Abstract
Vanillin is the substance responsible for the flavor and smell of vanilla, a widely used flavoring agent. Previous studies reported that vanillin is a good antimutagen and anticarcinogen. However, there are also some contradicting findings showing that vanillin was a comutagen and cocarcinogen. This study investigated whether vanillin is an anticarcinogen or a cocarcinogen in rats induced with azoxymethane (AOM). Rats induced with AOM will develop aberrant crypt foci (ACF). AOM-challenged rats were treated with vanillin orally and intraperitoneally at low and high concentrations and ACF density, multiplicity, and distribution were observed. The gene expression of 14 colorectal cancer-related genes was also studied. Results showed that vanillin consumed orally had no effect on ACF. However, high concentrations (300 mg/kg body weight) of vanillin administered through intraperitoneal injection could increase ACF density and ACF multiplicity. ACF were mainly found in the distal colon rather than in the mid-section and proximal colon. The expression of colorectal cancer biomarkers, protooncogenes, recombinational repair, mismatch repair, and cell cycle arrest, and tumor suppressor gene expression were also affected by vanillin. Vanillin was not cocarcinogenic when consumed orally. However, it was cocarcinogenic when being administered intraperitoneally at high concentration. Hence, the use of vanillin in food should be safe but might have cocarcinogenic potential when it is used in high concentration for therapeutic purposes.
Key Words: apoptosis, cell cycle, DNA repair, protooncogene, tumor suppressor gene, vanillin
Introduction
Vanillin is a substance widely used as flavoring agent in confectionaries and drinks. Numerous therapeutic properties of vanillin have been discovered, including (1) red blood sickle cell anemia inhibition, (2) sedative effects, and (3) antimutagenic property.1
Ohta et al.2 first studied the antimutagenic property of vanillin and found that vanillin could enhance recombinational repair in Escherichia coli. Sasaki et al.3,4 also reported that vanillin was an effective antimutagen toward X-ray–, ethyl methane sulfonate–, and N-ethyl-N′-nitro-N-nitrosoguanidine–induced mutations. By 1999, Santos et al.5 further reported that vanillin's antimutagenic effect was applicable to mitomycin-C- and methyl methane sulfonate–induced mutations as well.
As cancer is closely related to mutation accumulation, the antimutagenic property of vanillin makes it a potential anticarcinogen. An in vitro study demonstrated vanillin's cytostatic and cytolytic properties on human colorectal cancer cell line HT-29.6 However, the anticarcinogenic effect of vanillin in colorectal cancer has not been carried out in vivo.
Although there is abundant evidence that vanillin has anticarcinogenic properties, there are also some opposing opinions and evidence. Studies done by Kuroda and Inoue,7 Ferguson,8 and Fahrig9 showed that vanillin could sometimes become cocarcinogenic depending on the type of mutagen challenged and the type of model organism used in the study.
Hence, the purpose of this study was to investigate whether vanillin is an anticarcinogen or a cocarcinogen in azoxymethane (AOM)–induced aberrant crypt foci (ACF)–bearing rats. Besides that, the effects of vanillin on DNA repair mechanism, apoptosis, cell cycle, and tumor suppressor were also studied.
Materials and Methods
Materials
Vanillin purchased from MP biomedical was dissolved in 5% ethanol to increase its solubility. RNA extraction kit Ribopure and Turbo DNA-free kit were purchased from Ambion. Primers were purchased from First base and the GeXP kit was purchased from Beckman Coulter.
Methylene blue preparation
Methylene blue was prepared at a concentration of 0.2% (w/v). The solution must be sonicated and filtered before it is used to avoid precipitation of methylene blue powder while being viewed under a microscope.
AOM preparation
AOM was purchased from Sigma Aldrich. Two subcutaneous injections of 15 mg/kg body weight in subsequent weeks were needed to induce ACF in a rat.
Animals and treatments
Male Sprague–Dawley rats with body weights between 40 and 50 g were obtained from the Animal Unit, Faculty of Veterinary Medicine, Putra University, Malaysia (UPM). The rats were housed individually in plastic cages in a facility with a constant day–night cycle. Food equivalent to 10% of their body weight was provided and water was available ad libitum. The use of animals and the experimental protocol were approved by the Animal Ethics Committee of the UPM.
