Abstract
Heme plays fundamental roles as cofactor and signaling molecule in multiple pathways devoted to oxygen sensing and utilization in aerobic organisms. For cellular respiration, heme serves as a prosthetic group in electron transfer proteins and redox enzymes. Here we report that in the yeast Saccharomyces cerevisiae a heme-sensing mechanism translationally controls the biogenesis of cytochrome c oxidase (COX), the terminal mitochondrial respiratory chain enzyme. We show that Mss51, a COX1 mRNA-specific translational activator and Cox1 chaperone, which coordinates Cox1 synthesis in mitoribosomes with its assembly in COX, is a heme-binding protein. Mss51 contains two heme regulatory motifs or Cys-Pro-X domains located in its N-terminus. Using a combination of in vitro and in vivo approaches, we have demonstrated that these motifs are important for heme binding and efficient performance of Mss51 functions. We conclude that heme sensing by Mss51 regulates COX biogenesis and aerobic energy production.
Keywords: Mitochondrial oxidative phosphorylation, Cytochrome c oxidase, Heme-mediated regulation, Heme sensing, Mitochondrial translation, Translational regulation, Mss51
INTRODUCTION
Heme (iron protoporphyrin IX) plays essential roles in pathways devoted to oxygen sensing, transport and utilization in aerobic organisms (Mense and Zhang, 2006), including the generation of cellular energy in the form of ATP by the mitochondrial electron transport chain (ETC) and the oxidative phosphorylation (OXPHOS) system. Several hemes with different chemical structure (hemes B, C and A) are embedded in the components forming the ETC (Kim et al., 2012). Cytochrome c oxidase (COX), the single cellular enzyme that contains heme A, is the terminal ETC oxidase. COX is the primary site of cellular oxygen consumption and, as such, is central to OXPHOS and aerobic energy generation. COX is a mitochondrial inner membrane complex formed by three catalytic core subunits (Cox1, Cox2 and Cox3) encoded in the mitochondrial DNA (mtDNA) and additional nuclear DNA-encoded subunits (at least 8 in the yeast Saccharomyces cerevisiae) that provide stability and protection to the core (Soto et al., 2012). The catalytic core contains two heme moieties (a and a3) located in Cox1 and two copper centers, CuA in Cox2 and CuB in Cox1. In Cox1, the heme iron of cytochrome a3 together with CuB constitutes the binding site for oxygen. Four electrons from ferricytochrome c are sequentially transferred to CuA, heme a and subsequently to the binuclear heme a3-CuB center to reduce one O2 molecule to two H2O in a process involving four one-electron reduction reactions and the pumping of protons into the mitochondrial intermembrane space to drive ATP synthesis (Soto et al., 2012).
COX biogenesis is extensively regulated to facilitate the stoichiometric accumulation of its constitutive subunits, the sequential incorporation of subunits to a Cox1 assembly seed and the formation of the metal centers, while minimizing the buildup of reactive assembly intermediates. In addition to the structural subunits, a growing list of more than 30 ancillary factors, usually conserved from yeast to human, assists all the steps of COX biogenesis (Soto et al., 2012).
Synthesis, membrane insertion and maturation of Cox1 determine the rate of COX assembly. The importance of coordination of these steps with COX complex formation is highlighted by the existence in S. cerevisiae of at least two regulatory mechanisms pacing Cox1 synthesis (Barrientos et al., 2004) and hemylation (Barros and Tzagoloff, 2002) to its assembly into COX. Cox1 synthesis is under the control of a negative feedback that is depending on the availability of its assembly partners. Briefly, Cox1 synthesis requires two translational activators, Pet309 and Mss51. Although both interact with the COX1 mRNA 5’-UTR to promote translation, Mss51 plays additional chaperoning roles by coordinating Cox1 synthesis and assembly (Barrientos et al., 2004; Perez-Martinez et al., 2003; Zambrano et al., 2007). During Cox1 synthesis on the mitoribosomes, Mss51 interacts with newly synthesized Cox1. The translational complex is stabilized by the COX assembly factors Cox14 (Barrientos et al., 2004) and Cox25/Coa3 (Fontanesi et al., 2011; Mick et al., 2010). It additionally contains the mitochondrial Hsp70 chaperone Ssc1 and its co-chaperone Mdj1 (Fontanesi et al., 2010), which could facilitate the proper folding of Cox1 while it is co-translationally inserted into the inner membrane. Subsequently, a 450 kDa Ssc1-Mss51-Cox1-Cox14-Cox25/Coa3 pre-assembly complex remains stable until Cox1 proceeds to downstream assembly steps. This complex, abundant in wild-type cells, represents a reservoir of stable Cox1 ready to be matured and/or to progress in the COX assembly process when required. We and others have postulated that Mss51 interactions within the translational and pre-assembly complexes down-regulate Cox1 synthesis when COX assembly is impaired by trapping Mss51 and limiting its availability for COX1 mRNA translation (Fontanesi et al., 2011; Fontanesi et al., 2010). The C-terminal residues of Cox1 are essential for Mss51 sequestration and to stabilize Ssc1-Mss51-Cox14-Cox25/Coa3 interaction (Shingu-Vazquez et al., 2010). According to the translational regulation model, the release of Mss51-Ssc1 from the pre-assembly complex to make Mss51 available for Cox1 synthesis occurs when Cox1 acquires its prosthetic groups or interacts with other COX subunits, a step possibly catalyzed by the COX assembly factors Shy1 and/or Coa1 (Barrientos et al., 2002; Fontanesi et al., 2008; Mick et al., 2007; Pierrel et al., 2007). When Mss51 is released from the pre-assembly complex it forms a stable 120 kDa heterodimeric complex with Ssc1. This complex constitutes a pool of Mss51 that is not involved in Cox1 chaperoning and may be the source of translationally competent Mss51 (Fontanesi et al., 2010). A second level of regulation of COX biogenesis implicates heme A, the prosthetic group contained in holoenzyme. The biosynthesis of heme A is also controlled by downstream events in the COX assembly process (Barros and Tzagoloff, 2002). The connections between heme availability, heme A biosynthesis and COX1 mRNA translation and assembly remain to be fully understood.
Heme does not only function as a prosthetic group in proteins and enzymes but also directly regulates the activity of signal transducers, transcriptional and translational regulators involved in oxygen sensing and utilization in bacteria, yeast and mammals (Mense and Zhang, 2006). In such proteins, heme exerts its regulatory function through binding to conserved Heme Regulatory Motifs (HRM) defined by a Cysteine-Proline-X (CPX) sequence. In our search for putative functional domains in Mss51, we detected the presence of two conserved CPX motifs located in its N-terminus. In the work described here, we have used in vivo and in vitro approaches to address the role of the CPX motifs in heme binding to Mss51 and characterize the requirement of heme binding for efficient performance of Mss51 functions. The identification of a COX1 mRNA-specific translational activator that senses heme provides a key element for a regulatory mechanism that coordinates assembly of COX with heme and oxygen availability for respiration and aerobic energy production.
