1. INTRODUCTION
In diagnostic radiology receiver-operating-characteristic (ROC) curves are commonly used to quantify the accuracy with which a reader (typically a radiologist) can discriminate between images from nondiseased (or normal) and diseased (or abnormal) cases. Although the ROC curve concisely describes the trade-offs between sensitivity and specficity, typically accuracy is summarized by a summary index that is a function of the ROC curve. Commonly used summary indices include the area under the ROC curve (AUC), the partial area under the ROC curve (pAUC), sensitivity for a given specificity, and specificity for a given sensitivity. See Zou et al [1] for a concise introduction to ROC analysis.
A common method for estimating the ROC curve is likelihood estimation under the assumption of a latent binormal model [2–5]; alternatively, a generalized linear model approach can also be used [6, 7] based on the binormal model assumption. Under the latent binormal model assumption the ROC curve can be described by two parameters. Except for the pAUC, analytic expressions have been routinely employed for expressing the indices previously mentioned as a function of the binormal ROC curve parameters. Presently it is generally believed that the pAUC, assuming a latent binormal model, cannot be expressed as an analytic expression. For example, Pepe [8, p 84] states: “Unfortunately, a simple analytic expression does not exist for the pAUC summary measure. It must be calculated using numerical integration or a rational polynomial approximation.” Similarly, Zhou et al [9, p 128] state: “This partial area as it is known, is evaluated by numerical integration (McClish, 1989).” Although these methods can be programmed, having a simple expression for the pAUC would be much more convenient.
It is generally not known that Pan and Metz [10] provided analytic expressions for the two forms of pAUC. However, the expressions they provided were incorrect and they did not provide proofs for their results. More importantly, it is generally not known that Thompson and Zucchini [11] provided a correct analytic expression for one form of pAUC, as well as the proof. In fact, we only became aware of this latter result during the final stage of submitting this paper. The purpose of this paper is to bring to the attention of the reader the result provided by Thompson and Zucchini, extend their result to the second form of pAUC, and illustrate use of both pAUC expressions with a real data set that compares the relative performance of single spin-echo magnetic resonance imaging (SE MRI) to cinematic presentation of MRI (CINE MRI) for the detection of thoracic aortic dissection. In addition, we provide proofs for both results which are more accessible to radiology researchers and clinicians than the proof given by Thomas and Zucchini.
2. MATERIALS AND METHODS
2.1. Two different pAUCs
Let FPF and TPF denote false and true positive fractions for a given classification threshold such that an image with a test result equal or greater than the threshold is classified as diseased, and otherwise nondiseased. That is, FPF is the probability that a test result for a non-diseased subject exceeds the threshold and TPF is the probability that a test result for a diseased subject exceeds the threshold. The ROC curve is a plot of TPF versus FPF for all possible thresholds. FPF and TPF are the same as 1–specificity and sensitivity, respectively.