After 1 week of adaptation, all the rats were divided into eight groups of eight rats. The first group did not receive any treatment. The second group received subcutaneous AOM injections (15 mg/kg body weight) for two consecutive weeks in order to induce ACF. The third and fourth groups received subcutaneous AOM injections and 5% ethanol orally and through intraperitoneal (IP) injection, respectively. The fifth and sixth groups received subcutaneous AOM injections and 150 mg/kg body weight vanillin dissolved in 5% ethanol orally and through IP injection, respectively. The seventh and eighth groups received subcutaneous AOM injections and 300 mg/kg body weight vanillin dissolved in 5% ethanol orally and through IP injection, respectively. Vanillin administration was carried out three times per week alternately and the doses of vanillin were chosen based on a previous study of vanillin toxicity.10 The treatments were continued for 14 weeks to allow the development of more advanced stage ACF.11 By week 15, rats were anesthetized with chloroform and the their final body weights were recorded. The weights of liver, kidney, lung, spleen, and heart were also measured and recorded. Colons were excised and rinsed with normal saline. After washing, the colons were sliced open, pinned on polystyrene board, and fixed with RCL-2 solution. The RCL-2 solution is a new fixing agent that could preserve the RNA in fixed sample with higher efficiency compared with the conventionally used formalin.12
It should be noted that IP injection of vanillin is not the common way for human to consume vanillin. IP injection of vanillin was used in this study because vanillin could be oxidized in the upper gastrointestinal tract into vanillin acid and subsequently loses its potential anticarcinogenic/cocarcinogenic effect. Therefore, in order to study its effects, vanillin had to be administered intraperitoneally to avoid oxidation.10
ACF density, multiplicity, and distribution
A total of six colons from each group were scored for their ACF density and distribution. In brief, colons excised from rats were rinsed with normal saline to wash away RCL-2. The length and width of the colons were measured and recorded. The colon was then placed in a Petri dish with mucosa facing upward. Normally, the side of the colon with mucosa is less shiny compared with the side without mucosa. Methylene blue was poured into the Petri dish until the colon was submerged. After 2 min, the stained colon was washed with normal saline again to wash away excessive methylene blue. The colon was then examined under light microscope with 150× magnification. The number of ACF was counted; the multiplicity and distribution were recorded.13 The ACF density of the colon was calculated with the following formula:
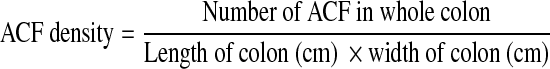 |
Tissue samples and RNA extraction
RNA from the colons previously used to evaluate ACF was extracted using Mirvana Paris kit from Ambion according to the instructions. First, the mucosa layer of the colon (including both proximal and distal colon) was scrapped with a blade. The mucosa layer appearance was different from the serosa layer as it was less shiny and usually easier to be scrapped off compared with the serosa layer. Cell disruption buffer was then added to the mucosa layer and the sample was homogenized. After cell disruption, RNA was released from the cells and thus 2× denaturing solution was added as soon as possible to avoid degradation of RNA by RNase. Next, acid-phenol-chloroform solution was added to the RNA lysate. After being vortexed, the solution was centrifuged at 10,000 g for 5 min to separate the solution into two phases. The upper aqueous phase containing the RNA was carefully removed and transferred into a new tube. Then, 100% ethanol was added to the aqueous phase and the solution was transferred to a filter cartridge. After centrifugation for 30 sec at 10,000 rpm, the flowthrough was discarded and the RNA was trapped at the filter cartridge. The RNA was then washed three times with washing buffer before being eluded with elution buffer.
The RNA solution obtained was then purified with Turbo DNA-free kit from Ambion to make sure it was free of DNA. The procedures were according to the manual. In brief, 10 μL of 10× turbo buffers and 1 μL of turbo DNase were added into the RNA solution and mixed well. After that, the solution was incubated at 37°C for 25 min to allow degradation of DNA. Next, 10 μL of inactivation reagent was added to halt the DNase activity. After 12,000 g centrifugation for 1.5 min, the supernatant (RNA) was transferred into a new tube.
The RNA concentration and quality were checked by using a nano-photometer (Implen).
