Abstract
Here we describe the embryonic CNS expression of 5,000 GAL4 lines made using molecularly defined cis-regulatory DNA inserted into a single attP genomic location. We document and annotate the patterns in early embryos when neurogenesis is at its peak, and in older embryos where there is maximal neuronal diversity and the first neural circuits are established. We note expression in other tissues such as the lateral body wall (muscle, sensory neurons, trachea) and viscera. Companion papers report on the adult brain and larval imaginal discs, and the integrated datasets are available online (www.janelia.org/flylight/gal4-gen1). This collection of embryonically-expressed GAL4 lines will be valuable for determining neuronal morphology and function; the 1862 lines expressed in small subsets of neurons (<20/segment) will be especially valuable for characterizing interneuronal diversity and function, as interneurons comprise the majority of all CNS neurons, yet their gene expression profile and function remain virtually unexplored.
Introduction
All model organisms can benefit from tools that allow targeted gene expression. In mouse, collections of Cre lines allow “floxed” or “flox-stop” transgenes to be expressed or inactivated, respectively (reviewed in Branda and Dymecki, 2004). In Drosophila and zebrafish, the yeast GAL4/UAS sytem has become a standard method for generating cell type specific gene expression (reviewed in Baier and Scott, 2009; Scott, 2009; Scott et al., 2007; Venken and Bellen, 2012). Drosophila is a powerful model system for studying cell biology, development, and neuroscience, primarily due to its facile genetics. Over the past two decades, genetic screens have identified key regulators of highly conserved cellular processes (e.g. cell cycle progression, cell death, cell migration), developmental events (e.g. Hox function, cellular memory, cell-cell signaling pathways), and neurobiological processes (e.g. axon pathfinding, learning and memory, behavior).
We are interested in characterizing the generation of cellular diversity within the embryonic central nervous system (CNS) and linking neuronal development with function and behavior. The CNS develops starting at stage 8 with the formation of the specialized ventral midline cells; at stage 9, the bilateral neuroectoderm generates an array of ~30 neural progenitors, called neuroblasts (Doe, 2008; Skeath and Thor, 2003). All ventral midline progenitors and neuroblasts are stereotyped and individually identifiable (Broadus et al., 1995; Wheeler et al., 2006). Each neuroblast divides asymmetrically to generate smaller ganglion mother cells and a self-renewed neuroblast (reviewed in Skeath and Thor, 2003). Each ganglion mother cell either divides once to generate a pair of sibling neurons (Pearson and Doe, 2003), divides more than once to generate a small number of glia (Akiyama-Oda et al., 1999), or directly differentiates into a neuron (Baumgardt et al., 2009). By stage 16, neuroblast divisions have largely ceased and each segment of the CNS typically contains 22 midline cells (Wheeler et al., 2006) and ~335 bilateral cells: ~35 motoneurons, ~35 glia, and ~ 260 interneurons or unknown cell types (Beckervordersandforth et al., 2008; Landgraf et al., 1999)(ESH and CQD, unpublished).
Embryonic motoneurons are relatively well characterized; most can be identified by the presence of nuclear phosphorylated Mothers against Dpp (pMad) protein or Zfh1 protein (Layden et al., 2006), and subsets can be identified that express the even-skipped (eve) gene, encoding a transcription factor that promotes axon targeting to dorsal muscle groups (Fujioka et al., 2003; Landgraf et al., 1999), or hb9 (Flybase: exex), which encodes a transcription factor promoting axon targeting to ventral muscles (Broihier and Skeath, 2002; Odden et al., 2002). GAL4 lines are available that are expressed in subsets of the Eve+ neurons (Fujioka et al., 1999) and the entire population of Hb9+ neurons (Broihier and Skeath, 2002). Similarly, glial cells can be identified based on position, molecular markers, and GAL4 line expression (Beckervordersandforth et al., 2008). Despite the work of many labs over the years, very little is known about the majority of interneurons within the CNS. There are few molecular markers for interneuronal subsets and even fewer GAL4 lines with well-characterized expression in subsets of interneurons. This is a barrier to understanding how interneurons develop, and how they participate in neural circuits regulating larval behavior.
A powerful tool for characterizing neuronal morphology and function in Drosophila is the GAL4/UAS system (Brand and Perrimon, 1993). GAL4 transgenes with distinct patterns have been made by random insertion of constructs containing a basal promoter and the GAL4 coding sequence, a variation of the “enhancer trapping” method (Bellen et al., 1989; Bier et al., 1989; O'Kane and Gehring, 1987) that has been widely used to identify cell type specific enhancers and their associated genes. The growing collection of Drosophila GAL4 transgenes have been useful for marking neuronal cell types or expressing UAS-RNAi constructs to assay gene function (see above), as well as in the Mosaic Analysis with Repressible Cell Marker (MARCM) system to generate positively-marked homozygous mutant neuronal clones (Lee and Luo, 2001). These are all powerful methods, but they each have limitations. First, most GAL4 lines have been generated by random insertion of a basal GAL4 transgene randomly within the genome (“enhancer traps”), making it difficult to generate additional lines with the same pattern (e.g. LexA versions of the line) and impossible to perform bioinformatic comparisons of the DNA sequences that confer different GAL4 expression patterns. Second, relatively few GAL4 lines are expressed in small subsets of the CNS, making it impossible to characterize the morphology and function of the vast majority of neurons.
Here we describe the embryonic CNS expression of 5,000 GAL4 lines made using molecularly defined cis-regulatory DNA inserted into a single attP genomic location using the PhiC31 integrase system (Bischof et al., 2007; Groth et al., 2004). This will facilitate production of non-GAL4 drivers that can be used in combination with GAL4 lines. It will also make it possible to use bioinformatics to look for cis-regulatory elements that are shared by GAL4 lines with co-expression in one or more neurons. We describe the embryonic CNS expression at stages 9–11 (when neurogenesis is maximal) and stage 16 (when neuronal diversity is maximal). We comprehensively annotate expression patterns in the CNS midline cells, neuroblasts, and neurons at both stages. We note expression in other embryonic tissues such as the lateral body wall (muscle, sensory neurons, trachea, etc.) and viscera. Companion papers report on the expression of the same collection of GAL4 lines in the adult brain (Jenett et al., accompanying paper) and imaginal discs (Jory et al., accompanying paper ), and all three datasets are publicly available on-line (www.janelia.org/flylight/gal4-gen1). The embryonic data derived from this collection of GAL4 lines will be valuable for determining the morphology and function of all neurons in the embryonic CNS, but especially for shedding light on the function of interneurons, which have only begun to be defined although they comprise the majority of all neurons in the CNS.