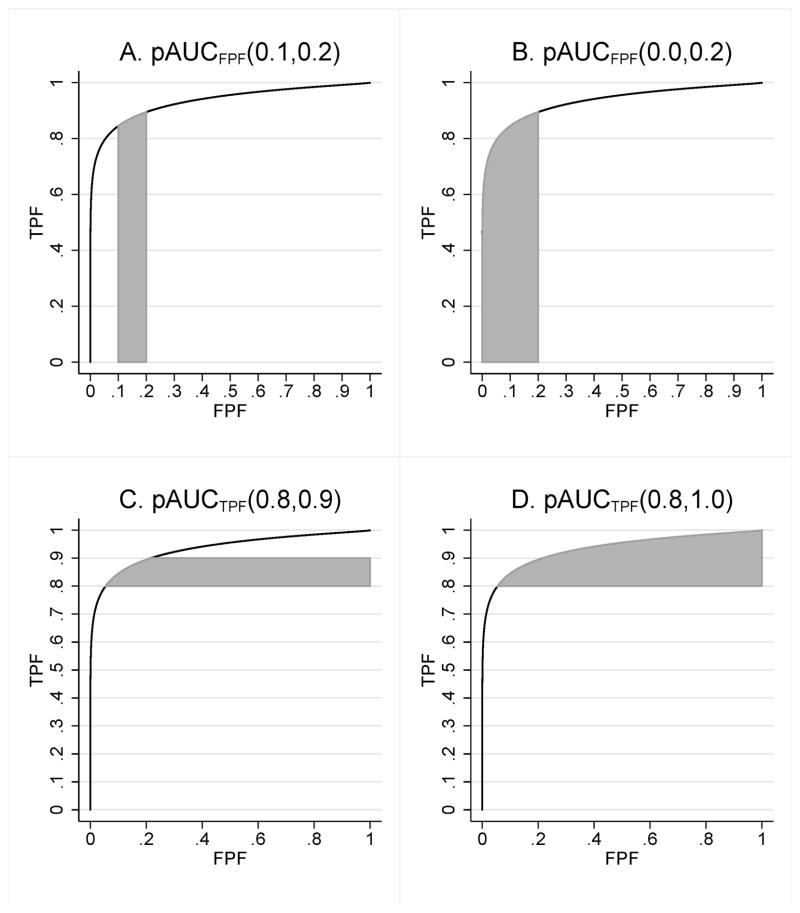
Two fundamentally different partial areas have been proposed [11–13]. One partial area corresponds to the area under an ROC curve over an interval (FPF1 < FPF2), which we denote by pAUCFPF (FPF1, FPF2). This pAUC is illustrated in Figures 1A and 1B. Often this pAUC is normalized by dividing by FPF2 – FPF1, which allows it to be interpreted as the average value of TPF over all values of FPF between FPF1 and FPF2. This partial area is typically useful when a clinical task demands high specificity; for this situation FPF1 = 0, FPF2 is small (e.g., .10 or .20), and thus it is pAUCFPF (0, FPF2) that we are interested in computing. Because pAUCFPF (FPF1, FPF2) = pAUCFPF (0, FPF2) – pAUCFPF (0, FPF1) for FPF1 < FPF2, it suffices to provide a a general formula only for pAUCFPF (0, FPF0). Walter [14] has discussed using this pAUC with summary ROC curves.
Figure 1.
Partial areas under the ROC curve
The other pAUC corresponds to the area to the right of the ROC curve in the interval (TPF1 < TPF2), which we denote by pAUCTPF(TPF1, TPF2). This pAUC is illustrated in Figures 1C and 1D. Often this pAUC is normalized by dividing by TPF2 – TPF1, which allows it to be interpreted as the average value of 1–FPF (i.e., specificity) over all values of TPF between TPF1 and TPF2. This pAUC is typically useful when a clinical ask demands high sensitivity: TPF1 is large, TPF2 = 1, and thus it is pAUCTPF (TPF1, 1) that we are interested in computing. Because pAUCTPF(TPF1, TPF2) = pAUCTPF (TPF1, 1) –pAUCTPF (TPF2, 1) for TPF1 < TPF2, it suffices to provide a a general formula only for pAUCTPF (TPF0, 1).
2.2. Analytic expressions for the pAUCs
In this section we present analytic expressions for the two forms of pAUC under the assumption of a latent binormal model. These expressions are the primary contribution of this paper. Corresponding proofs are presented in the Appendix.
2.2.1. Binormal model assumptions
Throughout we assume that the ROC curve is based on a latent binormal model. The latent binormal model assumes that the latent decision variable used to classify cases (or some unknown strictly increasing transformation of it) arises from a pair of normal densities corresponding to the nondiseased and diseased case populations, having generally different means and standard deviations. Because ROC curves are invariant under strictly increasing transformations of the decision variable, we can assume without loss of generality that the normal distribution for nondiseased cases has zero mean and unit standard deviation, whereas that for diseased cases has mean μ and standard deviation σ, where μ > 0 and σ > 0. Thus letting X and Y denote independent decision variables having the same distributions as the decision variable distributions for nondiseased and diseased cases, respectively, we are assuming that X ~ N (0, 1) and Y ~ N (μ, σ2).
2.2.2. Results for pAUCFPF (0, FPF0)
Let Φ (u) denote the standardized normal distribution function; i.e., Φ(u) = Pr (U < u) where U has a normal distribution with zero mean and unit variance. Let FBVN (z, u; ρ) denote the standardized bivariate normal distribution function with correlation ρ; i.e., FBVN (z, u; ρ) = Pr(Z < z and U < u), where Z and W jointly have a standardized bivariate normal distribution and ρ = corr(Z, U). This function is available in many statistical software programs, such as SAS, Stata, SPSS, and the freely available R program.