GeXP analysis of rat's colon gene expression
The Genome Lab GeXP genetic analysis system was used to analyze the gene expression of the rats' colons. The GeXP system could analyze up to 30 genes per reaction. It uses combined gene-specific, universal priming strategy that converts multiplexed polymerase chain reaction (PCR) to a two-primer process using universal primers. The gene ratio in RNA samples was maintained during the PCR process.
First, the GeXP primers were designed with eXpress Multiplex Designer software from Beckman Coulter. Primers were purchased from First base. The sample preparations were according to the manual provided by Beckman Coulter. Briefly, reverse primers of different genes were mixed together to become the reverse primer mix. RNA (300 ng) was then mixed with 1 μL of reverse transcriptase, 4000 ng of each primer, 4 μL of 5× reverse transcription buffer, and 9 μL of water resulting in a total volume of 20 μL. Reverse transcription PCR was run using the conditions as specified in the manual. After reverse transcription, the second PCR process was carried out to amplify the cDNA. About 4 μL of magnesium chloride was mixed with 4 μL of 5× PCR buffer, 0.7 μL of Taq polymerase, 2 μL of forward primer mixture (containing 200 ng/μL of each forward primer), and 9.3 μL of cDNA. The PCR process was run with conditions as specified in the manual. Lastly, 2 μL of the PCR product was mixed with 38.5 μL of sample loading solution and 0.5 μL of size standard and was ready to be run by the GeXP machine. The expression of each gene was compared with housekeeping gene cyclophilin A and relative difference was calculated in fold.
Statistical analysis
Data were expressed as mean±standard deviation (in table) and mean±standard error (in figure). Duncan's test was used to determine the significance of differences between groups. A P-value<.05 was considered to be significant.
Results
Final body weights and organ weights of rats
There were no significant differences in rats' food consumption, general well-being, final body weights, and organ weights (liver, kidney, lung, spleen, and heart) among the groups (Table 1).
Table 1.
Final Body Weight and Organ Weight of Rats
| Groups | Final body weight (gram) | Liver weight (gram) | Kidney weight (gram) | Lung weight (gram) | Spleen weight (gram) | Heart weight (gram) |
|---|---|---|---|---|---|---|
| No treatment | 370.78±58.12 | 12.83±2.40 | 2.36±0.51 | 1.89±0.22 | 0.64±0.09 | 1.15±0.11 |
| AOM | 311.00±58.12 | 9.13±1.57 | 1.85±0.24 | 1.83±0.46 | 0.55±0.09 | 1.07±0.17 |
| AOM+oral 5% ethanol | 359.11±42.89 | 11.93±1.53 | 2.52±0.41 | 2.03±0.26 | 0.62±0.19 | 1.11±0.14 |
| AOM+IP 5% ethanol | 274.89±60.28 | 9.88±2.32 | 2.13±0.31 | 1.76±0.40 | 0.64±0.17 | 1.04±0.16 |
| AOM+oral 5% ethanol+150 mg/kg vanillin | 337.50±23.90 | 11.06±0.68 | 2.58±0.30 | 2.05±0.36 | 0.56±0.28 | 1.16±0.19 |
| AOM+IP 5% ethanol+150 mg/kg vanillin | 326.50±64.37 | 11.07±2.35 | 2.19±0.48 | 2.18±0.44 | 0.66±0.18 | 1.07±0.21 |
| AOM+oral 5% ethanol+300 mg/kg vanillin | 339.44±106.04 | 11.93±2.80 | 2.13±0.71 | 1.96±0.77 | 0.55±0.16 | 1.12±0.38 |
| AOM+IP 5% ethanol+300 mg/kg vanillin | 315.89±50.28 | 9.88±2.31 | 2.26±0.38 | 2.07±0.41 | 0.65±0.28 | 0.95±0.33 |
The data (expressed as mean±standard deviation) had no statistical significance compared with controls (AOM+oral 5% ethanol and AOM+IP 5% ethanol).
AOM, azoxymethane; IP, intraperitoneal.
