Abstract
p63, p73 and p53 are transcription factors members of the p53 gene family involved in development, differentiation and cell response to stress. p53 gene is mutated in 50% of human cancer. Moreover, when p53 gene is not mutated then its tumour suppressor pathway is lost through interaction with abnormally expressed cellular protein or viral protein .Therefore p53 pathway inactivation is a common denominator to cancer. However, it is still difficult to associate in the clinic p53 status to cancer prognosis and diagnosis. Recent publications may have a profound impact on our understanding of p53 tumour suppressor activity. p63, p73 and p53 genes have a dual gene structure conserved in drosophila, zebrafish and man. They encode for multiple p63, p73 or p53 proteins containing different protein domains (isoforms) due to multiple splicing, alternative promoter and alternative initiation of translation. The interplay between p53, p63 and p73 isoforms are likely to be fundamental to our understanding of tumour formation.
Keywords: Splice, promoter, tumour, transcription, apoptosis, mutation, cell cycle
Introduction
The response to cellular damage is complex involving recognition and repair of the lesions in DNA to minimize the risk of genetic instability. Therefore, mutations or alterations of protein expression involved in this process predispose to genome instability, cancer and other pathologies [1]. A central player in protecting the integrity of the genome is p53. The importance of its role is exemplified by the facts that p53 activity is ubiquitously lost in human cancer either by p53 protein inactivation or by p53 gene mutation [2].
p53 protein is expressed at low levels under unperturbed conditions. However, the p53 pathway is activated by any cellular stresses that alter the normal cell cycle progression or can induce mutations of the genome leading to the transformation of a cell into a cancerous cell. Activated p53 protein stops the cell cycle or in many case switches ‘on’ the programmed cell death pathways (apoptosis) forcing the damaged cells to commit suicide. The p53 protein prevents therefore the multiplication of stressed cells that are more likely than undamaged cells to contain mutations and exhibit abnormal cellular growth. Hence, p53 protein is the guardian of the genome preventing cancer formation [3].
The mechanisms, by which p53 accomplishes its tumour suppressor activity are still not completely understood. The best described mechanism is the ability of p53 to modulate gene expression. p53 is a transcription factor that binds directly and specifically as a tetramer to sequences of DNA [4-6]. The ability of p53 to modulate gene expression is required for its tumour suppressor activity. Identification of the cyclin-dependent kinase inhibitor Waf as a p53-responsive gene helps to explain how p53 can induce cell cycle arrest [7, 8]. Recently, several p53-inducible genes that encode for proteins with apoptotic potential have been identified. However, the tumor suppressor p53 can trigger cell death independently of its transcriptional activity through subcellular translocation and activation of proapoptotic Bcl-2 family members. The regulation of such activity of endogenous p53 in response to stress remains largely unknown.
Two p53-related genes, p63 and p73, were identified in 1997 [9] [10]. The high level of sequence similarity in the DNA binding domain between p53 protein family members allows p63 and p73 to transactivate p53-responsive genes causing cell cycle arrest and apoptosis. However, p53, p63 and p73 proteins are not functionally entirely redundant as each p53-family transgenetic knockout mice develop distinct phenotypes, illustrating that p53, p63 and p73 have their own unique functions.
We recently published that the p53 gene family has a dual gene structure conserved in drosophila, zebrafish and man [11] [12]. Like most of the genes in the human genome [13], p53 gene family members express multiple mRNA variants due to multiple splicing and alternative promoters. Hence p53 gene family members express different forms of p53 protein containing different domain of the protein (isoforms).
p63 isoforms
The human and mouse p63 genes express at least 3 alternatively spliced C-terminal isoforms (α, β, γ) and can be transcribed from an alternative promoter located in the intron 3 (figure1a). The transactivating isoforms (TAp63) are generated by the activity of the promoter upstream of exon-1 while the alternative promoter in intron-3 leads to the expression of amino-terminal truncated p63 isoforms (ΔNp63) containing a different N-terminal domain. Although ΔNp63 isoforms lack the transactivation domain present in TAp63 isoform, they can transactivate through a different transactivation domain present in their distinct N-terminal end [14]. Altogether, the p63 gene expresses at least 6 mRNA variants which encode for 6 different p63 protein isoforms (TAp63α, TAp63β, TAp63γ, ΔNp63α, ΔNp63β, and ΔNp63γ) Fig. (1).