Assuming the binormal model described in Section 2.2.1, pAUCFPF (0, FPF0) is given by
| (1) |
In terms of the binormal parameters a = μ/σ and b = 1/σ, we can write Eq. 1 in the form
| (2) |
Equation 2 is also given by Thompson and Zucchini [11].
2.2.3. Results for pAUCTPF (TPF0, 1)
Assuming the binormal model described in Section 2.2.1, pAUCTPF (TPF0, 1) is given by
| (3) |
In terms of the binormal parameters a = μ/σ and b = 1/σ, we can write Eq. (3) in the form
| (4) |
2.3. Estimation and inference for pAUC
Because pAUC is a function of the binormal ROC curve parameters, estimation of pAUC involves estimating the parameters for an ROC curve under the assumption of a latent bi-normal model and then using Eqs. 1–4 with the ROC curve parameters replaced by estimates. A likelihood or generalized linear model approach can be used to estimate the parameters, as mentioned in the Introduction. The variance of the pAUC estimate for one test or for the difference of two tests can be estimated using a first-order Taylor series approximation (the “delta method”) [12, 13, 15], or by resampling methods such as the bootstrap and jackknife [16, 17]. For multireader studies, the methods proposed by Dorfman, Berbaum, and Metz (DBM) [18] and by Obuchowski and Rockette (OR) [19] can be used for variance estimation and inference. Confidence intervals, assuming approximate normality for the pAUC estimates, can be based on the variance estimates in the usual way.
2.4. Example data set
To illustrate use of pAUCs, we consider an example from Carolyn Van Dyke, MD, that we have analyzed in previous papers. The study [20] compared the relative performance of single spin-echo magnetic resonance imaging (SE MRI) to cinematic presentation of MRI (CINE MRI) for the detection of thoracic aortic dissection. There were 45 patients with an aortic dissection and 69 patients without a dissection imaged with both SE MRI and CINE MRI. Five radiologists independently interpreted all of the images using a five-point ordinal scale: 1 = definitely no aortic dissection, 2 = probably no aortic dissection, 3 = unsure about aortic dissection, 4 = probably aortic dissection, and 5 = definitely aortic dissection. We estimate the ROC curves using likelihood estimation based on a latent binormal model [2–4]. From the binormal ROC curve parameters we estimate pAUCs corresponding to two different FPF and two different TPF intervals and compute corresponding standard deviations using the jackknife.
We also analyze the pAUC outcomes from the example data set using the multireader data analysis method proposed by Dorfman et al (DBM) [18, 21] and updated as described by Hillis et al [22]. This analysis tests if the means of the AUCs differ between the modalities. It has been shown that the method proposed by Obuchowski and Rockette (OR) [19], updated by the degrees of freedom estimate proposed by Hillis [23], yields results identical to those of DBM when the jackknife is used to estimate the error covariances. Thus our analysis results can be considered to have been produced by either method.
Data analyses were performed using SAS [24]. The binormal AUC was computed in SAS using a dynamic link library (DLL), written in Fortran 90 by Don Dorfman and Kevin Schartz, which was accessed from within the IML procedure in SAS; this DLL can be downloaded from http://perception.radiology.uiowa.edu. The SAS program for implementing this analysis [25], as well as a user-friendly stand-alone program [26] for implementing it, can also be downloaded from http://perception.radiology.uiowa.edu.