ACF density, multiplicity, and distribution
ACF are colon cells that are mutated and could be considered as preneoplastic lesions of colorectal cancer. The higher density and multiplicity of ACF always indicate a higher chance of developing colorectal cancer.14 As anticipated, Table 2 shows that colon of rats without AOM did not contain ACF while all rats with AOM injection developed ACF. Besides, vanillin did not increase the ACF density or ACF multiplicity of AOM-injected rats when administered through oral gavages. However, when administered through IP injection at higher concentration (300 mg/kg body weight), vanillin could increase ACF density compared with the control group (AOM+IP 5% ethanol). AOM-induced rats that were treated with IP 5% ethanol and 300 mg/kg body weight of vanillin also exhibited significantly higher ACF multiplicity compared with the control group (AOM-induced+IP 5% ethanol). In addition, it was found that most ACF developed in the distal part of colon rather than mid- and proximal colon. Rats injected with 300 mg/kg body weight of vanillin were found to have significantly higher count of ACF in the distal compared with the control group (AOM-induced+IP 5% ethanol).
Table 2.
Aberrant Crypt Foci Density, Multiplicity, and Distribution
| Groups | ACF density | Lower multiplicity (ACF<3 crypt) | Higher multiplicity (ACF>3 crypt) | ACF count in proximal colon | ACF count in middle colon | ACF count in distal colon |
|---|---|---|---|---|---|---|
| No treatment | 0.006±0.013 | 0.20±0.45 | 0.00±0.00 | 0.00±0.00 | 0.00±0.00 | 0.20±0.45 |
| AOM | 1.752±0.911 | 33.83±17.93 | 12.67±7.31 | 8.17±4.62 | 4.67±3.44 | 33.67±17.59 |
| AOM+oral 5% ethanol | 2.866±0.957 | 55.80±21.59 | 77.00±25.43 | 18.00±11.60 | 6.00±1.87 | 52.00±20.02 |
| AOM+IP 5% ethanol | 1.431±0.895 | 33.25±20.19 | 7.00±5.90 | 10.25±6.40 | 3.70±3.50 | 26.25±16.56 |
| AOM+oral 5% ethanol+150 mg/kg vanillin | 2.362±0.542 | 48.86±14.17 | 19.29±6.47 | 17.00±11.24 | 5.57±2.88 | 45.57±16.27 |
| AOM+IP 5% ethanol+150 mg/kg vanillin | 2.556±0.560 | 53.14±12.16 | 20.14±6.82 | 13.29±6.24 | 5.00±2.45 | 55.00±15.34 |
| AOM+oral 5% ethanol+300 mg/kg vanillin | 2.019±0.772 | 47.00±18.57 | 11.00±5.50 | 13.43±6.92 | 5.43±1.81 | 39.14±17.80 |
| AOM+IP 5% ethanol+300 mg/kg vanillin | 2.626±0.682# | 39.00±11.20 | 25.00±5.10# | 11.25±3.86 | 4.50±3.32 | 48.25±10.21# |
The data (expressed as mean±standard deviation) showed statistical significance as indicated: #P<.05 compared with control (AOM+IP 5% ethanol). The data had no statistical significance compared with control (AOM+oral 5% ethanol).
ACF, aberrant crypt foci.
GeXP analysis of rat's colon gene expression
In the GeXP analysis of rat colon genes, we investigated a total of 14 genes responsible for different functions. Depending on the role of the genes, they were assigned into seven groups: (1) DNA repair, (2) colorectal cancer biomarkers, (3) protooncogene, (4) apoptosis, (5) cell cycle, and (6) tumor suppressor gene.
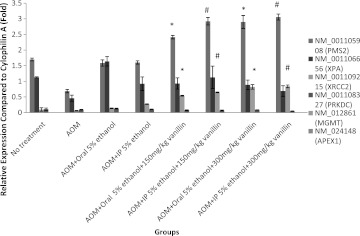
For some of the genes, the gene expression was high and the expression levels between groups could be easily compared. However, for the other genes, the expression level could not be detected because the expression was too low or not expressed at all. Figure 1 shows that vanillin could significantly increase the expression of PMS2 (mismatch repair gene) and XRCC2 (recombinational repair gene) but had no effect on the expression of XPA (nucleotide excision repair [NER] gene), protein kinase C (nonhomologous end joining [NHEJ] gene), MGMT (direct reversal gene), and Apex1 (Base excision repair [BER] gene). This shows that vanillin could only enhance MMR and recombinational repair genes.