RESULTS
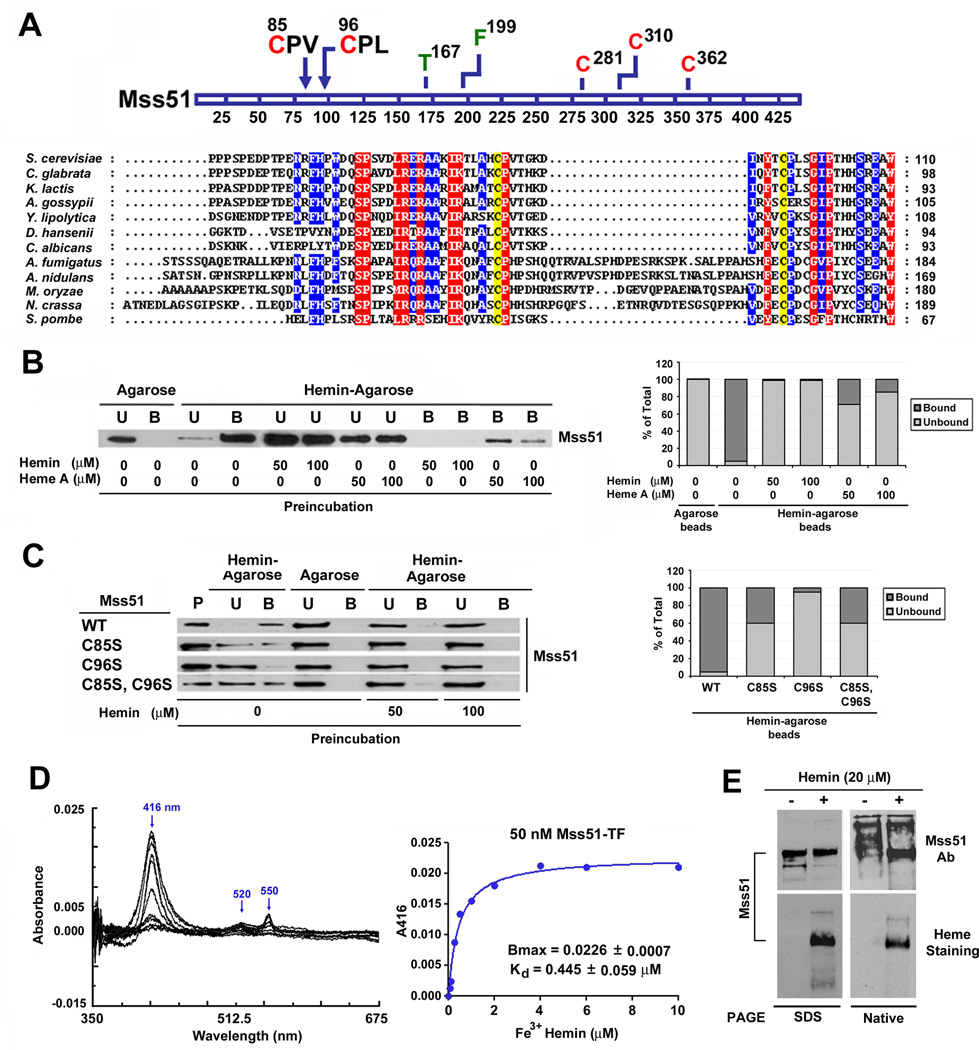
Recombinant Mss51 has the Ability to Bind Heme through CPX Heme Regulatory Motifs
Mss51 is a peripheral mitochondrial inner membrane protein that faces the matrix. Its N-terminal region contains a hydrophilic domain, which protrudes into the mitochondrial matrix (Zambrano et al., 2007). By sequence alignments we have identified in this region the presence of two Cys-Pro-X (CPX) motifs, which are conserved among fungi (Fig. 1A). Because these are potential heme regulatory motifs (Mense and Zhang, 2006), we tested whether Mss51 binds heme in vitro. For this purpose we purified recombinant Mss51 carrying a thrombin-cleavable Escherichia coli trigger factor N-terminal tag (Mss51-TF) to increase Mss51 solubility and facilitate its purification (Supplemental Fig. S1A). The purified protein was subsequently found to specifically bind hemin-agarose beads (Fig. 1B) whereas trigger factor alone did not (not shown). Competition assays performed by pre-incubating Mss51 with purified soluble heme species (heme A or hemin) prior to exposing the protein to the hemin-agarose beads showed hemin to be the preferred in vitro ligand (Fig. 1B).
Figure 1. Mss51 contains two conserved CPX motifs and binds heme in vitro.
(A) Partial N-terminal sequence alignment of Mss51 from 12 fungal species. Sequences were aligned using the CLUSTALW algorithm (Blosum62 scoring matrix) in Bioedit software. Amino-acids that are conserved in all sequences are shaded red and those conserved in at least 9 of 12 sequences are shaded blue. The conservation of two cysteines in the CPX motifs is marked in yellow. In the linear graph of the protein, all the cysteines (red) and other relevant residues for this study (green or black) are indicated.
(B and C) Immunoblot analyses of the binding to hemin-agarose beads of recombinant (B) wild-type Mss51 or (C) CPX mutant Mss51. Mss51 was pre-incubated or not with the indicated concentrations of either hemin or heme A prior to incubation with the beads. To quantify the signals, the images were digitalized and densitometric analyses performed using the histogram function of the Adobe Photoshop program. The graph represents the proportion of bound versus unbound protein for each sample. B, bound; U, unbound; P, protein alone.
(D) Difference spectroscopy titration of heme binding to Mss51. Difference absorption spectra and titration curves of wild-type Mss51 (50 nM) with increasing concentrations of Fe3+ hemin (from 0 to 10 µM) as indicated. The curves were generated from fits to an equation describing a single binding site using GraphPad Prism.
(E) Analyses of Mss51 and heme-bound-Mss51 by SDS- and Native-PAGE. The upper panel represents immunoblot analyses with an anti-Mss51 antibody. The lower panel represents in gel heme staining.
To assess whether heme binding by Mss51 is dependent on the integrity of the CPX motifs, we purified recombinant Mss51-TF variants in which cysteine residues from either or both motifs had been substituted by serines (C85S, C96S and C85SC96S) (Fig. S1A). The ability of these variants to bind hemin-agarose beads was severely attenuated when compared to wild-type Mss51 (>60% of the proteins remained unbound; Fig. 1C), indicating that heme binding to Mss51 in vitro is mediated by the CPX sequences.
Characterization of the Mss51-Heme Interaction in vitro
Heme binding to Mss51 was monitored by difference UV-visible spectroscopy as reported (Gupta and Ragsdale, 2011). Binding equilibrium was achieved within seconds because the Soret peak maxima absorbance did not increase with longer incubation times (not shown). When 50 nM recombinant Mss51 was titrated with hemin, the difference spectra at pH 8.0 revealed a Soret band at 416 nm and alpha and beta bands at 550 and 520 nm, respectively. (Fig. 1D). This profile is reminiscent of c-type cytochrome spectra and also hemoprotein spectra in which iron coordination involves at least one cysteine (Gupta and Ragsdale, 2011). Mss51 purified in reducing conditions binds Fe3+ (Fig. 1D) and Fe2+ hemin (not shown) with a similar affinity (Kd ~445 nM). Heme titration using the three recombinant cysteine variants of Mss51 (C85S, C96S and C85SC96S) resulted in several changes in the recorded spectra. First, we noted a slight shift of the Soret peak (from 414 or 412 nm) and a significant decrease in its amplitude. Moreover, a diffuse unresolved broad band in the 525–540 nm region replaced the 550 nm and 520 nm bands (Fig. S1B). Such spectral modifications suggest a heme ligand change in the mutant proteins. Accordingly, the C85S, C96S and C85SC96S variants have a 10-fold lower Bmax than wild-type Mss51 (Fig. S1B). The C85S variant binds Fe3+ hemin with affinity (Kd = 0.324 ± 0.074 µM) similar to the wild-type protein. However, for the C96S and C85SC96S variants binding was not strong enough to confidently calculate kinetic parameters. We repeated the experiment using increased amount of recombinant proteins (150 nM). The Kd values for the wild-type and C85S proteins were again similar although slightly higher that when using lower protein concentration (Fig. S1C and D). In this assay, the C96S protein bound hemin with a significantly lower Bmax but apparently higher affinity than the wild-type whereas hemin binding by C85SC96S never reached saturation suggesting nonspecific hemin binding (Fig. S1C and D). These results support the involvement of the Mss51 CPX motif cysteines in the regulation of heme binding, which is particularly influenced by C96.
To further test the properties of heme binding to Mss51, the hemin-reconstituted recombinant wild-type protein was separated in parallel by SDS- and Native-PAGE, and analyzed by immunoblotting and in-gel heme staining. In native conditions, only the heme-bound protein entered the gel and migrated properly as detected by immunoblotting (Fig. 1E). This observation suggests that unliganded Mss51 misfolds and/or oligomerizes and that heme-binding induces a conformational change in Mss51. In-gel assessment of heme-associated peroxidase activity (heme staining) efficiently detected the heme bound to Mss51 in both denaturing and non-denaturing conditions (Fig. 1E). Even boiling Mss51 for 30 min in 2% SDS and/or 6M urea failed to remove heme from the polypeptide (Fig. S1E). This result indicates that heme binding to Mss51 is resistant to denaturing conditions that typically destroy the non-covalent heme B-protein interactions therefore suggesting tight binding.