3. RESULTS
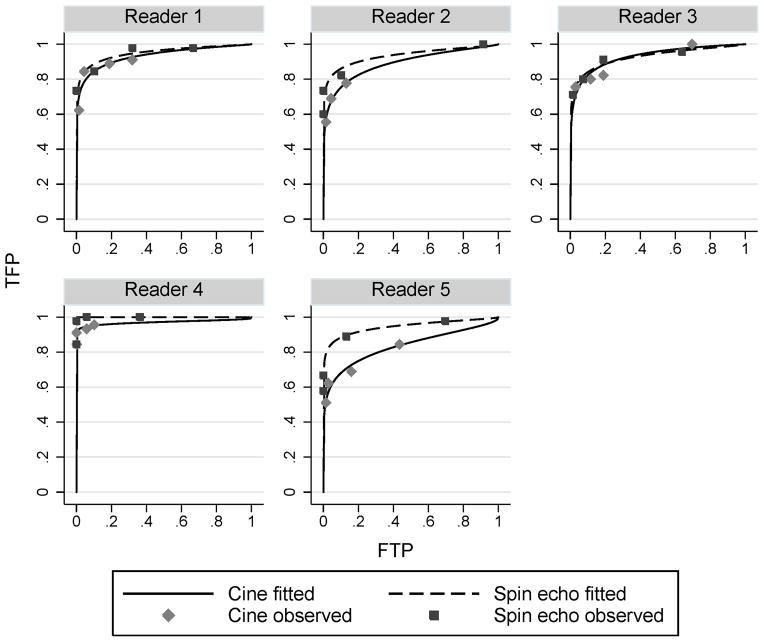
The ROC curves computed for the example data set are presented in Figure 2. Table 1 presents the corresponding binormal parameter estimates for a and b and estimates and standard errors for AUC, pAUCFPF for FPF intervals (0.0, 0.2) and (0.0, 0.1), and pAUCTPF for TPF intervals (0.8, 1.0) and (0.9, 1.0). The pAUCs have been normalized by dividing by the length of the defining interval; thus the pAUC values represent average sensitivity or specificity over the corresponding defining FPF or TPF interval. Having an analytic expression for the partial areas makes a table like Table 1 easy to construct, since the pAUCs can be computed directly from a and b. However, we note that the standard errors could not be directly computed from the ROC parameters, but rather they had to be computed separately using the jackknife.
Figure 2.
Binormal ROC curves for Van Dyke et al [20] data by reader.
Table 1.
VanDyke et al [13] binormal parameter estimates and corresponding summary indices. Notes: pAUCFPF(interval below) is the area under the ROC curve with the given FPF interval; sens(spec below) is sensitivity for the stated specificity; pAUCTPF(interval below) is the area to the right of the ROC curve within the stated TPF interval. pAUCs have been normalized by dividing by the length of the defining interval. Standard errors, computed using the jackknife, are shown in parentheses.
| modality | reader | a | b | AUC | pAUCFPF(interval below)
|
pAUCTPF(interval below)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (0,0.2) | (0,0.1) | (0.8,1) | (0.9,1) | ||||||||||
|
|
|
|
|
|
|
||||||||
| Cine | 1 | 1.7022 | 0.5368 | 0.93 | (0.03) | 0.82 | (0.07) | 0.77 | (0.10) | 0.69 | (0.17) | 0.49 | (0.30) |
| 2 | 1.4033 | 0.5607 | 0.89 | (0.06) | 0.73 | (0.07) | 0.66 | (0.09) | 0.52 | (0.27) | 0.31 | (0.34) | |
| 3 | 1.7408 | 0.6346 | 0.93 | (0.02) | 0.79 | (0.07) | 0.73 | (0.09) | 0.68 | (0.08) | 0.51 | (0.10) | |
| 4 | 1.9255 | 0.2015 | 0.97 | (0.02) | 0.95 | (0.03) | 0.94 | (0.03) | 0.85 | (0.12) | 0.70 | (0.24) | |
| 5 | 1.0630 | 0.4635 | 0.83 | (0.05) | 0.66 | (0.08) | 0.60 | (0.09) | 0.32 | (0.17) | 0.12 | (0.14) | |
| Spin echo | 1 | 1.8501 | 0.5030 | 0.95 | (0.02) | 0.87 | (0.04) | 0.83 | (0.05) | 0.76 | (0.12) | 0.58 | (0.21) |
| 2 | 1.6552 | 0.4473 | 0.93 | (0.02) | 0.84 | (0.05) | 0.80 | (0.06) | 0.68 | (0.10) | 0.46 | (0.14) | |
| 3 | 1.6220 | 0.4878 | 0.93 | (0.03) | 0.82 | (0.06) | 0.77 | (0.07) | 0.66 | (0.15) | 0.44 | (0.23) | |
| 4 | 7.1233 | 0.8806 | 1.00 | (0.00) | 1.00 | (0.00) | 1.00 | (0.00) | 1.00 | (0.00) | 1.00 | (0.00) | |
| 5 | 1.7329 | 0.4221 | 0.94 | (0.03) | 0.87 | (0.05) | 0.84 | (0.06) | 0.73 | (0.13) | 0.52 | (0.22) | |
Table 2 presents the results of the DBM/OR analyses. For our discussion we assume alpha = .05. AUC did not show a significance difference (p = 0.14). In contrast, pAUCFPF(0.0, 0.2) almost reached significance (p = .06) and tests based on pAUCFPF (0.0, 0.1) and pAUCFPF (0.0, 0.05) were both significant (p = .0399 and 0.0278, respectively). These results suggest that partial AUC provides a more powerful test than AUC for these data. Sensitivity for a fixed specificity also resulted in more significant results than AUC. Results for the horizontal-band pAUCs was similar to that of AUC for two intervals (p = .14 and .15 for TPF > .8 and .9, respectively), but somewhat less for the third interval, defined by TPF > 0.95.