FIG. 1.
Relative expression of DNA repair genes. Bar on column represents the standard error for each group (rat-to-rat variance). Certain genes' expression was too low to be detected and thus could not be shown. Statistical significance is indicated: *P<.05 compared with control (azoxymethane [AOM]+oral 5% ethanol); #P<.05 compared with control (AOM+intraperitoneal [IP] 5% ethanol).
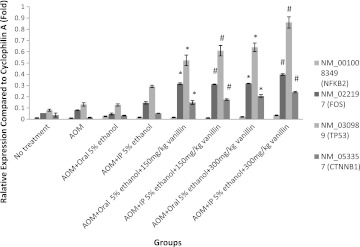
For colorectal cancer biomarker and protooncogene, Figure 2 shows that beta-catenin (CTNNB1) and FOS were both regulated significantly by vanillin administration compared with the control group (AOM+IP 5% ethanol). However, the expression level of apoptotic gene NFκB was low and no significant difference was observed between groups.
FIG. 2.
Relative expression of apoptosis, protooncogene, tumor suppressor gene, and colorectal cancer biomarker gene. Bar on column represents the standard error for each group (rat-to-rat variance). Statistical significance is indicated: *P<.05 compared with control (AOM+oral 5% ethanol); #P<.05 compared with control (AOM+IP 5% ethanol).
The tumor suppressor gene p53 (TP53) expression level was also upregulated by vanillin (Fig. 2). The expression level of the group with 300 mg/kg body weight of IP vanillin injection was three times higher than the expression level of control group (AOM-induced+IP injection 5% ethanol+no vanillin). Besides that, the expression level was also dose dependent.
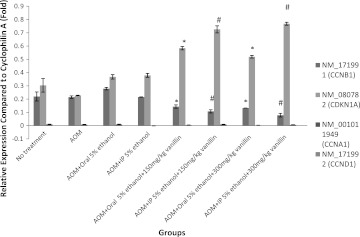
For cell-cycle-related genes, Figure 3 shows that cyclin A (CCNA1) and cyclin D (CCND1) expression was too low to be detected. However, p21 (CDKN1A) and cyclin B (CCNB1) expression was influenced by vanillin administration. p21 expression was significantly increased by vanillin while cyclin B expression was significantly downregulated.
FIG. 3.
Relative expression of cell cycle gene expression. Bar on column represents the standard error for each group (rat-to-rat variance). Certain genes' expression was too low to be detected and thus could not be shown. Statistical significance is indicated: *P<.05 compared with control (AOM+oral 5% ethanol); #P<.05 compared with control (AOM+IP 5% ethanol).
Discussion
Due to its reported antimutagenic effects, vanillin is expected to be a good anticarcinogen. However, some studies also showed that vanillin could be comutagenic and a cocarcinogen. Hence, in this study the effect of vanillin against AOM-induced mutations was investigated. Apart from that, the effects of vanillin on colon gene expression (DNA repair, apoptosis, cell cycle, and tumor suppressor genes) were also studied.
In this study, AOM was used to induce ACF and colorectal cancer. ACF count and ACF multiplicity could reveal a tendency for increasing colorectal cancer occurrence14–16 and also function as measurements of the efficiency of vanillin as anticarcinogen/cocarcinogen. The doses of vanillin used were based on previous study about the toxicity10 and therapeutic dose range of vanillin.17 According to Abraham et al.,17 vanillin could be used as sedative agent only when its concentration was in the range of at least 200–300 mg/kg body weight. Table 2 shows that oral consumption of vanillin did not change the ACF density and multiplicity. However, when AOM-injected rats were injected with 300 mg/kg body weight of vanillin, the ACF density and multiplicity were significantly higher than the control group (AOM-injected+IP 5% ethanol). This indicates that IP injection of vanillin at the higher dose might be cocarcinogenic to AOM-induced colon. Besides, it should be noted that for all groups, the ACF were primarily found in the distal rather than mid- and proximal sections of colon. This coincides with the findings of a previous study that stated that rats induced with 2 injections of AOM would develop ACF in the distal regions for the 1st week through the 14th week, before the ACF become more advanced and appear more frequently on the proximal section.11
The results of ACF count and ACF multiplicity were further confirmed by the elevated expression of colorectal cancer molecular biomarker (beta-catenin)18 and protooncogene c-Fos (Fig. 2). c-Fos is a subunit of AP-1 that is important for the survival of cancerous cells. The AP-1 could restart arrested cell cycle in cancerous cells even with mutations present and thus could lead to unrepaired DNA mutations.19 These findings were consistent with a previous report from Akagi et al.20 reporting that vanillin might increase the occurrence of colorectal cancer.