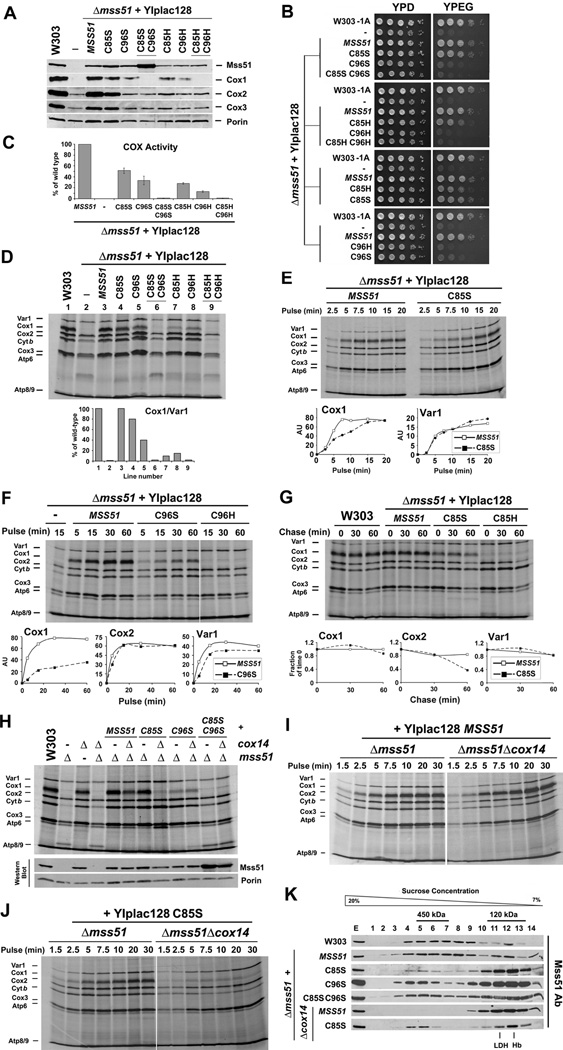
The Integrity of the CPX Motifs is Necessary for Full Mss51 Function in vivo
To assess the relevance of the CPX motifs for Mss51 function in vivo, we engineered Δmss51 yeast strains expressing cysteine to serine mutant forms of Mss51 from integrative or multi-copy plasmids. Additional strains were generated to express Mss51 variants in which the CPX cysteines were substituted by histidines (C85H, C96H and C85HC96H), a potential alternative residue for heme coordination.
The C85S and C96S mutations did not affect the stability of the mutant proteins in mitochondria and when expressed from single copy integrants, accumulated at wild-type steady-state levels. For the double mutant C85SC96S protein levels were unexpectedly two-fold higher (Fig. 2A). The three Cys to His variants accumulated at lower steady-state levels than the Cys to Ser variants (Fig. 2A). The overall stability of all these proteins in mitochondria allowed us to characterize the resulting phenotypes.
Figure 2. Mutations in Mss51 CPX motifs affect Cox1 synthesis and assembly.
(A) Immunoblot analyses of the steady-state levels of Mss51 and COX catalytic core subunits in a wild-type strain (W303) and a Δmss51 strain expressing the indicated wild-type or CPX mutant variants of Mss51.
(B) Growth test using serial dilutions of the indicated strains in complete media containing fermentable (glucose, YPD) or non-fermentable (ethanol-glycerol, YPEG) carbon sources. The plates were incubated at 30°C and the pictures taken after 2 days of growth.
(C) Cytochrome c oxidase (COX) activity measured spectrophotometrically in the indicated strains. Error bars represent the mean ± SD.
(D–J) In vivo mitochondrial protein synthesis in the indicated strains. Pulses were of 30 min (in D and H) or the indicated times. In (G), following a 15 min pulse, labeling was terminated by addition of 12 µg/ml puromycin + 80µM non-radioactive methionine and the products chased during the indicated times. The signals were quantified as in Fig. 1B and used to plot the graphs in the lower panels.
(K) Sucrose gradient sedimentation analyses of Mss51 in mitochondrial extracts prepared from the indicated strains. Fractions that contain Mss51-complex peaks are indicated. The gradients were calibrated with lactate dehydrogenase (LDH, 130 kDa) and hemoglobin (Hb, 67 kDa).
C85S-Mss51 largely complemented the respiratory growth defect of the Δmss51 strain, whereas C96S-Mss51 did not support respiration or did very poorly (Fig 2B). The entire set of mutations affected COX activity in a manner consistent with the respiratory growth phenotypes of the mutants (Fig. 2C). As previously reported, ~50% of COX activity is enough to support wild-type growth in complete respiratory media (Horn et al., 2008). In all mutant strains, the steady-state levels of the COX catalytic core subunits were decreased (Fig. 2A), consistently with their residual COX activity. The HPX variants were less effective than the Ser variants in complementing the respiratory growth and COX defects of the Δmss51 strain. This can be explained by the lower steady-state levels of the C85H, C96H and C85HC96H Mss51 proteins detected in mitochondria (Fig. 2A). Independently, our results indicate that although the cysteines in the two CPX motifs are required for full Mss51 function in vivo, modifications of C96 within the second CPX motif produced a more profound phenotype. The fact that Pro to Ser changes in either or both CPX motifs did not alter Mss51 function (not shown) indicates that the cysteines are the critical residues within these motifs.
To know whether the CPX mutations affect the ability of Mss51 to activate COX1 mRNA translation we performed in vivo mitochondrial protein synthesis assays. Pulses of 30 minutes showed Cox1 synthesis to be mildly attenuated in the C85S mutant compared to wild-type levels, more dramatically decreased in the C96S mutant and very poor but detectable in the double mutant C85SC96S (Fig. 2D). In the C85H and C96H mutants, Cox1 synthesis was significantly more attenuated than in the serine mutants (Fig. 2D), most probably as a result of the lower steady-state levels of the corresponding Mss51 variants in mitochondria as stated earlier. Consistent with this possibility, overexpression of all the single variants, including the C85H and C96H mutants, significantly improved Cox1 synthesis and respiratory growth (Fig. S2). When expressed in single copy, the effects of the C85S and C96S variants on Cox1 synthesis are better appreciated in time-course pulse-labeling experiments presented in Fig. 2E and F. Cox1 synthesis in the strain expressing C85S proceeds at a lower rate than in wild-type cells (Fig. 2E). Synthesis of Cox1 in the C96S mutant proceeds at an even lower rate (Fig. 2F), and only some traces of radiolabeled Cox1 were detected in the C96H mutant (Fig. 2F). For the C85S and C85H mutants, we assessed the stability of the newly synthesized Cox1 by pulse-chase. The amount of labeled Cox1 detected in these mutants was as stable as in the wild-type strain but Cox2 and Cox3 are less stable, presumably because COX assembly is compromised in these strains (Fig. 2G).
To test whether the decrease in Cox1 synthesis we observed in the cysteine mutants of Mss51 CPX motifs is directly due to a defect in translation and not the result of Cox1 synthesis down-regulation, we introduced a Δcox14 mutation in the strains expressing a single copy of the C85S, C96S and C85SC96S mutant variants. In most COX assembly mutants, the absence of Cox14 destabilizes the complex of sequestered Mss51 with newly synthesized Cox1 and renders Mss51 available for translation, thus bypassing the feedback regulatory mechanism (Barrientos et al., 2004). As shown in Fig. 2H, the absence of Cox14 did not alter the stability of the Mss51 variants. However, Δcox14 did not restore Cox1 synthesis in any of the mutant strains and it was even detrimental for the strain expressing the C85S variant (Fig. 2H–J). These results suggest that the absence of either cysteine decreases Mss51 stimulation of Cox1 translation, although the strain expressing the C85S mutation produces enough Cox1 to support respiratory growth.
In a wild-type strain, Mss51 mostly accumulates in a 450 kDa Cox1 preassembly complex that contains newly synthesized Cox1, and other proteins such as Cox14, Cox25/Coa3 and Ssc1 (Barrientos et al., 2004; Fontanesi et al., 2011; Mick et al., 2010) although a small portion of Mss51 is also detected in a 120 kDa heterodimer with Ssc1 (Fontanesi et al., 2010). By sucrose gradient sedimentation analyses of mitochondrial extracts, we detected the C85S, C96S and C85SC96S Mss51 variants mostly in the 120 kDa complex (Fig. 2K). A small portion of each variant was detected in the 450 kDa complex, consistent with the residual amount of Cox1 synthesis retained in the mutant strains. Unexpectedly, in the absence of Cox14, some C85S was trapped in the high molecular mass complex thus explaining the poor Cox1 synthesis in this strain (Fig. 2K).