Table 2.
ROC summary measure estimates for Van Dyke et al [20] data assuming a latent binormal model. P -value is for H0: the pAUC means are equal for cine and spin-echo MRI.
| Type of estimator | Specific estimator | Estimates
|
P-value | |
|---|---|---|---|---|
| Cine | Spin-echo | |||
| AUC | AUC | 0.911 | 0.952 | 0.1413 |
| pAUCFPF | pAUCFPF (0.0, 0.2) | 0.790 | 0.880 | 0.0600 |
| pAUCFPF (0.0, 0.1) | 0.740 | 0.848 | 0.0399 | |
| pAUCFPF (0.0, 0.05) | 0.691 | 0.817 | 0.0278 | |
| Sensitivity at fixed specificity | Sens (spec = 0.80) | 0.863 | 0.925 | 0.1265 |
| Sens (spec = 0.90) | 0.811 | 0.894 | 0.0778 | |
| Sens (spec = 0.95) | 0.760 | 0.862 | 0.0491 | |
| pAUCTPF | pAUCTPF (0.8, 1.0) | 0.613 | 0.765 | 0.1426 |
| pAUCTPF (0.9, 1.0) | 0.427 | 0.599 | 0.1534 | |
| pAUCTPF (0.95, 1.0) | 0.251 | 0.430 | 0.2277 | |
4. DISCUSSION
For the two types of pAUCs we derived analytic expressions under the assumption of a latent binormal model. Previously it was believed that analytic expressions did not exist, even though Thompson and Zucchini [11] had stated and proved Eq. 2, and thus numerical methods have been used to solve for pAUC values. The formulas presented in this paper greatly simplify computation of pAUCs.
We illustrated use of these expressions with a real data set where, using a multireader analysis, we found that pAUCFPF gave more significant results than did AUC. This example illustrates the ease with which pAUC measures can be computed using the expressions provided in this paper. It also suggests that partial areas can be more powerful for comparing modalities than AUCs under certain circumstances. While we recognize that it is generally thought that AUC provides a more powerful test than a partial area [15, 27], this example suggests that there may be situations where pAUCs will be more powerful and provides motivation for a closer examination of the relationship between pAUC and AUC with respect to power.
Although Thompson and Zucchini [11] provided a proof for Eq. 2, their proof requires a solid understanding of calculus and familiarity with the bivariate normal density function. In contrast, the proofs that we provide for the expressions for both pAUCs requires only basic knowledge of statistics and algebra. Thus we believe that our proofs will be more accessible to radiological researchers and clinicians.
Acknowledgments
We thank Carolyn Van Dyke, MD for sharing her data set for the example. We thank the reviewers for helpful suggestions that clarified the presentation. Stephen Hillis was supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), grants R01EB000863 and R01EB013667.
Appendix A. Appendix: Derivation of Eqs. 1–4 in Section 2.2.2
In this section we derive Eqs. 1–4 in Section 2.2.2. We assume an underlying binormal distribution, as discussed in Section 2.2.1. In particular, we assume that X and Y denote independent decision variables having the same distributions as the decision variable distributions for nondiseased and diseased cases, respectively, with X ~ N (0, 1) and Y ~ N (μ, σ2).