Figure 1. human p63.
a) Schema of the human p63 gene structure: Alternative splicing (α, β, γ) and alternative promoters (P1 and P2) are indicated.
b) p63 protein isoforms: TAp63 proteins encoded from promoter P1 contain the conserved N-terminal domain (FxxψW) of transactivation (TA). ΔNp63 proteins encoded from promoter P2 are amino-truncated proteins containing an N-terminal transactivating domain different from TAp63 proteins. Numbers indicate the exons encoding p63 protein isoforms. Black boxes indicate conserved domains.
p73 isoforms
Like p63, the p73 gene can be transcribed from an alternative promoter located in the intron 3. The p73 gene expresses at least 7 alternatively spliced C-terminal isoforms (α, β, γ, δ, ε, ζ, η) and at least 4 alternatively spliced N-terminal isoforms, which contain different parts of the transactivation domain. Altogether, the p73 gene expresses at least 35 mRNA variants which can encode theoretically 29 different p73 protein isoforms Fig. (2). p73 isoforms encoded by alternatively spliced exon2 and/or exon-3 mRNA variants are initiated at different ATG and contain therefore different part of the N-terminal domain, suggesting that they can have distinct protein interactions and specific activities.
Figure 2. human p73.
a) Schema of the human p73 gene structure: Alternative splicing (α, β, γ, ζ, δ, ε, η) and alternative promoters (P1 and P2) are indicated.
b) p73 protein isoforms:
TAp73 proteins encoded from promoter P1 contain the conserved N-terminal domain (FxxψW) of transactivation (TA). Ex2p73 proteins are due to alternative splicing of exon-2. They have lost the conserved N-terminal domain (FxxψW) of transactivation (TA) but still contain part of the transactivation domain (Exon-3). Ex2/3p73 proteins are due to alternative splicing of exons 2 and 3. They have entirely lost the transactivation domain (TA) and are initiated from exon-4.
ΔN’p73 variant is often overexpressed at the mRNA level in tumours. ΔN’p73 is due to alternative splicing of exon-3′ contained in intron-3. Theoretically, ΔN’p73 mRNA would encode either for a short p73 protein or p73 protein isoforms identical to ΔNp73. ΔN’p73 mRNA contains the normal initiation site of translation in exon 2 (ATG in perfect kozak sequence) and a stop codon in exon-3′. Therefore it could encode for a short p73 protein composed only of the transactivation domain (FxxψW). It is possible that translation of ΔN’p73 mRNA is initiated from the third ATG available present in exon-3′and leading to p73 protein identical to ΔNp73 protein isoforms.
ΔNp73 proteins encoded from promoter P2 are amino-truncated proteins containing an N-terminal domain different from TAp73 proteins. Numbers indicate the exons encoding p73 protein isoforms. Black boxes indicate conserved domains.
p53 isoforms
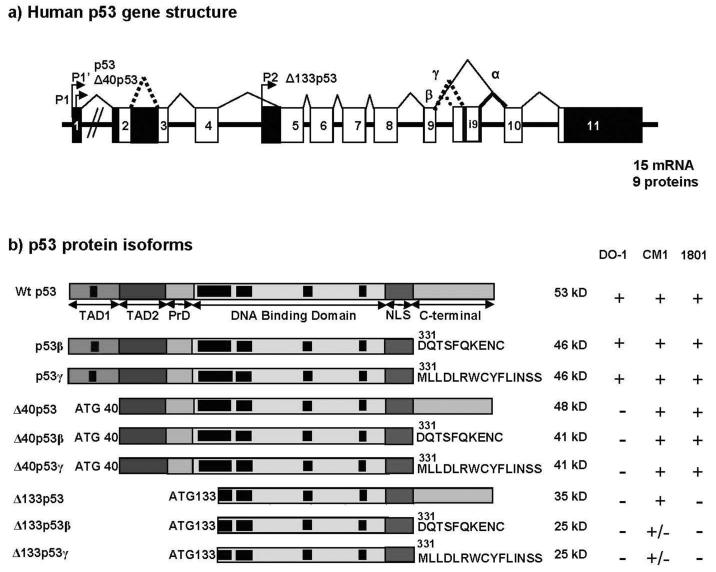
Until recently, our understanding of the p53 gene structure was simple. Only one promoter and three mRNA splice variants were described for p53, which would encode respectively full-length p53 (FLp53), p53i9 [15] [16] and Δ40p53 [17, 18]. However, this human p53 gene structure is not consistent with our current understanding of the evolution of the p53 gene family. Mammalian genomes contain three members of the p53 family while only one member has been identified in invertebrates, suggesting that the mammalian p53 family members are derived from the triplication of one ancestral gene. As p53 gene was discovered several years before PCR and modern molecular biology technologies, we revisited p53 gene expression in normal human tissue using the novel method of Generacer PCR [12]. We established then that human p53 gene has indeed a dual gene structure similar to p73 and p63 genes Fig. (3). We identified that p53 gene transcription can be initiated in normal human tissue from 2 distinct sites upstream of exon1 and from an internal promoter located in intron-4. The alternative promoter leads to the expression of an amino-terminally truncated p53 protein initiated at codon 133 (Δ133p53). The intron-9 can be alternatively spliced to produce 3 isoforms p53, p53β (identical to p53i9) and p53γ, where the p53β and p53γ isoforms lack the oligomerisation domain. Therefore, the human p53 gene can encode at least 9 different p53 protein isoforms, which we named accordingly to p63/p73 nomenclature p53, p53β, p53γ, Δ133p53, Δ133p53β and Δ133p53γ due to alternative splicing of the intron 9 and usage of the alternative promoter in intron 4, and also Δ40p53, Δ40p53β, Δ40p53γ due to alternative splicing of the intron 9 and alternative initiation of translation or alternative splicing of the intron2 [18] (figure 3). p53 variant mRNA are expressed in several normal human tissues in a tissue-dependent manner, suggesting that the internal promoter and the alternative splicing of p53 can be regulated. Interestingly, the dual gene structure of the p53 gene is conserved in drosophila [12], mouse (bourdon, unpublished data) and zebrafish [11] while the alternative splicing is species-specific. As the p53 family gene structure is conserved through evolution, it reveals an unforeseen complex regulation that may play a major role in controlling p53 activity.