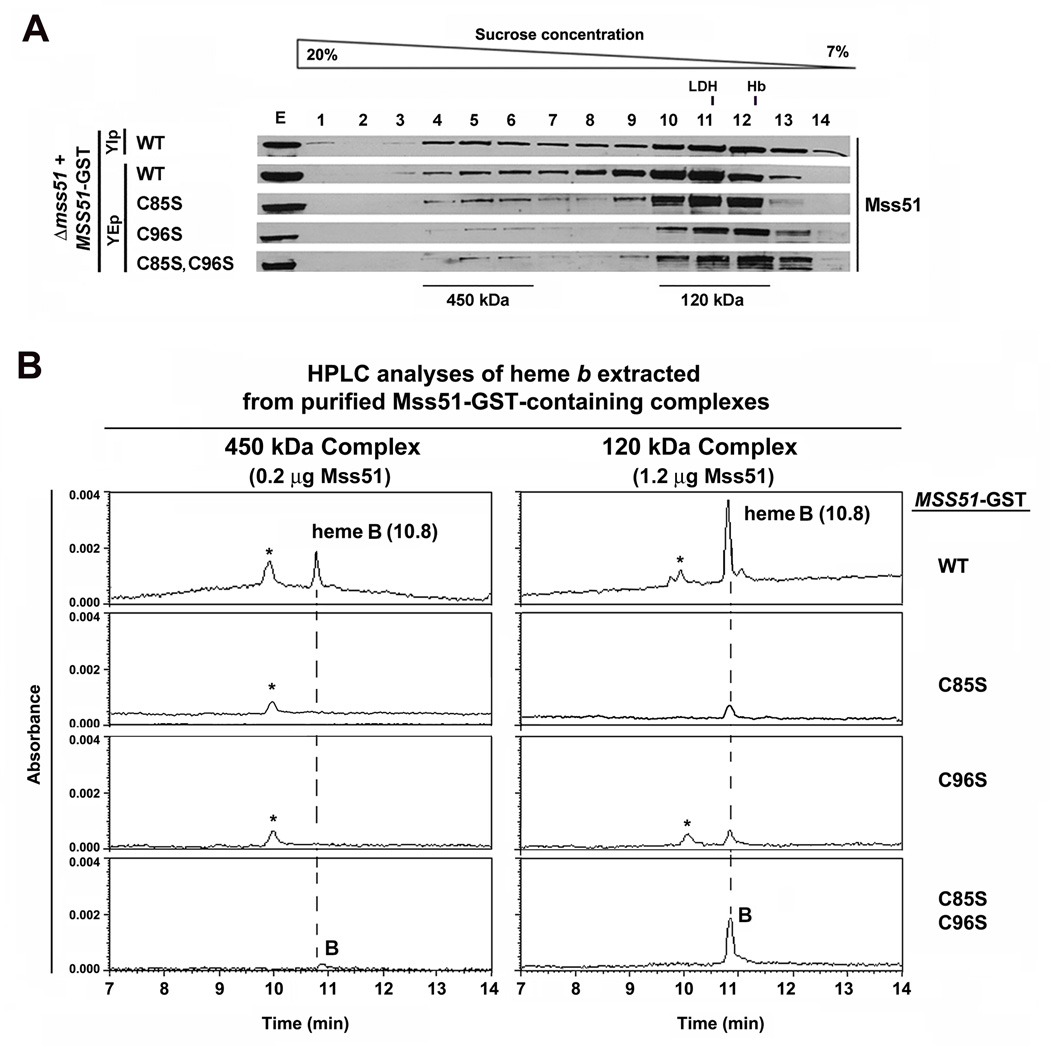
Mss51 Binds Heme B in vivo
To investigate whether Mss51 binds heme in vivo, we used a strain that expresses a fully functional C-terminal GST-tagged version of the protein (Barrientos et al., 2004). We generated additional strains expressing in single copy or overexpressing each of the Mss51 CPX variants. The Mss51-GST-tagged C85S and C96S variants were less stable than the wild-type protein but they accumulated when overexpressed (Fig. S3A), supported Cox1 synthesis partially (Fig. S3B) and complemented the respiratory growth of the Δmss51 strain as well as the untagged versions of these proteins (Fig. S3C).
The wild-type Mss51, C85S, C96S and C85SC96S GST-tagged proteins were extracted from isolated mitochondria and separated by sucrose gradient sedimentation as described earlier. The pattern of native complexes was similar to that observed in the strains expressing the untagged proteins (Fig. 3A), although in general, more Mss51 accumulated in the 120 kDa complex, which is typical of strains overexpressing the protein (Fontanesi et al., 2010). The 450 kDa and 120 KDa Mss51-GST-containing complexes were purified from each strain and the amount of Mss51 estimated by immunoblotting using a standard curve constructed with recombinant protein (not shown) as reported (Fontanesi et al., 2010). Approximately 0.2 µg of Mss51 in the 450 kDa complex and 1.2 µg of Mss51 in the 120 kDa complex purified from each strain were concentrated and treated with 250 mM HCl acetone for heme extraction and HPLC analysis. We detected heme B (elution 10.8 min) in both the 450 kDa and 120 kDa complexes purified from mitochondria expressing wild-type Mss51-GST. The fact that we were able to detect heme in the organic solvent following extraction with acidic acetone indicates that heme is not covalently bound to mitochondrial Mss51 (Fuhrhop and Smith, 1975). No traces of heme A (elution time 29 min, not shown in the graphs) were detected. As a negative control, no traces of heme were detected when using mitochondria expressing untagged Mss51. This result supports that the heme B content in Mss51-GST-containing complexes is specific to the presence of Mss51, and not the result of a non-specific interaction of heme with the glutathione beads.
Figure 3. Mitochondrial Mss51 binds heme B.
(A) Sucrose gradient sedimentation analyses of Mss51 in mitochondrial extracts prepared from the indicated strains. The indicated fractions containing Mss51-complex peaks were used for GST-pulldown purification of the complexes.
(B) Hemes were extracted from 450 kDa and 120 kDa Mss51-containing complexes purified from the indicated strains and analyzed by HPLC on a reverse phase C18 column. The peaks corresponding to heme B (B) are marked. The peak labeled * has not been identified but in some repetitions also appeared in the blank and could be part of the background. The amounts of heme B were calculated from the areas under the peaks.
The identification of heme B as the species binding to Mss51 in vivo does not support a role for Mss51 in handling heme A for insertion into newly synthesized Cox1. Furthermore, the absence of heme A in the 450 kDa complex suggests that the Cox1 protein is not hemylated, in agreement with recent observations (Bestwick et al., 2010). This result indicates that Cox1 hemylation does not occur co-translationally and that it is not required for Cox1 stability in the Mss51-containing pre-assembly complex as reported (Khalimonchuk et al., 2010). The presence of heme B in both complexes suggests that this moiety is necessary for Mss51 functions at different steps of Cox1 biogenesis.
The 120 kDa complexes purified from the strains expressing the partially functional C85S-GST and C96S-GST variants contained an amount of heme B 5–7-fold lower than the wild-type complexes (Fig. 3B), in agreement with the limited ability of these mutant proteins to bind hemin in vitro. In most 450 kDa complex preparations analyzed, the amounts of heme were below the detection limits (Fig. 3B) and only traces were detected in one out of three preparations containing C85S-GST (not shown). Interestingly, we detected a significant amount of heme B in the complexes containing the non-functional double mutant C85SC96S-GST protein, particularly in the more abundant 120 kDa complex. This observation is consistent with the in vitro data suggesting unspecific heme binding to the double mutant protein.
Quantitative analysis of the HPLC data, allowed us to estimate a Mss51/heme molecular ratio ~3:1 in the wild-type 450 kDa Mss51 complex and ~7:1 in the 120 kDa complex. Although our results clearly show the presence of heme in these complexes, the quantitative results could be underestimated if in vivo some heme interacts weakly or transiently with Mss51.