Results and Discussion
Overview of embryonic patterns and database features
We stained over 5,500 enhancer-gal4 lines generated at Janelia Farm Research Campus (JFRC) to determine their pattern of expression during embryogenesis (Supplemental Table 1), acquiring interpretable images for 5,000 lines representing 796 genes. The method for generating the lines has been described previously: each line contains an average of 3 kb cis-regulatory DNA from intergenic and intronic regions of genes known or suspected of being expressed in the adult brain, and each transgene was integrated into the same attP2 site on the second chromosome (Pfeiffer et al., 2008). We chose embryos at two stages for detailed analysis: germband elongated (GBE; embryonic stages 9–12) and germband shortened (stages 15–17; most embryos were at stage 16). We crossed each line to a line containing UAS-GFP:NLS (GFP with a nuclear localization signal) and/or cd8:GFP (membrane tethered GFP) and stained for GFP to determine the expression pattern of each line. We co-stained each line for the Even-skipped (Eve) transcription factor, which detects a subset of neurons and muscle precursors and anal pad (Frasch et al., 1987); these cells can be used as well-characterized landmarks to help annotate the GAL4 patterns. Our goal was to comprehensively annotate the gene expression patterns in neuroblasts (NBs), GMCs, neurons and glia, including the specialized neurons and glia at the ventral midline. We noted expression outside the CNS but did not attempt to annotate non-neural tissue patterns.
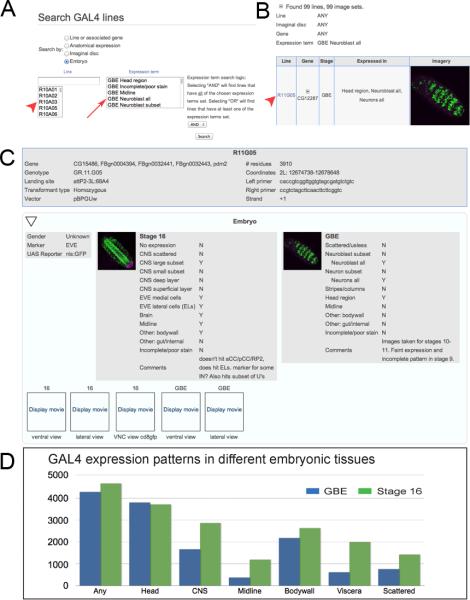
Our data are included in a publicly available database hosted by JFRC (www.janelia.org/flylight/gal4-gen1); see Figure 1A–C for screenshots describing how to search the database for embryonic patterns. Additional details of the website construction and database functions are described in an accompanying paper (Jenett et al., accompanying paper). The database integrates expression data from embryos (this work) as well as from the adult brain (Jenett et al., accompanying paper) and larval imaginal discs (Jory et al., accompanying paper). Our embryonic datasets include a maximum intensity projection of the most relevant portion of the CNS staining pattern, as well as at least one movie that steps through a z-stack of confocal images to illustrate all aspects of the pattern. Searching the database can be done using selected keywords (see Table 1 for GBE categories; Table 2 for stage 16 categories; and Table 3 for midline categories).
Figure 1. The GAL4 database interface.
(A) Searching the GAL4 database. Searches can be done by GAL4 line name (red arrowhead) or by tissue expression (red arrow).
(B) Search results. Searches using tissue expression criteria can return multiple hits (99 in this example). Each hit can be viewed by selecting the GAL4 line name (red arrowhead).
(C) Information for each GAL4 line. This includes the associated genes for the line, genotype, genomic insertion site, size of cis-regulatory DNA, primers used to PCR amplify the cis-regulatory DNA (top box). It also includes gene expression data from the adult brain and ventral nerve cord (see companion papers) and the embryonic CNS at germband elongated (GBE; stages 9–12) and stage 16. For the embryonic CNS, the annotated expression patterns are given (Y = expression observed; N = no expression observed), as well as both maximum intensity projections and QuickTime movies (click “Display movie” link to play movie). Higher resolution image datasets are available upon request.
(D) Histogram showing the number of GAL4 lines with the indicated expression pattern. Any = any expression in the entire embryo; Head = entire head region including brain and other cell types; CNS = ventral nerve cord from T1–A8; Midline = CNS midline cells; Bodywall = all cell types within the lateral body wall including sensory neurons, muscle, trachea, histoblasts; Viscera = internal organs excluding the CNS; Scattered = unpatterned cells in the CNS or elsewhere, including hemocytes.
Table 1.
Lines expressed in germband elongated embryos (stages 9–12)
| Expression pattern | # lines (%) | # genes (%) |
|---|---|---|
| Total | 5,000 (100%) | 796 (100%) |
| Head | 3,806 (76%) | 736 (92%) |
| Neuroblasts, subset | 662 (13%) | 313 (39%) |
| Neuroblasts, most or all | 99 (2%) | 69 (9%) |
| GMCs/Neurons, subset | 1,411 (28%) | 480 (60%) |
| GMCs/Neurons, most or all | 46 (1%) | 36 (5%) |
| Stripe or Column pattern | 224 (4%) | 140 (18%) |
| Other: body wall | 2,176 (44%) | 577 (72%) |
| Other: gut/internal | 613 (12%) | 336 (42%) |
| Scattered | 756 (15%) | 377 (47%) |
| Poor stain | 385 (8%) | 226 (28%) |
Table 2.
Lines expressed in stage 16 embryos.
| Expression pattern | # lines (%) | #genes (%) |
|---|---|---|
| Total | 5,000 (100%) | 796 (100%) |
| Head | 3,712 (74%) | 731 (92%) |
| Neurons, small subset | 1,862 (37%) | 553 (69%) |
| Neurons, most or all | 822 (16%) | 381 (48%) |
| EVE medial cells | 661 (13%) | 309 (39%) |
| EVE lateral cells | 785 (16%) | 366 (46%) |
| Other: body wall | 2,631 (53%) | 635 (80%) |
| Other: gut/internal | 1,997 (40%) | 619 (78%) |
| Scattered | 1,423 (28%) | 520 (65%) |
| Poor stain | 764 (15%) | 341 (43%) |
| No expression | 28 (1%) | 27 (3%) |
Table 3.
Lines expressed in the CNS midline.
| Expression pattern | # lines (%) | # genes (%) |
|---|---|---|
| Total | 5,000 (100%) | 796 (100%) |
| Midline expression | 1,285 (26%) | 471 (59%) |
| HS total | 438 (9%) | 253 (32%) |
| LS total | 847 (17%) | 387 (49%) |
| GBE midline expression | 389 (8%) | 244 (31%) |
| GBE HS | 137 (3%) | 104 (13%) |
| GBE HS all | 24 (1%) | 16 (2%) |
| GBE HS subset | 113 (2%) | 91 (11%) |
| GBE LS | 252 (5%) | 183 (23%) |
| St16 midline expression | 1,261 (25%) | 468 (59%) |
| St16 HS | 435 (9%) | 252 (32%) |
| St16 HS all | 26 (1%) | 18 (2%) |
| St16 HS midline glia | 109 (2%) | 83 (10%) |
| St16 HS midline glia (-all) | 83 (2%) | 69 (9%) |
| St16 HS VUMs | 89 (2%) | 66 (8%) |
| St16 HS MNB progeny | 26 (1%) | 23 (3%) |
| St16 HS neuron subset | 331 (7%) | 205 (26%) |
| St16 HS neuron subset –VUM -MNB | 223 (4%) | 156 (20%) |
| St16 LS | 826 (17%) | 383 (48%) |
| Dorsal median cells | 24 (1%) | 20 (3%) |
| Channel glia | 15 (<1%) | 14 (2%) |
| MM-CBG | 14 (<1%) | 13 (2%) |
HS – high specificity midline expression; LS – low specificity midline expression.