Figure 3. human p53.
a) Schema of the human p53 gene structure: Alternative splicing (α, β, γ) and alternative promoters (P1, P1′ and P2) are indicated.
p53 protein isoforms: p53, p53β and p53γ proteins encoded from P1 or P1′ promoters contain the conserved N-terminal domain (FxxψW) of transactivation (TA). Δ133p53 isoforms encoded from promoter P2 are amino-truncated proteins deleted of the entire transactivation domain and deleted of part of the DNA binding domain. Translation is initiated at ATG-133. Δ40p53 protein isoforms encoded from P1 or P1′ promoters are amino-truncated proteins due to alternative splicing of exon-2 and/or alternative initiation of translation at ATG-40). Δ40p53 protein isoform have lost the conserved N-terminal domain of transactivation (FxxψW) but still contain part of the transactivation domain. Black boxes indicate conserved domains.
It has recently been reported that alternative splicing of the exon7 of p53 leads to a p53 isoform deleted of the conserved box V in the DNA binding domain (Δp53) [19]. This splicing is quite remarkable as it is not consistent with any known rules of splicing, which are strictly conserved through eukaryotes (from yeast to human). Despite all our efforts, we could not detect by PCR, the variant Δp53 in 21 normal human tissue analysed (Brain, Heart, Lung, liver, Colon, Bone Marrow, Thymus, Spleen, Testis, Prostate, Uterus, Skeletal muscle, Stomach, Kidney, Placenta, Fetal brain, Fetal liver, Salivary gland, Adrenal gland, Thyroid, Breast) nor in numerous tumours of several human tissue origin. Moreover no specific antibodies have been produced to detect specifically Δp53 isoform proteins. Therefore, we wonder whether the Δp53 isoform is really expressed.
p53 isoforms biochemical and biological activities
We have shown using commonly available p53 antibodies that endogenous p53 isoforms are expressed at the protein level. However, such antibodies cannot identify specifically the p53 isoforms. It is only by raising a specific anti-p53β antibody that we demonstrated expression of the endogenous p53β and Δ133p53β protein isoforms. This implies that p53γ, Δ133p53γ and Δ133p53 are expressed at the protein level. Specific antibodies to these p53 isoforms will be shortly available.
Importantly, like p63 or p73 isoforms, p53 isoforms can have distinct biochemical activities. The presence/absence of p53β at p53 responsive promoters is involved in the cellular response to p53 activation. p53β binds preferentially the p53-responsive promoters p21 and Bax rather than Mdm2, while p53 binds preferentially to Mdm2 and p21 rather than Bax promoters. p53β can form a protein complex with p53 and can specifically enhance p53 transcriptional activity at the Bax promoter, while it has no effect on the p21 promoter. Co-transfection of p53 with p53β increases slightly p53-mediated apoptosis, while co-transfection of p53 with Δ133p53 strongly inhibits p53-mediated apoptosis in a dose dependent manner. This indicates that wild type p53 activity may be modulated in the presence of p53 isoforms, and thus that regulation of p53 function in normal and tumor tissues in man is likely to be more complex than has been hitherto appreciated. Moreover, each p53 protein isoform may have specific biological activities independent of full-length p53. This may explain how p53 can be involved in the regulation of so many biological functions (i.e cell-cycle arrest, apoptosis, differentiation, replication, DNA repair, meiosis, mitosis, etc... ).