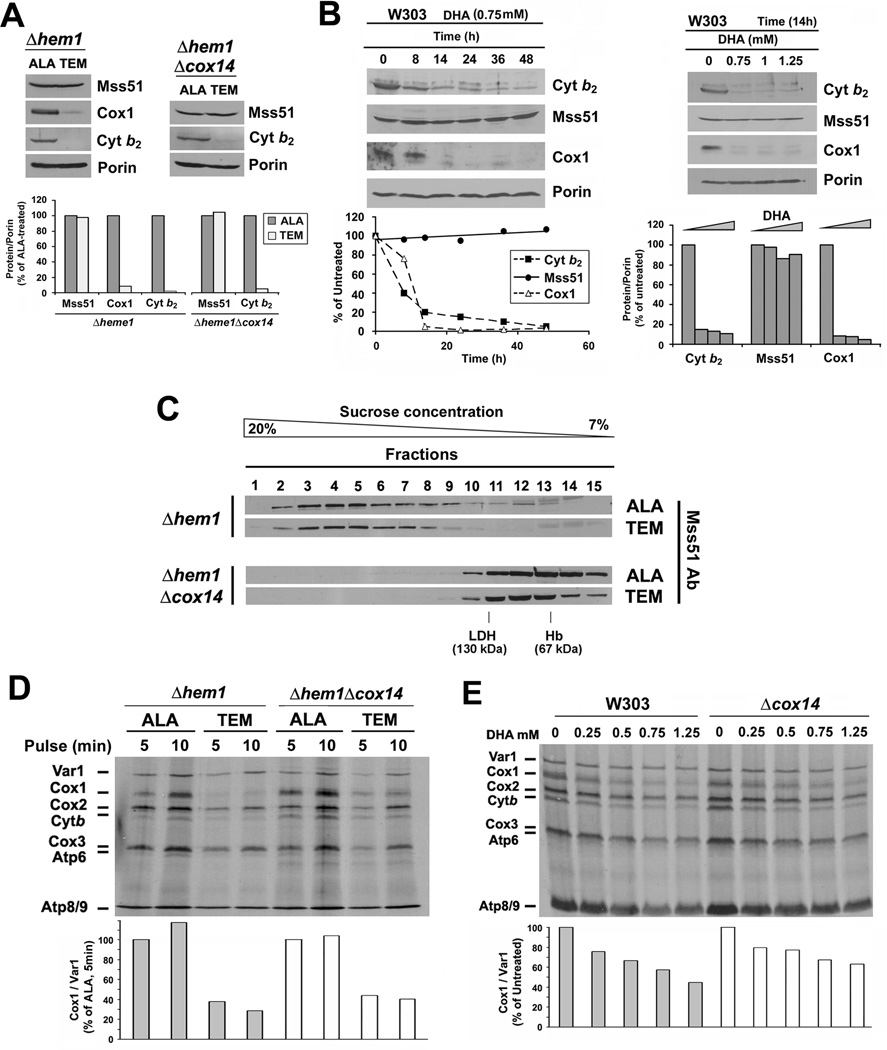
Mss51 is Stable in Heme-Depleted Cells
To further investigate the role of heme in regulating Cox1 synthesis we analyzed a strain carrying a null mutation in HEM1 which encodes the 5-aminolevulinate acid synthase, a protein located in the mitochondrial matrix, where it catalyses the first step in heme biosynthesis. A Δhem1 strain can only survive in fermentable media supplemented with a source of oleic acid (Tween 80), Ergosterol and Methionine (TEM), whose biosynthesis is mediated by hemoproteins (Gollub et al., 1977). In contrast, the Δhem1 strain recovers respiratory capacity when grown in the presence of a heme precursor such as aminolevulinic acid (ALA) to support heme synthesis. The mutant phenotype can be induced in TEM-containing medium by depriving the cells of ALA for 16 hours at which time heme levels were barely detectable (less than 1% as detected in mitochondrial extracts by HPLC) compared to the cells grown in ALA-supplemented medium (Fig. S4). Heme depletion readily affected the accumulation of heme containing proteins such as Cox1 (Fig. 4A) and also cytochrome b2, which expression is transcriptionally-controlled by the heme-dependent regulator Hap1 and by the HAP transcriptional complex (Lodi and Guiard, 1991). However, MSS51 expression is not regulated by the HAP complex (Fontanesi et al., 2008) and Mss51 levels were similar in heme-containing and heme-depleted wild-type and Δcox14 cells (Fig. 4A). As an independent approach to deplete heme we exposed wild-type cells to the heme synthesis inhibitor 4,6-dioxoheptanoic acid (DHA) in the presence of TEM for 14 hours. Following this treatment, Cox1 and cytochrome b2 failed to accumulate but Mss51 levels remained unchanged (Fig 4B). These results indicate that in the absence of heme, Mss51 steady-state levels remains stable.
Figure 4. Heme is not required for Mss51 stability but for Mss51 function.
(A) Immunoblot analyses of the steady-state levels of Mss51, Cytochrome b2 and Cox1 in Δhem1 and Δhem1Δcox14 strains grown in the presence of ALA or TEM. The signals were quantified as in Fig. 1B, normalized to porin signal and plotted in the lower panel.
(B) Same as in 4A but here a wild-type strain (W303) was treated with the heme biosynthesis inhibitor DHA at the indicated concentrations and periods of time in the presence of TEM. The signals were quantified as in Fig. 1B and plotted in the lower panels.
(C) Sucrose gradient sedimentation analyses of Mss51 in mitochondrial extracts prepared from the indicated strains as in Fig. 2I.
(D, E) In vivo mitochondrial protein synthesis in the indicated strains and growth conditions performed as in Fig. 2D. The signals were quantified as in Fig. 1B and used to plot the graphs in the lower panels.
Sucrose gradient sedimentation analyses of Mss51 extracted from the Δhem1 strain grown in the presence of ALA showed a wild-type Mss51 distribution, with most Mss51 in the 450 kDa complex and approximately 10% in the 120 kDa complex (Fig. 4C). Mss51 extracted from the Δhem1 strain grown in the presence of TEM accumulated exclusively in the 450 kDa complex. This is consistent with the lack of heme A as observed in Cox1 hemylation mutants, although in these mutants Mss51 additionally accumulates complexed with the COX1 mRNA translational machinery (Fontanesi et al., 2010). Mss51 extracted from the Δhem1Δcox14 strain showed a similar pattern when grown in the presence of ALA or TEM, with all Mss51 accumulated in the 120 kDa complex. In conclusion, the absence of heme from Mss51 does not alter the stability of Mss51 or its accumulation within high molecular mass complexes.
Cox1 Synthesis is Attenuated in Heme-Depleted Cells
To explore whether the absence of heme from Mss51 affects the function of Mss51, we performed in vivo mitochondrial protein synthesis analyses in Δhem1 strains. Detection of [35S]-methionine accumulation into newly synthesized Cox1 and cytochrome b was strongly attenuated in heme-depleted Δhem1 cells (Fig. 4D). The poor Cox1 synthesis is not the result of translational downregulation due to a COX assembly defect but rather is predominantly due to a specific Cox1 translational defect in the absence of heme, because Cox1 synthesis recovered only partially in heme-depleted Δhem1 cells expressing an additional Δcox14 mutation (Fig. 4D).
As for the Δhem1 mutant, DHA-treated wild-type cells displayed a significant attenuation on [35S-methionine] incorporation into newly synthesized Cox1 and cytochrome b, which in the case of Cox1 was only partially restored in the absence of Cox14 (Fig. 4E). Whether cytochrome b synthesis is down-regulated in the absence of heme or its turnover is enhanced in the absence of its prosthetic group is an open question out of the scope of this manuscript.
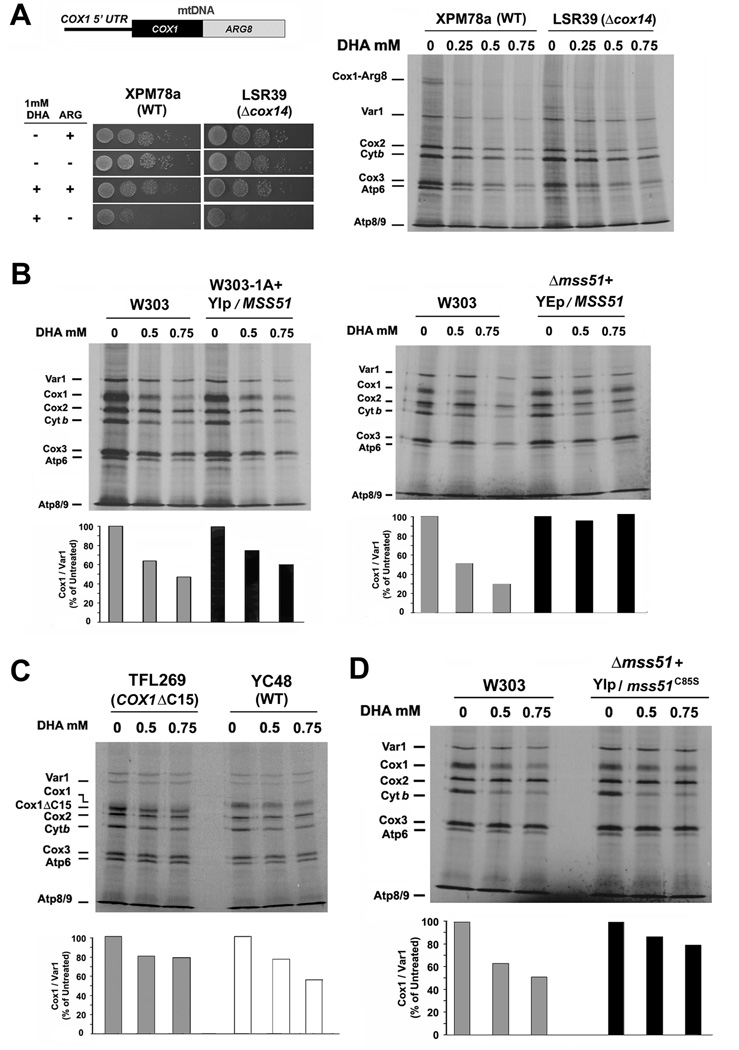
In the case of Cox1, to further distinguish between synthesis defect and increased turnover, we treated with DHA a strain carrying a reporter gene for COX1 mRNA translation. The system is based on the insertion of a recoded version of the nuclear gene ARG8 into the mtDNA of a Δarg8 strain (Steele et al., 1996). Mitochondrial ARG8 expression in the Δarg8 strain confers arginine prototrophy. We used the strain XPM78a (Perez-Martinez et al., 2003), in which expression of ARG8 fused to COX1 was placed under the control of the COX1 5’UTR (Fig. 5A) and the derivative strain LSR39, which carries an additional Δcox14 mutation (Shingu-Vazquez et al., 2010). It is known that following synthesis, the Cox1-Arg8 fusion protein is cleaved and the two proteins are functional (Steele et al., 1996). In both DHA-treated XPM78a and LRS39 cells, synthesis of the fusion protein was significantly decreased (Fig. 5A). DHA-treated XPM78a and LRS39 cells failed to grow robustly in fermentable media lacking arginine (Fig. 5A). Because arginine-dependent growth reports COX1 mRNA translation, such results strongly support a direct heme-dependent regulation of Cox1 synthesis, presumably by a mechanism based on inhibition of the translational activation activity of Mss51 when the protein does not bind heme.