We found that nearly all of the 5,000 lines imaged had expression in both young GBE embryos (4,289; 86%) and older stage 16 embryos (4,672; 93%) (Figure 1D); this is an underestimate of the GBE expression because we did not image this stage in 410 lines (see methods). Thus, most of the lines contained active cis-regulatory modules (CRMs or enhancers). The most common pattern was head expression (GBE, 72%; stage 16, 74%; Figure 1D), possibly because the genes expressed in the adult brain were preferentially used to generate the GAL4 lines (Jenett et al., accompanying paper). It should be noted, however, that head expression was observed in 59% of the 500 lacZ “enhancer trap” lines (Bellen et al., 1989), so the head may utilize a large number of CRMs. The next most common patterns were CNS expression (GBE, 33%; stage 16, 57%) and body wall expression (GBE, 43%; stage 16, 53%; Figure 1D); this may be due to the complexity of cell types in these tissues. We saw relatively fewer lines expressed in the midline, viscera, and in a scattered pattern; but for each of these we observed nearly double the number of expressing lines in older embryos (Figure 1D), perhaps due to the increasing complexity of these tissues as development proceeds. We observed one expression pattern common to most of the 5,000 lines assayed: a pattern of four neuronal clusters in the stage 16 subesophageal CNS (Figure S1). This gene expression pattern is presumably derived from a DNA sequence shared by all GAL4 constructs (see Discussion).
Germband elongated patterns (embryonic stages 9–12)
At this stage of neurogenesis the neuroectoderm covers the ventral surface of the embryo, and just internal to these cells is the neuroblast array. Progressively more internal are GMCs and newly born neurons (reviewed in Skeath and Thor, 2003). We used the Even-skipped (Eve) protein as a fiduciary marker (Figure 2A). Eve is detected in the first-born GMC from NB1-1 and NB4–2; the first five GMCs from NB7–1, and a segment-specific number of GMCs from NB3-3 (five in T1–T2, six in T3, and ten in A1–A7). Each of these GMCs divides to generate an Eve+ neuron and an Eve− sibling (Skeath and Doe, 1998). In addition, Eve is detected in pericardial cells at the dorsal surface of the embryo and the anal pad (Frasch et al., 1987) (Figure 2A).
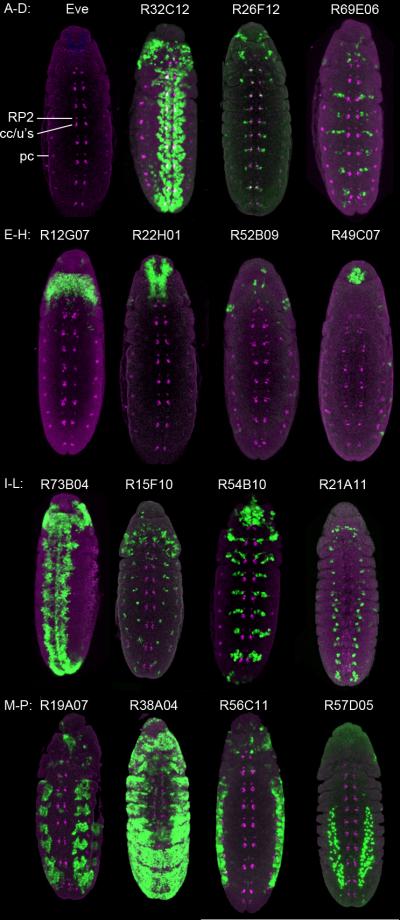
Figure 2. Representative GAL4 expression patterns in germband elongated embryos.
(A) The Even-skipped (Eve) pattern at germband elongated stages. RP2 = RP2 motoneuron; cc/u's = aCC/pCC/U1/U2 (either GMCs or neurons); pc = pericardial precursors.
(B–D) Lines expressed in most neuroblasts (B) or subsets of neuroblasts (C–D). (B) Line R32C12, gene stg; (C) Line R26F12, gene grn; (D) Line R69E06, gene gsb-n.
(E–H) Lines expressed in most GMCs/neurons (E) or subsets of GMCs/neurons (F–H). (E) Line R73B04, gene Sox21b; (F) Line R15F10, gene ss; (G) Line R45B10, gene pnt; (H) Line R21A11, gene eya.
(I–L) Lines expressed in the head. (I) Line R12G07, gene kn; (J) Line R22H01, gene fru; (K) Line R52B09, gene gol; (L) Line R49C07, gene beat-IIIc.
(M–P) Lines expressed in non-neural tissues. (M) Body wall muscle; line R19A07, gene vvl; (N) Epithelial; line R38A04, gene rst; (O) Abdominal body wall; line R56C11, gene lin-28; (P) Internal cell types; Line R57D05, gene HLH54F.
All panels show ventral views of the CNS of the indicated GAL4 UAS-nGFP lines stained for Eve (magenta) and nuclear GFP (green). Anterior, top.
We found 4320 lines expressed at GBE (Table 1; Figure 1D). We were interested in pan-neuroblast lines, with the rationale that the associated genes may show neuroblast-specific gene expression. We identified NBs based on their relatively large size and position just internal to the ventral neuroectoderm; despite the lack of a definitive NB marker such as Deadpan or Miranda (Doe, 2008), we feel confident that size and position are sufficient criteria to identify NBs. About 2% (99) of the GBE+ lines were detected in most or all NBs (Figure 2B). We were also interested in identifying GAL4 lines that label subsets of neuroblasts, particularly single neuroblasts that could be used for lineage analysis. Over 15% (662) of the GBE+ lines were detected in a subset of NBs, with some being in just a few neuroblasts per hemisegment (Figure 2C,D). Additional experiments will be necessary to map these GAL4+ neuroblasts onto the “neuroblast map” (Broadus et al., 1995), and this may be best achieved using new semiautomated methods for creating gene expression atlases (Long et al., 2009; Peng et al., 2010; Qu et al., 2011). The lines expressed in subsets of neuroblasts should be useful as neuroblast markers to study the specification of unique neuroblast identity; they could also be used to drive yeast Flippase to excise an FRT-stop-FRT cassette (del Valle Rodriguez et al., 2012) to allow permanent tracing of the neuroblast and its progeny beyond embryonic stages; this would help align embryonic neuroblast identity with larval neuroblast identity (Broadus et al., 1995; Truman and Bate, 1988) as well as help determine the contribution of specific neuroblasts to the adult CNS.