p53 isoforms in cancer
Deregulation of p53 isoforms expression may play a role early in tumor formation since attenuation of the WT p53 response would render the cells more susceptible to further genetic damage and therefore to neoplastic transformation and tumor progression. Tumors with abnormal p53 isoform expression would have a predicted phenotype of WT p53 by sequence but compromised p53 activity. Such hypothesis is consistent with our RT-PCR analysis of p53 isoforms expression on 30 breast cancer. p53β expression is frequently lost and Δ133p53 is frequently overexpressed on breast tumours, while only 25% of breast tumours express mutant p53. Similarly in Acute Myeloid Leukemia (AML) where p53 is mutated only in 10%, we observed abnormal expression of p53 isoform. This strongly suggests that the differential expression of p53 isoforms could disrupt the p53 response and contribute to tumour formation [20]. Moreover, it may provide some explanation to the difficulties in many clinical studies to link p53 status to the biological properties and drug sensitivity of human cancers as p53 status is determined either by sequencing and/or immunohistochemistry.
p53 mutation analysis has to be re-evaluated in cancer and Li-Fraumeni syndrome in light of p53 isoform expression. p53 isoforms are encoded by exons different from full-length p53, mutations occuring upstream of codon 133 (exon-5) or downstream of codon 331 (exon-9), would affect some p53 isoforms but not others. This may lead to the loss of some p53 biological activities, keeping others unaffected.
p53 immunostaining on tumour sections should also be carefully interpreted as commonly available p53 antibodies can detect some p53 isoforms but do not identify them specifically. The mouse monoclonal antibody DO-1 but also Bp53-12, DO-7 recognise p53, p53β and p53γ but not the other p53 isoforms, while polyclonal p53 antibodies raised against recombinant full-length p53 protein will recognize all p53 isoforms (although Δ133p53β and Δ133p53γ are weakly recognized by polyclonal antibodies as these isoforms have lost most immunogenic domains of p53). Only future specific p53 isoforms antibodies will allow a clear identification of the p53 isoforms. Meanwhile, RT-PCR method is the only alternative available to determine p53 isoform expression in tumours. Unfortunately, RT-PCR method can not be performed on paraffin-embedded tumour section as paraffin treatment destroys low abundant mRNA. For best results, mRNAs have to be extracted immediately after surgical resection or on tumour samples nitrogen-frozen immediately after surgical resection as mRNAs are rapidly degraded in absence of blood supply (half-life of 15 min).
Another complexity in p53 pathways is that p53/p63/p73 family members can interact with each other in many ways involving direct or indirect protein interactions, regulation of same target gene promoter and regulation of each other’s promoters. The p53 family members and their isoforms can bind differentially to promoters. The ratio between isoforms can thus be an important cell fate determinant. Further work on the interplay between the p53/p63/p73 proteins, will be essential in understanding their individual and collective roles. The changes upon stimuli of the balance and interactions between the isoforms are likely to be fundamental to our understanding in the transition between normal cell cycling and the onset of tumour formation.
Conclusion
Based on our current understanding of the p53 pathway, an integrated analysis of p53/p63/p73 isoform expressions associated with p53 sequence analysis and p53 target gene expression (p21, Bax, Scotin, ....) seems to be the best method to establish a link between p53 family gene and cancer treatment.
References
- [1].Kops GJ, Weaver BA, Cleveland DW. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- [2].Oren M. Cell Death Differ. 2003;10:431–42. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- [3].Lane DP. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- [4].El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45–9. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- [5].Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. Mol Cell Biol. 1992;12:2866–71. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. Oncogene. 1997;14:85–94. doi: 10.1038/sj.onc.1200804. [DOI] [PubMed] [Google Scholar]
- [7].El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- [8].Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- [9].Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- [10].Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Cell. 1997;90:809–19. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- [11].Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Genes Dev. 2005;19:2900–11. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. Genes Dev. 2005;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- [14].Helton ES, Zhu J, Chen X. J Biol Chem. 2006;281:2533–42. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- [15].Flaman J-M, Waridel F, Estreicher A, Vannier A, Limacher J-M, Gilbert D, Iggo R, Frebourg T. Oncogene. 1996;12:813–8. [PubMed] [Google Scholar]
- [16].Chow VT, Quek HH, Tock EP. Cancer Lett. 1993;73:141–8. doi: 10.1016/0304-3835(93)90256-9. [DOI] [PubMed] [Google Scholar]
- [17].Yin Y, Stephen CW, Luciani MG, Fahraeus R. Nat Cell Biol. 2002;4:462–7. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- [18].Ghosh A, Stewart D, Matlashewski G. Mol Cell Biol. 2004;24:7987–97. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- [20].Anensen N, Oyan AM, Bourdon JC, Kalland KH, Bruserud O, Gjertsen BT. Clin Cancer Res. 2006;12:3985–92. doi: 10.1158/1078-0432.CCR-05-1970. [DOI] [PubMed] [Google Scholar]