Appendix A.1. Derivation of pAUCFPF (0, FPF0) results (Eqs. 1–2)
It has been shown [8, 28] that
| (A.1) |
where SX, defined by SX (x) = Pr (X > x), is the complement of the cumulative distribution distribution function of X. This is a general result that holds even if the conditional distributions are not normal. Noting that , where FPF0 = Pr(X > ξ0), it follows from Eq. A.1 that
| (A.2) |
We will use Eq. (A.2) to derive pAUCFPF (0, FPF0) below in terms of the latent binormal decision-variable distribution parameters.
From Eq. A.2 we have
| (A.3) |
where . It is easy to show that Z ~N (0, 1) and . To show the correlation result, note X and that Y are independent and corr (Z, X) = cov (Z, X) because both Z and X have unit standard deviation. Thus . It follows that (Z, X) has a standardized bivariate normal distribution with correlation . Thus
| (A.4) |
where FBVN (z, x; ρ) is the standardized bivariate normal distribution function with correlation ρ as discussed in Section 2.2.2. Because FBVN (z, x; ρ) = 1 − FBVN (−z, −x; ρ), it follows from Eq. A.4 that
Using the relationship −ξ0 = Φ−1(FPF0) we have
| (A.5) |
In terms of the binormal parameters a = μ/σ and b = 1/σ, we can write Eq. A.5 in the form
| (A.6) |
Appendix A.2. Derivation of pAUCTPF (TPF0, 0) results (Eqs. 3–4) in Section 2.2.2
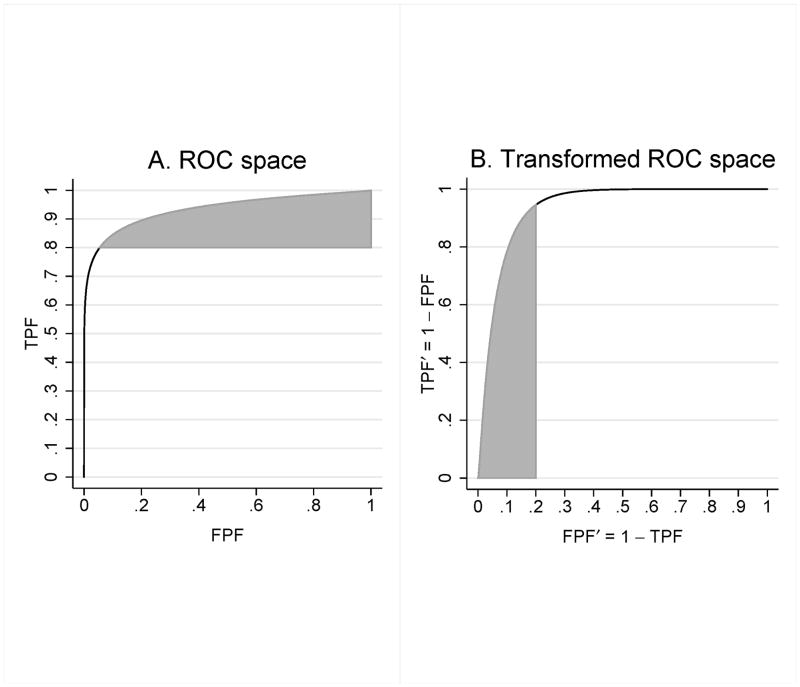
Our strategy for finding pAUCTPF (TPF0, 1) is to express pAUCTPF (TPF0, 1) in the form pAUCFPF (0, FPF0) for an appropriate binormal distribution and FPF0 value. Consider part A of Figure A1, which shows the ROC curve and the shaded region corresponding to pAUCTPF (TPF0, 1), with TPF0 = .8. Define
Part B shows the ROC curve and shaded portion after transformation to the coordinate system with FPF′ on the x-axis and TPF′ on the y-axis. We see that the resulting plot looks like an ROC plot with FPF′ and TPF′ as false and true positive fractions; below we prove this to be the case. Moreover, it is easy to show that area of the shaded region remains constant under the transformation.