We detected 46 lines expressed in most or all GMCs/neurons (Figure 2E) and 1,411 lines expressed in a subset of GMCs/neurons (Figure 2F–H). We did not try to distinguish GMCs from neurons, as these populations are intermingled at this stage and have the same cell size. Some of the expression patterns were regionally restricted, e.g. in stripes at different anterior/posterior positions in a segment; in medial or lateral columns along the length of the CNS; or segment-specific (thoracic only or abdominal only). These are annotated in the database and can be retrieved by checking the “stripe/column” box (quantified in Table 1). The GAL4 lines with regional expression (stripe or column) may be used to identify flanking genes that are involved in spatial patterning of the CNS, or as markers to study the effect of earlier patterning genes on the specification of regional neuronal identity. They may also be useful for designing split GAL4 transgenes (Luan et al., 2006; Pfeiffer et al., 2010) to restrict GAL4 expression to a very small subset of cells (e.g. using a columnar enhancer to drive the GAL4 DNA-binding domain and the stripe enhancer to drive the GAL4 VP16 transactivation domain). These experiments are straightforward because the enhancer and attP integration sites are defined, and thus new split GAL4 transgenes should have highly predictable expression patterns.
A large fraction of lines with GAL4 patterns showed expression in the head (3,806/4,320; 88%); we did not try to map the identity of these cells to neuroblasts, neurons, glia, or non-neuronal cell types of the head due to the complexity of the pattern (Figure 2I–L). A bias towards head expression may be because the most cis-regulatory DNA was selected based on its proximity to genes expressed in the adult brain.
We found many lines expressed in non-neural tissues (Figure 2M–P), but did not comprehensively annotate these patterns. We note that 2,176 lines were expressed in the body wall, which could include trachea, muscles, sensory neurons, histoblasts, or other cell types (e.g. Figure 2M–O). We also observed expression of 613 lines in the viscera, termed “internal/gut” in the database (Figure 2P). 756 lines showed “scattered” expression (data not shown; retrieve from database by checking “scattered” box); this could reflect stochastic expression of the line within a reproducible pattern of cells, expression in hemocytes that have variable positions due to their migratory nature, or unknown cell types that have a variable pattern.
Stage 16 CNS patterns
At stage 16 the ventral nerve cord is composed of ~335 bilateral cells, including ~35 motoneurons and ~35 glia (Beckervordersandforth et al., 2008; Landgraf et al., 1999)(ESH and CQD, unpublished); most of the remaining 260 cells are presumed to be interneurons, but in fact are mostly uncharacterized. A major goal of this study was to identify GAL4 lines that are expressed in subsets of these presumed interneurons to allow further morphological and functional characterization. We detected 4,672 lines expressed at stage 16 (Figure 1D). At stage 16, the CNS shows a segmentally reiterated group of medial Eve+ neurons consisting of the RP2 motoneuron, the aCC/pCC motoneuron/interneuron siblings, and the U1–U5 motoneurons (Skeath and Doe, 1998). In addition, there is a cluster of Eve-lateral (EL) interneurons (Figure 3A). We identified 1,862 lines (37%) that are expressed in small subsets of neurons per hemisegment (<30 neurons per hemisegment, or about 10% of total cells; examples shown in Figure 3B–E). The more specific of these will be especially useful for characterizing neuronal morphology and function. In contrast, we identified just 822 lines (16%) expressed in most or all neurons in the CNS; some are specific to the CNS and others have additional expression outside the CNS (examples shown in Figure 3F,G). Interestingly, previous “enhancer trap” experiments showed a similar or lower percentage of lines with “all CNS” expression – 7/49 (14%) (O'Kane and Gehring, 1987) and 42/3768 (1%) (Bier et al., 1989), despite each insertion presumably querying a broader cis-regulatory region. It is somewhat surprising how few lines are expressed in a “pan-neuronal” pattern; this suggests that there may be relatively few cis-regulatory modules (CRMs) devoted to pan-neuronal expression, and that genes expressed widely may utilize multiple, dispersed CRMs to achieve pan-neuronal expression.
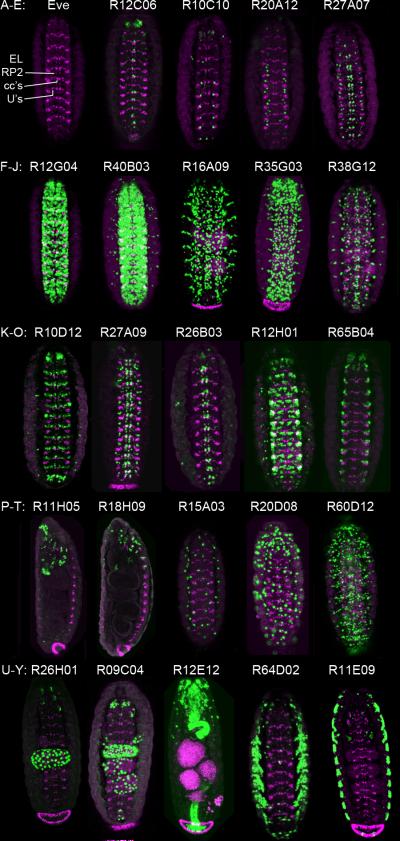
Figure 3. Representative GAL4 expression patterns in the stage 16 CNS.
(A) The Even-skipped (Eve) pattern at stage 16. RP2 = RP2 motoneuron; cc's = aCC/pCC and U1/U2 neurons; U's = U3–U5 neurons; EL = Eve lateral cluster.
(B–E) Lines expressed in a small subset of neurons. (A) Line R23C02, gene Adar; (B) Line R12C06, gene Oli; (C) Line R10C01, gene bi; (D) Line R20A12, gene CG16805; (E) Line R27A07, gene Fur1.
(F,G) Lines expressed in large subsets of neurons. (F) Line R12G04, gene Oli; (G) Line R40B03, gene CG2672.
(H–J) Lines expressed in glia. (H) Line R16A09, gene ey; (I) Line R35G03, gene Lag1; (J) Line R38G12, gene Nrt.
(K–O) Lines expressed in Eve-positive neurons: K–M, in medial Eve neurons (aCC, pCC, RP2 or U1–U5); N–O, in lateral Eve neurons. (K) Line R10D12, gene Fas2; (L) Line R27A09, gene grn; (M) Line R26B03, gene grn; (N) Line R12H01, gene kn; (O) Line R65B04, gene Tk.