Figure 5. Heme is required for efficient COX1 mRNA translation.
(A) In vivo mitochondrial protein synthesis in the indicated strains treated or not with DHA. The wild-type (XPM78a) and Δcox14 (LSR39) strains carry an engineered COX1-ARG8 reporter gene in their mitochondrial DNA (upper panel). The synthesized Cox1-Arg8 fusion protein is indicated. The lower panel shows a serial dilution growth test of the strains in synthetic media supplemented or not with DHA and arginine.
(B–D) In vivo mitochondrial protein synthesis in the indicated strains treated or not with DHA. The signals were quantified as in Fig. 1B and used to plot the graphs in the lower panels.
Several mechanisms known to by-pass feedback regulation of COX1 mRNA translation also failed to induce total recovery of Cox1 synthesis, albeit that at different degrees, in heme-depleted cells. This was true for strains carrying an additional copy of wild-type MSS51 (Fig. 5B). And for strains with increased amounts of Mss51 available for translation because Mss51 sequestration in the 450 kDa complex is prevented, such as in strains expressing a C-terminal truncated form of Cox1 (strain TFL269, (Shingu-Vazquez et al., 2010) (Fig. 5C) or a Δcox25/coa3 mutation (Fig. 6A). These results confirmed that although some of the Cox1 synthesis defect observed upon heme-depletion is probably down-regulation of its translation due to defective COX assembly, the absence of heme from Mss51 causes an additional robust decrease in COX1 mRNA translation. Only Mss51 overexpression supported Cox1 synthesis in heme-depleted cells (Fig. 5B). Overall, these results are compatible with the view that Mss51 can perform its role in COX1 mRNA translation with low levels of heme, but heme is necessary in order to accomplish this function efficiently.
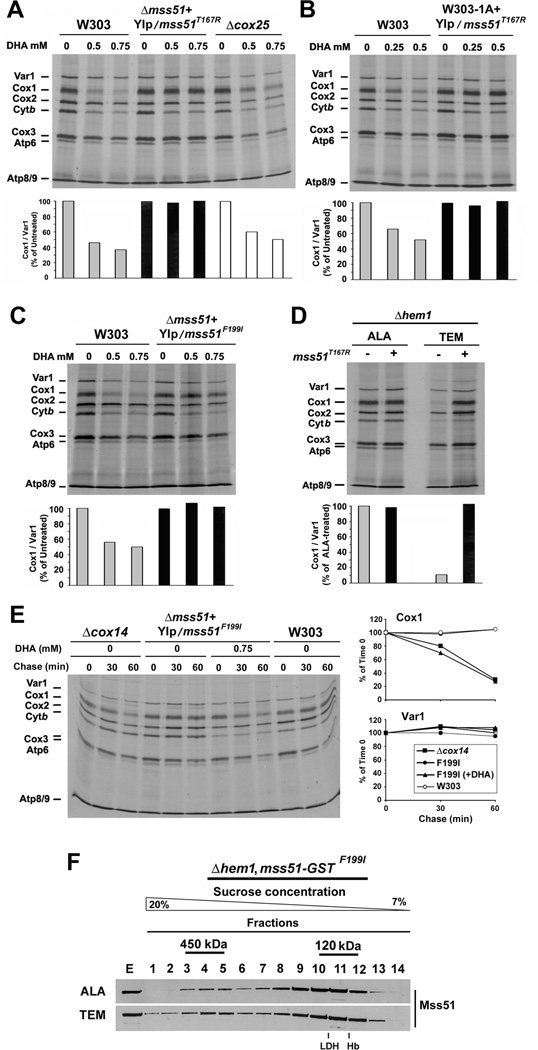
Figure 6. Mss51 mutants bypass the requirement of heme for efficient COX1 mRNA translation.
(A–E) In vivo mitochondrial protein synthesis in the indicated strains treated or not with DHA in the presence of TEM or in (D) the Δhem1, mss51T167R strain incubated in the presence of either ALA or TEM. In (E), following a 10 minute pulse, the labeled proteins were chased for 30 and 60 min. The signals were quantified as in Fig. 1B and used to plot the graphs in the lower or side panels.
(F) Sucrose gradient sedimentation analyses of GST-tagged Mss51F199I in mitochondrial extracts prepared from the indicated strains as in 2I.
We predicted that the C85S mutant that binds heme only poorly (Fig. 3B) yet sustains ~60% of Cox1 synthesis (Fig. 2E) should be less sensitive to heme-depletion. The results presented in Fig. 5D are compatible with this prediction and further underscore the role of CPX motifs in heme binding.
Mutations in Mss51 Suppress the Sensitivity of Cox1 Synthesis to Heme Depletion
We previously reported that a single copy of the mutant alleles mss51T167R and mss51F199I or additional copies of wild-type MSS51 partially suppress the respiratory defect of mutants of SHY1, a COX assembly factor, by increasing Cox1 synthesis (Barrientos et al., 2002). In most COX-deficient strains, mutant or additional copies of MSS51 do not suppress the COX assembly defect but significantly increase Cox1 synthesis bypassing the negative feedback regulation (Barrientos et al., 2004). We have proposed that the suppression of the COX1 mRNA translation defect by Mss51T167R and Mss51F199I is due to their increased intrinsic capacity to avoid the trap within the 450 kDa complex and to accumulate mainly in the 120 kDa complex with Ssc1, the complex that serves as a reservoir for translationally-competent Mss51 (Fontanesi et al., 2010).
Notably, expression of the T167R or F199I variants of Mss51 fully suppressed any sensitivity of Cox1 synthesis to DHA-induced heme depletion either when expressed in a wild-type or a Δmss51 strain (Fig. 6A–C). Similarly, a Δhem1 strain expressing Mss51T167R efficiently synthesized Cox1 both when grown in the presence of ALA or TEM (Fig. 6D). As expected, newly synthesized Cox1 in these strains grown in the presence of DHA is as unstable as in a Δcox14 strain (Fig. 6E). This is due to the lack of Cox1 hemylation and COX assembly in the absence of heme. A GST-tagged version of mss51F199I fully complemented a Δmss51 strain (Fig. S5A) and was used to purify the 450 kDa and the 120 kDa Mss51-containing complexes (Fig. S5B). HPLC analyses detected only traces of heme B in the 120 kDa complex (Fig. S5C). The distribution of Mss51F199I in the 450 kDa and 120 kDa complexes was similar in the presence and absence of heme, although in heme depleted cells more Mss51 was detected in the 450 kDa complex and in higher molecular mass material (Fig. 6F) as reported in Cox1 hemylation mutants (Fontanesi et al., 2010). Altogether, the observations presented here strongly link Mss51 to heme sensing and heme-mediated regulation of COX1 mRNA translation.
