;32:581–590. doi: 10.1016/j.tips.2011.06.008. doi:10.1016/j.tips.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manglik A, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012 doi: 10.1038/nature10954. doi:10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012 doi: 10.1038/nature10939. doi:10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavkin C, Goldstein A. Specific receptor for the opioid peptide dynorphin: structure--activity relationships. Proc Natl Acad Sci U S A. 1981;78:6543–6547. doi: 10.1073/pnas.78.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipkowski AW, Tam SW, Portoghese PS. Peptides as receptor selectivity modulators of opiate pharmacophores. J Med Chem. 1986;29:1222–1225. doi: 10.1021/jm00157a018. [DOI] [PubMed] [Google Scholar]
- 6.Satoh M, Minami M. Molecular pharmacology of the opioid receptors. Pharmacol Ther. 1995;68:343–364. doi: 10.1016/0163-7258(95)02011-x. [DOI] [PubMed] [Google Scholar]
- 7.Mollereau C, et al. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- 8.Cox Brian M., et al. Opioid receptors, introductory chapter. 2009 < http://www.iuphardb.org/DATABASE/FamilyIntroductionForward?familyId=50.>.
- 9.Portoghese PS, Sultana M, Nagase H, Takemori AE. Application of the message-address concept in the design of highly potent and selective non-peptide delta opioid receptor antagonists. J Med Chem. 1988;31:281–282. doi: 10.1021/jm00397a001. [DOI] [PubMed] [Google Scholar]
- 10.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. doi:10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George SR, et al. Oligomerization of mu- and delta-opioid receptors. Generation of novel functional properties. J Biol Chem. 2000;275:26128–26135. doi: 10.1074/jbc.M000345200. [DOI] [PubMed] [Google Scholar]
- 12.Portoghese PS, Larson DL, Sayre LM, Fries DS, Takemori AE. A novel opioid receptor site directed alkylating agent with irreversible narcotic antagonistic and reversible agonistic activities. J Med Chem. 1980;23:233–234. doi: 10.1021/jm00177a002. [DOI] [PubMed] [Google Scholar]
- 13.Bonner G, Meng F, Akil H. Selectivity of mu-opioid receptor determined by interfacial residues near third extracellular loop. Eur J Pharmacol. 2000;403:37–44. doi: 10.1016/s0014-2999(00)00578-1. [DOI] [PubMed] [Google Scholar]
- 14.Meng F, et al. Creating a functional opioid alkaloid binding site in the orphanin FQ receptor through site-directed mutagenesis. Mol Pharmacol. 1998;53:772–777. doi: 10.1124/mol.53.4.772. [DOI] [PubMed] [Google Scholar]
- 15.Eguchi M. Recent advances in selective opioid receptor agonists and antagonists. Med Res Rev. 2004;24:182–212. doi: 10.1002/med.10059. [DOI] [PubMed] [Google Scholar]
- 16.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2011 doi: 10.1038/nature10753. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valant C, et al. A novel mechanism of G protein-coupled receptor functional selectivity. Muscarinic partial agonist McN-A-343 as a bitopic orthosteric/allosteric ligand. J Biol Chem. 2008;283:29312–29321. doi: 10.1074/jbc.M803801200. doi:10.1074/jbc.M803801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzger TG, Paterlini MG, Ferguson DM, Portoghese PS. Investigation of the selectivity of oxymorphone- and naltrexone-derived ligands via site-directed mutagenesis of opioid receptors: exploring the “address” recognition locus. J Med Chem. 2001;44:857–862. doi: 10.1021/jm000381r. [DOI] [PubMed] [Google Scholar]
- 19.Xue JC, et al. Differential binding domains of peptide and non-peptide ligands in the cloned rat kappa opioid receptor. J Biol Chem. 1994;269:30195–30199. [PubMed] [Google Scholar]
- 20.Pepin MC, Yue SY, Roberts E, Wahlestedt C, Walker P. Novel “restoration of function” mutagenesis strategy to identify amino acids of the delta-opioid receptor involved in ligand binding. J Biol Chem. 1997;272:9260–9267. doi: 10.1074/jbc.272.14.9260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.