Let ξ denote a threshold value. Corresponding values for FPF′ and TPF′ are given by
Defining
we have
Furthermore, it follows that X′ ~ N (−μ, σ2) and Y ′ ~ N (0, 1). Thus the plot shown in Figure 3 is the ROC curve corresponding to the binormal distribution defined by nondiseased and diseased decision variables X′ and Y′. Noting that TPF= 1 is mapped to FPF′ = 0 and TPF= TPF0 is mapped to FPF′ = 1 − TPF0 for TPF0 < 1, it follows that pAUCTPF (TPF0, 1) for the binormal distribution with nondiseased and diseased distributions X and Y is equal to pAUCFPF (0, 1 − TPF0) for the binormal distribution for nondiseased and diseased distributions X′ and Y ′.
In order to use Eqs. 1–2, we rescale X′ and Y ′ so that rescaled X′, denoted by X̃, has zero mean and unit standard deviation:
Since the same transformation is applied to both X̃ and Ỹ, the ROC curve remains unchanged. It follows that
and the standard binormal parameters for the (X̃, Ỹ) binormal distribution are
| (A.7) |
From Eq. 1 it follows that, for the binormal distribution defined by X̃ and Ỹ, pAUCFPF (1 − TPF0, 0) is given by
| (A.8) |
Note that in Eq. A.5 that FPF0, μ, and σ were replaced by 1 − TPF0, μ/σ, and 1/σ, respectively, to yield Eq. A.8. Equivalently, in terms of a and b it follows from Eqs. A.7 and A.8 that
| (A.9) |
Note that Eqs. A.8 and A.9 are the same as Eqs. 3 and 4 in Section 2.2.2.
Appendix B
Figure A1.
ROC curve and pAUCTPF (.8, 1) shaded area in ROC space (A) and after transformation (B) to the coordinate system defined by FPF′ = 1 − TPF on the x-axis and TPF′ = 1 − FPF on the y-axis. In the original ROC space (A) the nondiseased and diseased decision variables X and Y define the ROC curve; in the transformed ROC space (B) the ROC curve is defined by nondiseased and diseased decision variables X′ = −Y and Y ′ = −X, with the shaded area equal to pAUCFPF (0, .2).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Stephen L. Hillis, Departments of Radiology and Biostatistics, University of Iowa; Comprehensive Access and Delivery Research and Evaluation (CADRE) Center, Iowa City VA Health Care System.
Charles E. Metz, Department of Radiology, University of Chicago.
References
- 1.Zou KH, O’Malley AJ, Mauri L. Receiver-Operating Characteristic Analysis for Evaluating Diagnostic Tests and Predictive Models. Circulation. 2007;115:654–657. doi: 10.1161/CIRCULATIONAHA.105.594929. [DOI] [PubMed] [Google Scholar]
- 2.Dorfman DD, Alf E., Jr Maximum likelihood estimation of parameters of signal-detection theory and determination of confidence intervals–rating method data. Journal of Mathematical Psychology. 1969;6:487–496. [Google Scholar]
- 3.Dorfman DD. RSCORE II. In: Swets JA, Pickett RM, editors. Evaluation of Diagnostic Systems: Methods from Signal Detection Theory. Academic Press; San Diego, CA: 1982. pp. 212–232. [Google Scholar]
- 4.Dorfman DD, Berbaum KS. Degeneracy and discrete receiver operating characteristic rating data. Academic Radiology. 1995;2:907–915. doi: 10.1016/s1076-6332(05)80073-x. [DOI] [PubMed] [Google Scholar]
- 5.Metz CE, Herman BA, Shen JH. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Statistics in Medicine. 1998;17:1033–1053. doi: 10.1002/(sici)1097-0258(19980515)17:9<1033::aid-sim784>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Pepe MS. An interpretation for the ROC curve and inference using GLM procedures. Biometrics. 2000;56:352–359. doi: 10.1111/j.0006-341x.2000.00352.x. [DOI] [PubMed] [Google Scholar]
- 7.Alonzo TA, Pepe MS. Distribution-free ROC analysis using binary regression techniques. Biostatistics. 2002;3:421–432. doi: 10.1093/biostatistics/3.3.421. [DOI] [PubMed] [Google Scholar]
- 8.Pepe M. The statistical evaluation of medical tests for classification and prediction. New York: Oxford University Press; 2003. [Google Scholar]