The effects of vanillin on DNA repair mechanisms were also studied. O6-methylguanine induced by AOM can lead to GC→AT transition.21 This mutation is commonly repaired by direct reversal using the enzyme O6-methylguanine-DNA methyltransferase (MGMT).22 However, it was shown in this study that (Fig. 1) AOM-injected rats' MGMT expression level was too low to be detected. The failure of vanillin to enhance the production of MGMT might cause vanillin to be unable to prevent AOM-induced ACF. This result was consistent with the increased ACF count and ACF multiplicity discussed previously.
Mismatch repair (MMR) genes would trigger apoptosis23,24 and cell cycle arrest (S and G2/M checkpoint)25,26 when encountering O6-methylguanine. Defects in MMR genes are known to associate with hereditary nonpolyposis colon cancer.27,28 Figure 1 shows that the MMR gene PMS2 was effectively increased by vanillin injection and thus we should expect a higher degree of apoptosis and cell cycle arrest. However, it should be remembered that even though PMS2 genes were enhanced, it might not be the rate-limiting protein in the control of MMR genes. Hence, even though PMS2 was enhanced the MMR genes could still be unaffected.
BER, NER, recombinational repair, and NHEJ genes have never been reported to be able to repair O6-methylguanine.29 As shown in Figure 1, neither NER genes nor NHEJ genes were enhanced by vanillin. However, vanillin could significantly increase the expression of recombinational repair genes. This result was in agreement with the findings of other researchers.2 As O6-methylguanine could not be repaired by recombinational repair, the enhanced recombinational repair provides no benefits in reducing mutations. Further, the excessive recombination might contribute to the loss of heterozygosity that leads to the cocarcinogenic property of vanillin.30
Apoptosis gene NFκB was not affected by vanillin introduction (Fig. 2), whereas p21, cyclin B, and p53 were affected by vanillin administration. As shown in Figure 3, p21 (a cyclin-dependent kinase inhibitor) expression was upregulated by vanillin. When p21 expression increases, cells would be arrested at the G2/M phase of the cell cycle.31 On the other hand, cyclin B was downregulated by vanillin. Cyclin B expression is crucial for a cell to progress through G2/M checkpoint.32 When cyclin B expression was low, cells would be arrested at G2/M phase. p53 is also responsible for governing the G1 and G2/M phase of the cell cycle when DNA damage is detected.33 Hence, gene expression results of p21, cyclin B, and p53 showed that vanillin could arrest cells at G2/M phase. Normally, this should indicate a reduction in cancer cases. However, on the contrary, the ACF density and multiplication of vanillin-treated rats increased rather than decreased. This might be due to the mutations on p53. p53 mutations were frequently found in ACF that would lead to p53 overexpression.34 p53 overexpression will not inhibit colon carcinogenesis but instead play a significant role in the development of ACF.35
In conclusion, it was summarized that (1) vanillin might enhance recombinational repair and MMR as it enhanced certain recombinational repair and MMR genes, (2) vanillin was cocarcinogenic for AOM-induced mutation, (3) vanillin did not induce apoptosis in ACF-bearing colon, and (4) vanillin might induce G2/M cell cycle arrest in ACF-bearing colon. However, the cocarcinogenic effect would only occur at high concentration through IP injection; therefore, normal oral consumption of vanillin should be safe. In this study, the cocarcinogenic effects of vanillin were only tested on AOM-induced ACF. Vanillin might be anticarcinogenic/cocarcinogenic on cancer induced by other carcinogen. Further study should be carried out to determine vanillin anticarcinogenic/cocarcinogenic properties using other carcinogens.