(P–Q) Lines expressed in the brain. (P) Line R11H05, gene norpA; (Q) Line R18H09, gene wb.
(R–T) Lines expressed in a scattered pattern. (R) Line R15A03, gene Syt1; (S) Line R20D08, gene cib; (T) Line R60D12, gene arm.
(U–Y) Lines expressed in the gut (U–W) or lateral body wall (X–Y). (U) Line R26H01, gene lab; (V) Line R9C04, gene dac; (W) Line R12E12, gene h; (X) Line R64D02, gene 14-3-3zeta; (Y) Line R11E09, gene bi.
All panels show ventral views of the stage 16 CNS, except S–U are lateral views. Eve (magenta), nuclear GFP (green), anterior, top.
In the future, the GAL4 lines expressed in <10 neurons per hemisegment can be used for many experiments. We are currently characterizing about 100 of these lines in more detail, with the goal of mapping each of the GAL4+ neurons into a CNS atlas of gene expression which would allow us to register each GAL4+ neuron to known neurons or each other (Heckscher et al., in preparation). The lines can be used to express axon or dendrite markers to determine their morphology, possibly using GRASP (Feinberg et al., 2008) to identify candidate synaptic partners. The lines can be used to express Ca++ sensors to identify neurons with rhythmic activity matching the periodicity of larval body wall contractions during locomotion; they can be used to express neuronal silencers to screen for behavioral defects; they can be used to express Channelrhodopsin to allow the neurons to be light activated. The “pan-neuronal” lines will also be quite useful, particularly those without additional expression. Surprisingly few GAL4 lines are known to be pan-neuronal without additional expression; the most commonly used pan-neuronal GAL4 line is Elav-GAL4, but this line is expressed in neuroblasts that give rise to glia (Berger et al., 2007), and thus is not appropriate for driving neuron-specific expression. We have also observed lines expressed in subsets or all embryonic glia, such as enhancers near the glial cells missing gene (data not shown); some examples of glial patterns are shown in Figure 3H–J.
Previously our lab and others have characterized the genetic regulatory network that specifies the Eve+ RP2 motoneuron (McDonald and Doe, 1997; McDonald et al., 2003); similar work has been done on the Eve+ U1–U5 neurons (Grosskortenhaus et al., 2006; Isshiki et al., 2001; Kohwi et al., 2011; Tran and Doe, 2008; Tran et al., 2010), and EL neurons (Tsuji et al., 2008). Identification of GAL4 lines expressed by Eve+ neurons would be useful in providing tools to manipulate gene expression in these neurons, and the flanking genes may be important for Eve+ neuron development or function. Thus, we searched for lines co-expressed by medial or lateral Eve+ neurons. We found 661 lines expressed in the medial Eve+ neurons, and 785 lines expressed in the Eve+ lateral (EL) neurons (Table 2; some examples of each shown in Figure 3K–O). In some cases many other cells express the line, which is not very useful, but in other cases the lines are fairly specifically expressed in the Eve+ neurons (Figure 3M,O). These lines should help define the genetic regulatory network used to generate a specific neuron from an identified precursor, as the Eve+ neurons are among the best characterized in the CNS.
We also detected 3,712 lines expressed in the head (Figures 1D, 3P,Q); although this number may be artificially high due to the “four cluster of neurons” general background pattern in nearly all lines (Figure S1). Many lines were also expressed in non-neural tissues such as the gut (e.g. Figures 1D, 3U–W; quantified in Table 2), body wall (Figures 1D, 3X,Y) which include somatic body wall muscles, trachea, sensory neurons, and undefined cell types, as well as in in scattered cells that may include hemocytes (e.g. Figure 3R–T).
CNS midline patterns
The ventral nerve cord contains a specialized set of neurons and glia that lie along the midline. By virtue of their appearance as a stripe down the midline of the embryo (Figure 4), it is relatively easy to identify expression of genes or transgenic lines that are midline-expressed. Consequently, the midline cells are an attractive system for studying gene regulation, and this provides one rationale for our annotation of the JFRC GAL4 line collection for embryonic midline expression patterns.
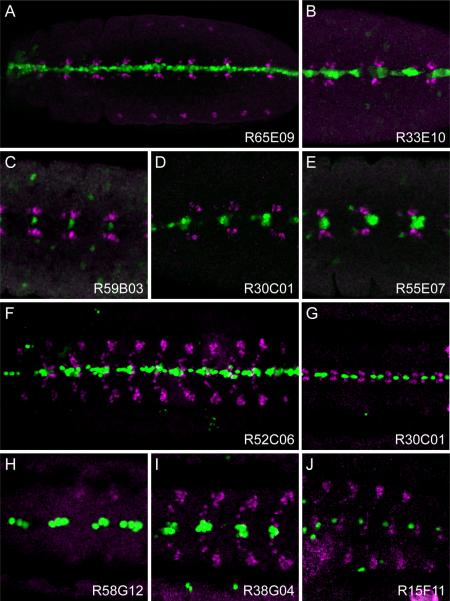
Figure 4. Representative GAL4 expression patterns in CNS midline cells.
(A–E) Images of GAL4 lines at GBE stages showing midline primordium staining of (A) all midline cells: Line R65E09, gene Tk; (B) a large subset: R33E10, gene ct; (C) a single cell: R59B03, gene CG8861; (D) a small subset of likely midline glia: Line R30C01, gene netB; and (E) a small subset of neural precursors or neurons: Line R55E07, gene CG13253.
(F–J) Images of stage 16 embryos showing (F) all midline cells: Line R52C06, gene cdi; (G) midline glia: Line R30C01, gene netB; (H) iVUMs: Line R58G12, gene Gad1; (I) MNB progeny: Line R38G04, gene sca; and (J) a single neuron: Line 15F11, gene Syt1.
All panels show ventral views of the CNS of the indicated GAL4 UAS-nGFP lines stained for Eve (magenta) and nuclear GFP (green). Ventral midline, arrowhead; anterior is to left.
There are two developmentally distinct groups of midline cells (Kearney et al., 2004). The cells generally considered “midline cells” are derived from single-minded+ mesectodermal cells, and consist of the midline glia that ensheath the axon commissures and a diverse set of neurons. The second group is referred to as midline accessory cells. They reside at the midline but are either mesodermal cells residing internal to the midline (dorsal-median cells) or are glia that arise from lateral neuro-glioblasts and migrate to the midline (channel glia and medial—most cell body glia, MM-CBG). The ~22 mesectodermal-derived mature midline cells/segment at stage 16 are diverse, consisting of: (1) ~3 midline glia, (2) 2 MP1 peptidergic neurosecretory cells, (3) H-cell, a dopaminergic interneuron, (4) H-cell sib, a glutamatergic interneuron, (5) 3 mVUM glutamatergic and octopaminergic motoneurons, (6) 3 GABAergic iVUM interneurons, and (7) the median neuroblast (MNB) and its ~8 interneuronal progeny (Wheeler et al., 2006). Large-scale in situ hybridization analysis documented midline expression of 286 genes (Kearney et al., 2004) – these data are publically available at the Drosophila CNS Midline Gene Expression Database (MidExDB) web-based database (Wheeler et al., 2009). Confocal microscopic analysis of ~100 genes has yielded detailed maps of midline cell expression at each embryonic stage, so that each midline cell type can be identified at each stage of development (Wheeler et al., 2006; Wheeler et al., 2008).