DISCUSSION
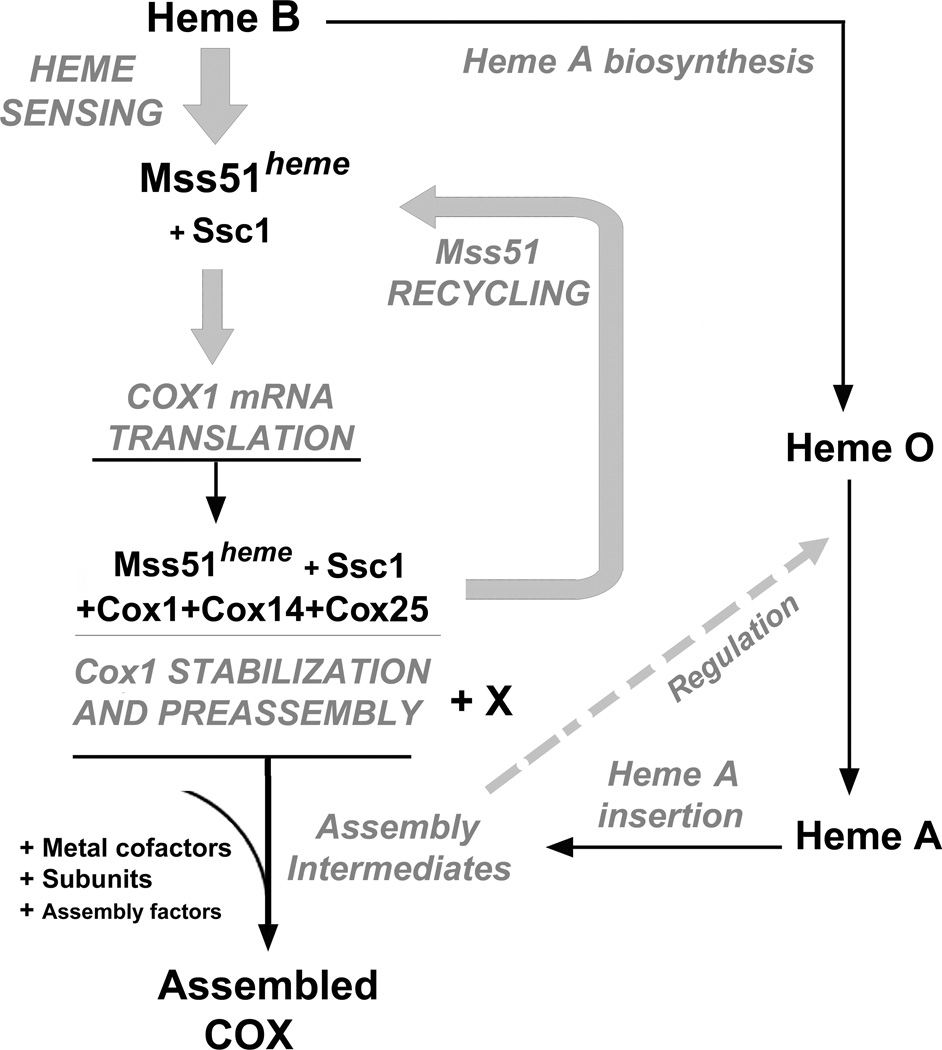
Although heme has been shown to play roles in regulation of nuclear gene expression (both transcription and translation), the possible role/s of heme in regulating mitochondrial gene expression are largely unexplored. Here we report a regulatory mechanism by which the function of the mitochondrial COX1 mRNA translational activator Mss51 is regulated by heme. The N-terminus of Mss51 contains two heme responsive CPX motifs. Our in vivo and in vitro data strongly support a role of these motifs in heme binding to Mss51. Mss51 is the central element of an auto-feedback translational repression mechanism in mitochondria that couples Cox1 synthesis with its assembly in COX (Fig. 7). These findings significantly enrich our understanding of the physiological role of Mss51 in the regulation of COX biogenesis and aerobic energy production by revealing an additional level of mitochondrial translational regulation involving heme sensing.
Figure 7. Model of Heme sensing by Mss51 to coordinate Cox1 translation and assembly.
See explanation in the Discussion section.
The observation that Mss51 binds heme B involving CPX motifs suggested heme could play a regulatory role in Mss51 functions. CPX motifs generally participate in transient heme binding to proteins and are known to control processes related to iron and oxidative metabolism in prokaryotes and eukaryotes (Mense and Zhang, 2006). For example, in S. cerevisiae, heme synthesis is regulated in response to oxygen levels (Hon et al., 2003). The heme activator protein Hap1 binds heme though CPX motifs and functions either as a transcriptional repressor or activator, depending on heme levels, thus mediating oxygen sensing and heme signaling (Zhang and Guarente, 1995). In mammals, CPX motifs are present, for example, in the transcriptional repressor Bach1, that regulates globin expression, and in the HRI kinase which coordinates globin protein synthesis and heme availability in reticulocytes (Mense and Zhang, 2006). With few exceptions, the molecular mechanisms of the “heme-regulated” proteins remain unclear due to the lack of their structural characterizations. In the bacterium Bradyrhizobium japonicum, the iron response regulator (Irr) mediates the iron-dependent transcriptional regulation of heme synthesis (Mense and Zhang, 2006) through heme binding to two sites, one of them consisting of the cysteine in a CPX motif (Ishikawa et al., 2011). In the case of the mammalian Rev-erbβ, a heme-binding nuclear hormone receptor that represses a broad spectrum of target genes involved in regulating metabolism and the circadian cycle, a thiol/disulfide redox switch involving the cysteines in two CPX motifs, controls heme binding and dissociation (Gupta and Ragsdale, 2011). In this case, heme iron is hexacoordinated by a CPX motif Cys and a His residue. The difference absorption spectra of heme-reconstituted recombinant Reverbβ displays a profile with Soret peak at 416 and α and β peaks at 537 and 567 respectively. Upon oxidation of the sulfhydryl to a disulfide, heme coordination switches and the binding affinity decreases (Gupta and Ragsdale, 2011). Heme coordination by Mss51 somehow resembles that of Rev-erbβ. The difference spectra revealed a Soret band at 416 nm and in the alpha and beta bands at 550 and 520 nm, respectively. We were able to extract heme B from mitochondrial Mss51 using acidic acetone, which indicates non-covalent heme-binding to the polypeptide. However, heme binding to recombinant Mss51 was resistant to denaturing conditions such as 2% SDS and 6M urea, which points to a tight interaction between Mss51 and heme as previously reported for other proteins (Lechardeur et al., 2010). In Mss51, the absence of C96 in the second CPX motif affects more severely heme binding and Mss51 function in vivo than the absence of C85 in the first CPX motif. The protein contains three additional cysteines (Fig. 1), mutation of each to serine does not affect respiratory growth (not shown). The identification of the residues directly involved in the heme coordination is the focus of ongoing efforts in our laboratory.
For most heme-regulated proteins, in vivo heme binding has not been demonstrated. This is not a trivial task given the transient interaction most of these proteins are predicted to have with heme. We have been able to detect heme B in Mss51 purified within physiological high molecular mass complexes isolated from mitochondria and to correlate heme binding and Mss51 function to the integrity of the CPX motifs. It is remarkable that Mss51 associates with the mitochondrial Hsp70 chaperone Ssc1 (Fontanesi et al., 2010) in a manner similar to other heme-regulated proteins that also interact with multichaperone complexes containing Hsp70 and Hsp90. In yeast Hap1, CPX motifs arbitrate heme-dependent formation of protein complexes with Hsp70 and 90 chaperones that allow Hap1 to bind DNA and activate transcription (Lan et al., 2004). In mammals, the HRI kinase also associates with Hsp70 and 90 chaperones to mediate heme-regulated translation inhibition and provide one major mechanism that ensures balanced synthesis of globins and heme as stated earlier (Mense and Zhang, 2006). Regulatory heme binding and release from Mss51 and other heme-regulated proteins would be expected to induce conformational changes in the protein that could be facilitated by their interacting chaperones.
Heme binding is not essential for Mss51 stability in vivo but it does regulate its function as a COX1 mRNA translational activator. Several pieces of information support this conclusion. (i) Translation of an ARG8 reporter gene placed under the control of the COX1 promoter is dependent on heme. (ii) Cox1 synthesis requires heme, even in strains in which the negative feedback translational regulation exerted by Mss51 is suppressed, namely strains carrying mutations in cox14, cox25/coa3 or expressing a C-terminal truncation in Cox1. (iii) Cox1 synthesis is attenuated in CPX mutant strains, particularly those expressing C96 variants. (iv) Cox1 synthesis in a strain expressing C85S is less sensitive to heme depletion, and (v) some mutant forms of Mss51, namely the T167R and F199I mutations, render the protein completely insensitive to heme regulation. This collection of observations strongly links both the CPX motifs in Mss51 to heme binding and the Mss51-heme binding to regulation of COX1 mRNA translation. Whether Mss51 requires heme binding to adopt the proper conformation that allows for efficient interaction with the COX1 mRNA and what is the contribution of Ssc1 in this process are outstanding questions that warrant future investigation.
The translational regulatory system described here was believed to be restricted to fungi because: (i) Mammalian mitochondrial mRNAs lack 5’UTR and how translation is activated in mammalian mitochondria remains to be understood and (ii) orthologs of Mss51, Cox14 and Cox25 have not been identified. However, mutations in proteins with similarity to Cox14 (C12Orf62) and Cox25 (CCDC56) have been recently recognized to cause COX-specific defects in humans and fly (Peralta et al., 2012; Weraarpachai et al., 2012). Although no clear Mss51 ortholog is encoded in mammalian genomes, the mammalian MSS51/ZMYND17 protein exhibits significant similarity (Szklarczyk et al., 2012). ZMYND17 contains a zinc finger domain formed by cysteines that in alignments with yeast Mss51 overlap with the CPX motifs cysteines (Fig. S6). The ability of the human protein to bind heme and its hypothetical role in COX assembly are of high biological and biomedical interest and also warrant future investigation.