Acknowledgment
The authors wish to merit Research University Grant Scheme (vote 91087), University Putra Malaysia for providing financial support to carry out the research project.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bythrow JD. Vanilla as a medicinal plant. Semin Integr Med. 2005;3:129–121. [Google Scholar]
- 2.Ohta T. Watanabe M. Watanabe K. Shirasu Y. Inhibitory effects of flavorings on mutagenesis induced by chemicals in bacteria. Food Chem Toxicol. 1986;24:51–54. doi: 10.1016/0278-6915(86)90264-4. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki YF. Imanishi H. Ohta T. Shirasu Y. Effects of vanillin on sister-chromatid exchanges and chromosome aberrations induced by mitomycin C in cultured Chinese hamster ovary cells. Mutat Res. 1987;191:193–200. doi: 10.1016/0165-7992(87)90153-9. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki YF. Ohta T. Imanishi H. Watanabe M. Matsumoto K. Kato T. Shirasu Y. Suppressing effects of vanillin, cinnamaldehyde, and anisaldehyde on chromosome aberrations induced by x-rats in mice. Mutat Res. 1990;243:299–302. doi: 10.1016/0165-7992(90)90146-b. [DOI] [PubMed] [Google Scholar]
- 5.Santos JH. Graf U. Reguly ML. Andrade HHR. The synergistic effects of vanillin on recombination predominate over its antimutagenic action in relation to MMC-induced lesions in somatic cells of Drosophila melanogaster. Mutat Res. 1999;444:355–365. doi: 10.1016/s1383-5718(99)00101-1. [DOI] [PubMed] [Google Scholar]
- 6.Ho K. Yazan LS. Ismail N. Ismail M. Apoptosis and cell cycle arrest of human colorectal cancer cell line HT-29 induced by vanillin. Cancer Epidemiol. 2009;33:155–160. doi: 10.1016/j.canep.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Kuroda Y. Inoue T. Antimutagenesis by factors affecting DNA repair in bacteria. Mutat Res. 1988;202:387–391. doi: 10.1016/0027-5107(88)90200-x. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson LR. Antimutagens as cancer chemopreventive agents in the diet. Mutat Res. 1994;307:395–410. doi: 10.1016/0027-5107(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 9.Fahrig R. Anti-mutagenic agents are also co-recombinogenic and can be converted into co-mutagens. Mutat Res. 1996;350:59–67. doi: 10.1016/0027-5107(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 10.Ho K. Yazan LS. Ismail N. Ismail M. Toxicology study of vanillin on rats via oral and intra-peritoneal administration. Food Chem Toxic. 2011;49:25–30. doi: 10.1016/j.fct.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Bird RP. Good CK. The significance of aberrant crypt foci in understanding the pathogenesis of colon cancer. Toxic Lett. 2000;112–115:395–402. doi: 10.1016/s0378-4274(99)00261-1. [DOI] [PubMed] [Google Scholar]
- 12.Masir N. Ghoddoosi M. Mansor S. Abdul-Rahman F. Florence CS. Mohamed-Ismail NA. Tamby MR. Md-Latar NH. RCL-2, a potential formalin substitute for tissue fixation in routine pathological specimens. Histopathology. 2012;60:804–815. doi: 10.1111/j.1365-2559.2011.04127.x. [DOI] [PubMed] [Google Scholar]
- 13.Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: Preliminary findings. Cancer Lett. 1987;37:147–151. doi: 10.1016/0304-3835(87)90157-1. [DOI] [PubMed] [Google Scholar]
- 14.Bird RP. Role of aberrant crypt foci in understanding the pathogenesis of colon cancer. Cancer Lett. 1995;93:55–71. doi: 10.1016/0304-3835(95)03788-X. [DOI] [PubMed] [Google Scholar]
- 15.Hong MY. Chapkin RS. Wild CP. Morris JS. Wang N. Carroll RJ. Turner ND. Lupton JR. Relationship between DNA adduct levels, repair enzyme, and apoptosis as a function of DNA methylation by azoxymethane. Cell Growth Differ. 1999;10:749–758. [PubMed] [Google Scholar]
- 16.Xiao R. Carter JA. Linz AL. Ferguson M. Badger TM. Simmen FA. Dietary whey protein lowers serum C-peptide concentration and duodenal SREBP-1c mRNA abundance, and reduces occurrence of duodenal tumours and colon aberrant crypt foci in azoxymethane-treated male rats. J Nutrbiochem. 2006;17:626–634. doi: 10.1016/j.jnutbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Abraham DJ. Harris LS. Meade BJ. Munson AE. Swerdlow PS. Patrick GA. Method of calming or sedating an animal with a hydroxyl benzaldehyde compound. Sep 16, 1997. U.S. Patent 5668182.