Analysis of horizontal views of GBE and stage 16 GAL4 transgenic embryos using anti-Eve staining was particularly useful for identifying lines with midline GFP expression, since medial Eve+ neurons, including aCC, pCC, and RP2 cells bracket midline-expressing cells (Figure 4A–J). At the GBE stage, midline cells are just acquiring their fates, and cannot easily be distinguished except by staining with cell-type specific markers. Consequently, we designate GBE midline-expressing enhancers as expressed in either (1) all midline cells or (2) a subset of midline cells. In contrast, at stage 16 it is often possible to make judgments as to the identity of the midline cell type, based on position along the apical-basal axis and morphology (Wheeler et al., 2006; Wheeler et al., 2008). In this manner, midline glia, VUM neurons, and progeny of the MNB can often be determined. In contrast, without appropriate markers, it is not possible to identity the other midline neurons. In annotating midline-expressed lines, we have chosen to liberally assign midline GFP+ cells to a specific midline cell type.
In our analysis, 5,000 lines were analyzed for midline expression corresponding to 796 genes (Table 3; Supplemental Table 2). We identified 1285 lines that had midline expression at either the GBE stage (389 lines) or at stage 16 (1261 lines). Overall, 26% of all lines examined had midline expression, corresponding to 59% of the genes analyzed. Of the 1,285 lines with midline expression we subdivided them into two classes: high specificity (HS) lines and low specificity (LS) lines. The low specificity lines (847; 66% of total) had midline expression, but usually also had expression in a large number of neurons in the lateral CNS. In contrast, the high-specificity lines (438 lines; 34% of total) generally showed strong midline expression, and were not generally expressed in large numbers of CNS cells. Nevertheless, many HS lines had expression in other embryonic and CNS cell types. The HS specificity lines corresponded to 253 genes, and 111 of these genes had multiple HS lines with an average of 2.7 lines/gene. Generally, when a gene had multiple lines with the same midline pattern, they were overlapping (and thus, likely contained the same CRM). In contrast, when a gene had multiple lines with distinct midline expression patterns, they were in non-overlapping DNA segments and must represent different CRMs. Only rarely were lines with identical patterns of expression in the same gene found in non-overlapping DNA segments; sometimes referred to as “shadow enhancers” (Barolo, 2012).
Of the HS genes, many are known to be expressed in midline cells. For example MidExDB currently contains expression data on 286 genes expressed in CNS midline cells. Of the 253 HS genes, 56 are listed in MidExDB. Another 20 HS genes are expressed in midline cells, based on published accounts. Consequently, while a significant fraction of the HS gene enhancers reside within known midline-expressed genes, new midline-expressed genes may be identified based on our discovery of new midline enhancers. However, there are examples of midline enhancers that reside within well-studied genes that do not have corresponding strong midline expression. For example, the similar (Drosophila Hypoxia Inducible Factor ortholog) bHLH-PAS gene, which is ubiquitously expressed (Nambu et al., 1996), has two intronic overlapping midline-expressed lines (R14D11, R14E09). Consequently, the role of these enhancers in vivo remains unknown. Many of the midline enhancers reside in 5'-flanking regions, 3'-flanking regions, and introns, often within relatively close distance to a midline-expressed gene. However, there are examples of midline enhancers that may act as relatively long distances. For example, the overlapping lines R25F10 and R27A10 reside ~15 kb upstream of the charlatan gene, which is broadly expressed in the CNS, but does not have prominent midline expression (Escudero et al., 2005; Yamasaki et al., 2011). However, the midline-expressed hibris gene is located 74 kb upstream of charlatan (Artero et al., 2001; Dworak et al., 2001), and the enhancer, which is ~58 kb downstream of hibris may control hibris midline expression.
The HS midline-expressed lines include a variety of midline patterns that are listed in Table 3, Supplemental Table 2, and Figure 4. At the GBE stage, 24 lines were expressed in all midline primordium cells (Figure 4A) and 113 in subsets (Figure 4B–E). The subsets included large subsets (Figure 4B), and subsets with as few as a single cell/segment (Figure 4C). In some cases, the subsets of midline primordium cells can be tentatively assigned to midline glia or neural precursors based on the identity of the stage 16 midline cell types. For example, R30C01 (netrin-B) is expressed only in midline glia at stage 16 (Figure 4G) and is found in a subset of midline cells (likely midline glia) in GBE embryos (Figure 4D).
At stage 16, the midline cells can in many cases be distinguished as to midline cell type. In 26 HS lines, expression was present in all midline cells (Figure 4F); these were generally the same lines with GBE all midline cell expression. Stage 16 expression in all midline cells may reflect perdurance from expression at the earlier midline primordium stages, since few genes, if any, are prominently expressed in all midline cells at stage 16, or this expression could be due to an incomplete CRM lacking a late repression element. Many HS lines were expressed in subsets of midline cells (Figure 4G–J); in some cases expression was present in a single midline cell type and in other cases expression was present in multiple midline cell types. We categorized HS lines expressed in specific cell types as: midline glia, VUMs, MNB progeny, and small subsets of neurons. The midline glia can generally be identified based on characteristic dorsal position, number, and elongated nuclear morphology. We annotated 83 HS lines with midline glia expression, excluding lines expressed in all midline cells. Of these 83 lines, there are 27 in which the only midline cell type is midline glia (Figure 4G). The VUM neurons can also be identified based on their characteristic ventral position and relatively large nuclei (Figure 4H). In the small number of cases in which we assign a VUM subset to either iVUMs or mVUMs, this is based on the existing knowledge that the corresponding gene is expressed in either iVUMs or mVUMs. For example, Gad1 is expressed in the GABAergic iVUMs (Wheeler et al., 2006), making it likely that line R58G12 whose fragment resides in an intron of Gad1 is expressed in iVUMs (Figure 4H). There are 89 HS lines expressed in the VUM neurons, many in VUM subsets, likely either iVUMs or mVUMs. The median neuroblast progeny tend to form a cluster of small neuronal nuclei (Figure 4I), and were the predominant midline cell type in 26 HS lines. The neural subset category consists of 223 HS lines that express GFP in one or a small number of neuronal nuclei, but could not be assigned to a specific midline neuron without staining with cell type-specific markers. In those rare cases in which a GFP+ neuron is assigned to a specific type (e.g. H-cell; R29A07), this is because the corresponding gene is known to be expressed in that cell type (pale) (Wheeler et al., 2006). The last class of annotated midline lines (includes HS and LS) corresponds to the midline accessory cells: the dorsal median cells (24 lines), channel glia (15 lines) and MM-CBG (14 lines). The large number of GAL4 lines corresponding to each midline cell type has potential to be useful for functional studies of midline cell types, as well analysis of enhancer function and the regulation of midline gene expression.