- 9.Zhou X-H, Obuchowski NA, McClish DK. Statistical methods in diagnostic medicine. New Jersey: Wiley; 2011. [Google Scholar]
- 10.Pan XC, Metz CE. The ”proper” binormal model: parametric receiver operating characteristic curve estimation with degenerate data. Academic Radiology. 1997;4:380–389. doi: 10.1016/s1076-6332(97)80121-3. [DOI] [PubMed] [Google Scholar]
- 11.Thompson ML, Zucchini W. On the statistical analysis of ROC curves. Statistics in Medicine. 1989;8:1277–1290. doi: 10.1002/sim.4780081011. [DOI] [PubMed] [Google Scholar]
- 12.McClish DK. Analyzing a portion of the ROC curve. Medical Decision Making. 1989;9:190–195. doi: 10.1177/0272989X8900900307. [DOI] [PubMed] [Google Scholar]
- 13.Jiang YL, Metz CE, Nishikawa RM. A receiver operating: Characteristic partial area index for highly sensitive diagnostic tests. Radiology. 1996;201:745–750. doi: 10.1148/radiology.201.3.8939225. [DOI] [PubMed] [Google Scholar]
- 14.Walter SD. The partial area under the summary ROC curve. Statistics in Medicine. 2005;24:2025–2040. doi: 10.1002/sim.2103. [DOI] [PubMed] [Google Scholar]
- 15.Obuchowski NA, McClish DK. Sample size determination for diagnostic accuracy studies involving binormal ROC curve indices. Statistics in Medicine. 1997;16:1529–1542. doi: 10.1002/(sici)1097-0258(19970715)16:13<1529::aid-sim565>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 16.Dongshen JT. The Jackknife and Bootstrap. Springer-Verlag; New York: 1995. [Google Scholar]
- 17.Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- 18.Dorfman DD, Berbaum KS, Metz CE. Receiver operating characteristic rating analysis: generalization to the population of readers and patients with the jackknife method. Investigative Radiology. 1992;27:723–731. [PubMed] [Google Scholar]
- 19.Obuchowski NA, Rockette HE. Hypothesis testing of the diagnostic accuracy for multiple diagnostic tests: an ANOVA approach with dependent observations. Communications in Statistics: Simulation and Computation. 1995;24:285–308. [Google Scholar]
- 20.Van Dyke CW, White RD, Obuchowski NA, Geisinger MA, Lorig RJ, Meziane MA. Cine MRI in the diagnosis of thoracic aortic dissection. 79th RSNA Meetings; Chicago, IL. November 28 – December 3, 1993. [Google Scholar]
- 21.Dorfman DD, Berbaum KS, Lenth RV, Chen YF, Donaghy BA. Monte Carlo validation of a multi-reader method for receiver operating characteristic discrete rating data: factorial experimental design. Academic Radiology. 1998;5:591–602. doi: 10.1016/s1076-6332(98)80294-8. [DOI] [PubMed] [Google Scholar]
- 22.Hillis SL, Berbaum KS, Metz CE. Recent developments in the Dorfman-Berbaum-Metz procedure for multireader ROC study analysis. Academic Radiology. 2008;15:647–661. doi: 10.1016/j.acra.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillis SL. A comparison of denominator degrees of freedom methods for multiple observer ROC analysis. Statistics in Medicine. 2007;26:596–619. doi: 10.1002/sim.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SAS for Windows, Version 9.3, copyright (c) 2002–2010. SAS Institute Inc; Cary, NC, USA: [Google Scholar]
- 25.Hillis SL, Schartz KM, Berbaum KS. [Accessed December 29, 2011.];OR/DBM MRMC procedure for SAS 3.0 (computer software) Available for download from http://perception.radiology.uiowa.edu.
- 26.Berbaum KS, Schartz KM, Hillis SL. OR/DBM MRMC (Version 2.4) [Computer software] Iowa City, IA: The University of Iowa; 2012. Available from http://perception.radiology.uiowa.edu. [Google Scholar]
- 27.Hanley JA. Receiver operating characteristic (ROC) methodology - the state of the art. Critical Reviews in Diagnostic Imaging. 1989;29:307–335. [PubMed] [Google Scholar]
- 28.Dodd LE, Pepe MS. Partial AUC estimation and regression. Biometrics. 2003;59:614–623.12. doi: 10.1111/1541-0420.00071. [DOI] [PubMed] [Google Scholar]