- 18.Markowits SD. Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straaten FV. Muller R. Curran T. Beveren CV. Verma IM. Complete nucleotide sequence of a humanc-onc gene: Deduced amino acid sequence of the human c-fos protein. Proc Natl Acad Sci USA. 1983;80:3183–3187. doi: 10.1073/pnas.80.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akagi K. Hirose M. Hoshiya T. Mizoguchi Y. Ito N. Shirai T. Modulating effects of ellagic acid, vanillin and quercetin in a rat medium term multi-organ carcinogenesis model. Cancer Lett. 1995;94:113–121. doi: 10.1016/0304-3835(95)03833-i. [DOI] [PubMed] [Google Scholar]
- 21.Cohen FO. Liveneh Z. In vitro UV mutagenesis associated with nucleotide excision-repair gaps in Escherichia coli. J Biol Chem. 1994;269:4953–4958. [PubMed] [Google Scholar]
- 22.Olsson M. Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Boil Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- 23.Hickman MJ. Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takagi Y. Takahashi M. Sanada M. Ito R. Yamaizumi M. Sekiguchi M. Roles of MGMT and MLH1 proteins in alkylation-induced apoptosis and mutagenesis. DNA Repair. 2003;2:1135–1146. doi: 10.1016/s1568-7864(03)00134-4. [DOI] [PubMed] [Google Scholar]
- 25.Hawn MT. Umar A. Carethers JM. Marra G. Kunkel TA. Boland CR. Koi M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 26.Stojic L. Mojas N. Cejka P. Pietro MD. Ferrari S. Marra G. Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel TA. Slippery DNA and diseases. Nature. 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 28.Leach FS. Nicolaides NC. Papadopoulos N. Liu B. Jen J. Parsons R. Peltomaki P. Sistonen P. Aaltonen LA. Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 29.Bjelland S. Bjoras M. Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylases I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnaudeau C. Miranda ET. Jenssen D. Helleday T. Inhibition of DNA synthesis is a potent mechanism by which cytostatic drugs induce homologous recombination in mammalian cells. Mut Res. 2000;461:221–228. doi: 10.1016/s0921-8777(00)00052-5. [DOI] [PubMed] [Google Scholar]
- 31.Vansant G. Pezzoli P. Saiz R. Birch A. Duffy C. Ferre F. Monforte J. Gene expression analysis of troglitazone reveals its impact on multiple pathways in cell culture: A case for in vitro platforms combined with gene expression analysis for early (idiosyncratic) toxicity screening. Int J Toxicol. 2006;25:85–94. doi: 10.1080/10915810600605690. [DOI] [PubMed] [Google Scholar]
- 32.Ito M. Factors controlling cyclin B expression. Plant Mol Boil. 2000;43:677–690. doi: 10.1023/a:1006336005587. [DOI] [PubMed] [Google Scholar]
- 33.He G. Siddik ZH. Huang Z. Wang R. Koomen J. Kobayashi R. Khokhar AR. Kuang J. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene. 2005;24:2929–2943. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- 34.Shivapurkar N. Huang L. Ruggeri B. Swalsky PA. Bakker A. Finkelstein S. Frost A. Silverberg S. K-ras and p53 mutations in aberrant crypt foci and colonic tumours from colon cancer patients. Cancer Lett. 1997;115:39–46. doi: 10.1016/s0304-3835(97)04709-5. [DOI] [PubMed] [Google Scholar]
- 35.Ghavam-Nasiri M. Rezaei E. Ghafarzadegan K. Seilanian-Toosi M. Malekifard H. Expression of p53 in colorectal carcinoma: Correlation with clinicopathologic features. Arch Iranian Med. 2007;10:38–42. [PubMed] [Google Scholar]