In conclusion, S. cerevisiae Mss51 is the first heme binding protein described to play a role in mitochondrial translational activation. By binding heme B, Mss51 could sense oxygen levels (required for heme biosynthesis) and heme availability for heme A biosynthesis (Fig. 7) to modulate Cox1 synthesis and subsequent assembly into COX, the major oxygen-consuming mitochondrial enzyme, essential for aerobic ATP production. Finally, on the light of the mechanism uncovered here, it is tempting to speculate that heme B could serve as a more general sensing element capable of activating not only COX assembly but also the biogenesis of other cytochrome-containing respiratory-chain components.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
All S. cerevisiae strains used are listed in supplemental Table S1 and the composition of the standard culture media used is defined in the supplemental material.
In vivo Mitochondrial Protein Synthesis
Mitochondrial gene products were labeled with [35S]-methionine (7 mCi/mmol, Perkin Elmer) in whole cells at 30°C in the presence of 0.2 mg/ml cycloheximide to inhibit cytoplasmic protein synthesis (Barrientos et al., 2002). Equivalent amounts of total cellular proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane and exposed to Kodak X-OMAT X-ray film.
Purification of Wild-type and CPX Mutant Variants of Mss51
Wild-type and Mss51 CPX mutant versions fused to a thrombin-cleavable trigger factor tag (TF-Mss51, TF-Mss51C85S, TF-Mss51C96S, TF-Mss51C85SC96S) were expressed as His6-tagged proteins from the pColdTF vector (Genescript) in the BL21 Star protein expression strain of E. coli (Invitrogen, Carlsbad, CA), extracted and purified by metal affinity using a Cobalt Sepharose HiTrap HP affinity column (Amersham) as described in the supplemental material.
Spectrophotometric Properties of Heme-Titrated Recombinant Mss51
Heme titrations were performed by difference spectroscopy aerobically and at room temperature using a double beam UV-2401PC Shimadzu spectrophotometer as described (Gupta and Ragsdale, 2011) and are fully explained in the supplemental material. To determine the binding parameters, the data obtained from the heme titrations were plotted and fit to an equation describing a single site binding using the GraphPad Prism software.
Heme content determination in mitochondrial Mss51-containing complexes
To obtain material for heme content determination by HPLC, we extracted 30 mg of mitochondria purified from strains expressing GST-tagged versions of Mss51, sedimented the extracts in multiple sucrose gradients, pooled the three fractions around the 120 kDa and 450 kDa peaks and performed GST-pull-down assays as described (Fontanesi et al., 2010) and explained in the supplemental material. The pull-down elutes were concentrated using Vivaspin 500 columns (Sartorius Stedim Biotech, Germany), extracted with 250 mM HCl Acetone and 50% acetonitrile and the samples analyzed using a C18 column as described (Barros and Tzagoloff, 2002). The amount of Mss51 in each sample was estimated for each experiment by immunoblotting using a standard curve generated with purified recombinant protein as detailed (Fontanesi et al., 2010). Heme concentration (µM) was calculated using the absorbance maxima at 10.8 minutes of elution and an extinction coefficient of 180 mM−1×cm−1 at 400 nm.
Miscellaneous procedures
In vitro Mss51-heme binding assays were performed using purified proteins and hemin-conjugated agarose beads (Sigma). The heme-associated peroxidase activity (heme staining) of PAGE-separated heme-bound Mss51 was revealed directly in-gel by the enhanced chemiluminescence method from Pierce (Supersignal Femto). Purification of mitochondria from yeast strains and mitochondrial respiratory chain enzymatic measurements were performed as reported (Barrientos et al., 2002). All miscellaneous methods are detailed in the supplemental material.
Statistical analysis
All experiments were done at least in triplicate. All data are presented as means ± SD of absolute values or percent of control. Values were analyzed for statistical significance by Student’s t-test. P < 0.05 was considered significant.
Supplementary Material
HIGHLIGHTS.
-
►
COX biogenesis is translationally regulated by heme availability.
-
►
The COX1 mRNA translational activator Mss51 is a heme binding protein.
-
►
Mss51 binds heme through two N-terminal Cys-Pro-X heme regulatory motifs.
-
►
Heme binding to Mss51 is required for efficient Cox1 synthesis.
ACKNOWLEDGEMENTS
We thank M. Tigano for technical assistance. We thank Dr. D. Winge (University of Utah, UT) for critical reading of the manuscript. We thank Dr. B. Guiard (CNRS, Paris, France), Dr. X. Perez-Martinez (Universidad Autónoma de Mexico, Mexico) and Dr. D. Winge for providing reagents. This research was supported by an NIH-RO1 GM071775-06 (to AB), MDA Research Grants (to AB and to PH), MDA Development Grant (to FF), funds from the Developmental Center for AIDS Research P30 AI 073961 (to RSM) and NIH F31 fellowship GM081975 (to IS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes 6 Figures, 1 Table and Supplemental Experimental Procedures.
REFERENCES
- Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A. Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 2002;516:119–123. doi: 10.1016/s0014-5793(02)02514-0. [DOI] [PubMed] [Google Scholar]
- Bestwick M, Khalimonchuk O, Pierrel F, Winge DR. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol Cell Biol. 2010;30:172–185. doi: 10.1128/MCB.00869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Jin C, Tzagoloff A, Barrientos A. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum Mol Genet. 2008;17:775–788. doi: 10.1093/hmg/ddm349. [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrhop J-H, Smith KM. Laboratory methods in porphyrin and metalloporphyrin research. Amsterdam: Elsevier Scientific Publishing; 1975. pp. 48–51. [Google Scholar]
- Gollub EG, Liu KP, Dayan J, Adlersberg M, Sprinson DB. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- Gupta N, Ragsdale SW. Thiol-disulfide Redox Dependence of Heme Binding and Heme Ligand Switching in Nuclear Hormone Receptor Reverb{beta} J Biol Chem. 2011;286:4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem. 2003;278:50771–50780. doi: 10.1074/jbc.M303677200. [DOI] [PubMed] [Google Scholar]
- Horn D, Al-Ali H, Barrientos A. Cmc1p is a conserved mitochondrial twin Cx9C protein involved in cytochrome c oxidase biogenesis. Mol Cell Biol. 2008;28:4354–4364. doi: 10.1128/MCB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Nakagaki M, Bamba A, Uchida T, Hori H, O'Brian MR, Iwai K, Ishimori K. Unusual heme binding in the bacterial iron response regulator protein: spectral characterization of heme binding to the heme regulatory motif. Biochemistry. 2011;50:1016–1022. doi: 10.1021/bi101895r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Khalimonchuk O, Smith PM, Winge DR. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 2012 Apr 24; doi: 10.1016/j.bbamcr.2012.04.008. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C, Lee HC, Tang S, Zhang L. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J Biol Chem. 2004;279:27607–27612. doi: 10.1074/jbc.M402777200. [DOI] [PubMed] [Google Scholar]
- Lechardeur D, Fernandez A, Robert B, Gaudu P, Trieu-Cuot P, Lamberet G, Gruss A. The 2-Cys peroxiredoxin alkyl hydroperoxide reductase c binds heme and participates in its intracellular availability in Streptococcus agalactiae. J Biol Chem. 2010;285:16032–16041. doi: 10.1074/jbc.M109.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodi T, Guiard B. Complex transcriptional regulation of the Saccharomyces cerevisiae CYB2 gene encoding cytochrome b2: CYP1(HAP1) activator binds to the CYB2 upstream activation site UAS1-B2. Mol Cell Biol. 1991;11:3762–3772. doi: 10.1128/mcb.11.7.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense SM, Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta S, Clemente P, Sanchez-Martinez A, Calleja M, Hernandez-Sierra R, Matsushima Y, Adan C, Ugalde C, Fernandez-Moreno MA, Kaguni LS, Garesse R. Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function. J Biol Chem. 2012;287:24174–24185. doi: 10.1074/jbc.M112.343764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu-Vazquez M, Camacho-Villasana Y, Sandoval-Romero L, Butler CA, Fox TD, Perez-Martinez X. The carboxyl-terminal end of Cox1 is required for feedback-assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J Biol Chem. 2010;285:34382–34389. doi: 10.1074/jbc.M110.161976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Liu J, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2012;1817:883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci U S A. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Wanschers BF, Cuypers TD, Esseling JJ, Riemersma M, van den Brand MA, Gloerich J, Lasonder E, van den Heuvel LP, Nijtmans LG, Huynen MA. Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome. 2012;13:R12. doi: 10.1186/gb-2012-13-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weraarpachai W, Sasarman F, Nishimura T, Antonicka H, Aure K, Rotig A, Lombes A, Shoubridge EA. Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am J Hum Genet. 2012;90:142–151. doi: 10.1016/j.ajhg.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:523–535. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.