Spatial and temporal changes in gene expression during development
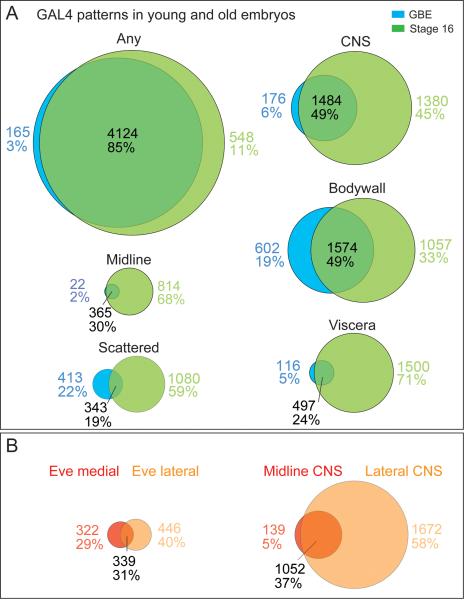
We identified over 4,000 GAL4 lines with expression in early embryos (germband elongated) or in late embryos (stage 16), allowing us to determine whether there are different CRMs for each tissue at each stage of development, or whether individual CRMs give persistent tissue specific expression over time. We found that the vast majority of GAL4 lines have some embryonic expression at both early and late embryos (85%), with only 3% early only and 12% late only (Figure 5A). Within the CNS there is more stage specific expression: 6% early-specific, 45% late-specific, and 49% at both stages (Figure 5A). The increased number of lines specifically expressed in the stage 16 CNS is likely due to the greater complexity of cell types present in the CNS at this stage. Similar results were observed for the midline, body wall, and viscera patterns (Figure 5A), reflecting the increased cellular complexity in these tissues. Most tissues had a relatively small number of early embryo specific patterns (6% CNS, 2% midline, 5% viscera; Figure 5A). This suggests that once a CRM is active in young embryos, often it may remain active as the embryo develops. Although we can't rule out the perdurance of GAL4 or the GFP reporter proteins as contributing to the observed persistent expression from young to old embryos, we note that some tissues do, in fact, show a much higher percentage of early embryo specific expression (19% body wall, 22% scattered; Figure 5A), which shows that GAL4 and GFP can be turned-over during early embryogenesis.
Figure 5. Spatial and temporal changes in gene expression during development.
(A) Venn diagrams showing the number of GAL4 lines expressed in different tissues (defined in Figure 1D legend) within early embryos (germband elongated, stages 9–12) and late embryos (stage 16). Only lines that had data for both GBE and stage 16 embryos were included.
(B) Venn diagrams showing the number of GAL4 lines expressed in Eve medial and Eve lateral neurons (left) or midline and lateral CNS (right) within early embryos (germband elongated, stages 9–12) and late embryos (stage 16).
We were also interested in whether the GAL4 lines showed a bias towards any cell type or subset of the CNS. We chose to focus on the Eve+ neurons and the midline neurons, which were the most comprehensively annotated cell types. The Eve+ neurons can be divided into a medial group that is primarily motoneurons (7 motoneurons and 1 interneuron) and a lateral group that is all interneurons. Interestingly, we found no significant bias in gene expression between these two groups: lines were nearly equally divided among medial-specific, lateral-specific, and both (Figure 5B). This shows that the CRMs in this study appear equally likely to be expressed in the medial vs. lateral regions of the CNS, and within motoneurons vs. interneurons – with the caveat that Eve is only representing a subset of motoneuron and interneurons in the CNS. Analysis of midline and lateral CNS expression gave a different result: 58% of the lines were expressed in the lateral CNS only, 37% in both the midline and lateral CNS, and only 5% in the midline only (Figure 5B). This suggests that the lateral CNS has more gene regulatory complexity than the midline, a result which is consistent with the greater number of unique cell types in the lateral CNS (Bossing et al., 1996; Schmid et al., 1999; Schmidt et al., 1997; Wheeler et al., 2009).
Conclusions and future directions
We have characterized the embryonic expression of 5,000 GAL4 lines, and provided the images and cis-regulatory DNA sequence information in a public website. All GAL4 lines were made with molecularly defined cis-regulatory DNA inserted into a single attP site in the genome, and thus the observed patterns can be replicated using other gene products. For example, we have already used the R9D11 cis-regulatory DNA to make a direct fusion to RFP, to be used as a marker in combination with other GAL4/UAS transgenes (O. Bayraktar and CQD, unpublished). Other applications would use cis-regulatory DNA to drive LexA or FLPase, which can then be used in combination with another GAL4/UAS transgene or to express split GAL4 components. The collection of GAL4 lines will allow investigators to target many different neuroblast lineages or neuronal subsets using misexpression or UAS-RNAi screens. As mentioned above, perhaps the most powerful use of these lines is in neural circuit analysis; lines with GAL4 expression in <10 neurons per hemisegment may be further refined by using smaller fragments of cis-regulatory DNA or by split GAL4 methodology, allowing single neurons to be targeted. Combining GAL4 lines with highly restricted expression patterns with permanent-labeling transgenes will be a powerful method for linking embryonic neurons to their adult morphology and function. Lastly, it should be possible to perform bioinformatic analysis on the cis-regulatory DNA from lines with overlapping expression patterns to identify motifs that may drive cell-specific gene expression, as has been done for larval imaginal disc CRMs (Jory et al., accompanying paper).
Experimental Procedures
Generation of Fragment Enhancer-GAL4 Lines
Details of the design and manufacture of the enhancer-GAL4 constructs, transformant fly stocks and all related protocol have been previously described (Pfeiffer et al., 2008).
Drosophila Genetics
Males from each GAL4 expressing line were crossed to virgin females of y,w; UAS-GFP∷lacZ.nls (Bloomington Indiana stock #6452) which produces nuclear localized GFP. In a few cases, the male GAL4 line was crossed to virgin females of y,w; UAS-mCD8∷GFP, w+ (Bloomington Indiana stock #5137) that produces membrane bound GFP.
Antibody staining and image analysis
We performed 24 hour embryo collections at room temp (22.5°C) on apple agar caps with yeast paste. Standard methods were used to fix the embryos (Odden et al., 2002) and they were stored in 100% ethanol at −20°C until antibody staining was performed. Embryos were incubated in primary antibody for 1.5 hours at room temp (22.5°C), rinsed in phosphate buffered saline (PBS) with 0.1% Triton-X100, 3% bovine serum albumin, and 10mM glycine (PTBG) for 60 min, incubated in secondary antibody for 1.5 hours at room temp (22.5°C), rinsed twice for 10 min in PTBG, followed by a single 15 minute rinse in 10% glycerol in PBS on a rocker. Embryos were dehydrated by passing through a 25%, 50%, 90% glycerol series and stored overnight in mounting media (90% glycerol with 2% N-propyl gallate). We used the following primary and secondary antibodies: chicken anti-GFP (1:1000; Aves, Tigard OR), mouse anti-Eve 2B10 concentrate with a working concentration of 4 μg/ml, pre-incubated on fixed wild type embryos prior to use (Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242), donkey anti-chicken DyLight 488 and donkey anti-mouse DyLight 549 (1:400; Jackson Immunochemicals, West Grove PA). Reduction of rinsing time dramatically increased signal of Eve antibody. Embryos were imaged on a Bio-Rad Radiance 2100MP or a Zeiss 700 confocal microscope. We did not determine the gender of the imaged embryos. ImageJ was used to produce QuickTime movies and TIFF projections.
Data Collection
We collected z-stack images of the stage 16 ventral nerve cord in the T2–A3 segment region. For each line, our goal was to customize the region imaged to the extent of the staining. We also imaged germband elongated embryos (GBE; embryonic stages 9–12), typically imaging the entire embryo; we did not image GBE stages for 410 lines due to lack of appropriately staged embryos or poor staining quality. Most of the image stacks were collected at 20× + 1.5 zoom at 1.0 μm z-steps; the number of z-steps depended on the extent of the staining. Eve protein was a marker for the location and general dimensions of the CNS and allowed accurate orientation and staging of the embryos.
Supplementary Material
Highlights
5,000 Drosophila genomic fragments were analyzed for embryonic CNS expression
>1500 lines in small subsets of neurons
The lines provide markers and functional access to >95% of CNS neurons
An online searchable image database is available (www.janelia.org/gal4-gen1)
Acknowledgements
We thank R. Mann for comments on the manuscript; Tallon Lamoreaux for assistance with fly stocks; and Janet Hanawalt for administrative support. This work was supported by an NRSA postdoctoral award to JCP (NICHD), NIH grants R01 NS64264 (NINDS) and R37 RD25251 (NICHD) to STC, and NIH grant HD27056 and the HHMI (CQD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama-Oda Y, Hosoya T, Hotta Y. Asymmetric cell division of thoracic neuroblast 6–4 to bifurcate glial and neuronal lineage in Drosophila. Development. 1999;126:1967–1974. doi: 10.1242/dev.126.9.1967. [DOI] [PubMed] [Google Scholar]
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–4264. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Baier H, Scott EK. Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight. Curr Opin Neurobiol. 2009;19:553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays. 2012;34:135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mechanisms of development. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Berger C, Renner S, Luer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Developmental dynamics. 2007;236:3562–3568. doi: 10.1002/dvdy.21372. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Developmental biology. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mechanisms of development. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nature methods. 2012;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Dworak HA, Charles MA, Pellerano LB, Sink H. Characterization of Drosophila hibris, a gene related to human nephrin. Development. 2001;128:4265–4276. doi: 10.1242/dev.128.21.4265. [DOI] [PubMed] [Google Scholar]
- Escudero LM, Caminero E, Schulze KL, Bellen HJ, Modolell J. Charlatan, a Zn-finger transcription factor, establishes a novel level of regulation of the proneural achaete/scute genes of Drosophila. Development. 2005;132:1211–1222. doi: 10.1242/dev.01691. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. The EMBO journal. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes & development. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin G, Ngo T, Shepard D, Murphy C, Dionne H, Pfeiffer B, Hibbard K, Cavallaro A, Hall D, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. doi: 10.1016/j.celrep.2012.09.011. accompanying paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jory A, Estella C, Giorgianni M, Slattery M, Laverty T, Rubin G, Mann R. A survey of 6300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophiila melanogaster. doi: 10.1016/j.celrep.2012.09.010. accompanying paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138:1727–1735. doi: 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron. 1999;22:43–52. doi: 10.1016/s0896-6273(00)80677-7. [DOI] [PubMed] [Google Scholar]
- Layden MJ, Odden JP, Schmid A, Garces A, Thor S, Doe CQ. Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Developmental biology. 2006;291:253–263. doi: 10.1016/j.ydbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Long F, Peng H, Liu X, Kim SK, Myers E. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nature methods. 2009;6:667–672. doi: 10.1038/nmeth.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JA, Doe CQ. Establishing neuroblast-specific gene expression in the Drosophila CNS: huckebein is activated by Wingless and Hedgehog and repressed by Engrailed and Gooseberry. Development. 1997;124:1079–1087. doi: 10.1242/dev.124.5.1079. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Fujioka M, Odden JP, Jaynes JB, Doe CQ. Specification of motoneuron fate in Drosophila: integration of positive and negative transcription factor inputs by a minimal eve enhancer. Journal of neurobiology. 2003;57:193–203. doi: 10.1002/neu.10264. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Chen W, Hu S, Crews ST. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1 alpha and Drosophila single-minded. Gene. 1996;172:249–254. doi: 10.1016/0378-1119(96)00060-1. [DOI] [PubMed] [Google Scholar]
- O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odden JP, Holbrook S, Doe CQ. Drosophila HB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:9143–9149. doi: 10.1523/JNEUROSCI.22-21-09143.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nature biotechnology. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Long F, Liu X, Kim S, Myers E, Peng H. Simultaneous recognition and segmentation of cells: application in C.elegans. Bioinformatics. 2011;27:2895–2902. doi: 10.1093/bioinformatics/btr480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Developmental biology. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Scott EK. The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits. J Neurochem. 2009;110:441–456. doi: 10.1111/j.1471-4159.2009.06161.x. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Current opinion in neurobiology. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3–1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Developmental biology. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Hasegawa E, Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–3869. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and PhiC31 integrase. Methods Mol Biol. 2012;859:203–228. doi: 10.1007/978-1-61779-603-6_12. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. Multiple Notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development. 2008;135:3071–3079. doi: 10.1242/dev.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. MidExDB: a database of Drosophila CNS midline cell gene expression. BMC Dev Biol. 2009;9:56. doi: 10.1186/1471-213X-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki Y, Lim YM, Niwa N, Hayashi S, Tsuda L. Robust specification of sensory neurons by dual functions of charlatan, a Drosophila NRSF/REST-like repressor of extramacrochaetae and hairy. Genes Cells. 2011;16:896–909. doi: 10.1111/j.1365-2443.2011.01537.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.