Abstract
The periodontal pathogen Actinobacillus actinomycetemcomitans produces a leukotoxin that is considered a primary virulence factor in localized juvenile Periodontitis (LJP). Select strains of the bacterium contain a 530-bp deletion in the promoter region of the leukotoxin gene operon which results in enhanced transcription of the leukotoxin. DNA hybridization and polymerase chain reaction (PCR) were used to examine genetic variants of A. actinomycetemcomitans in 24 LJP-susceptible children from 21 families having a history of the disease and 34 control children from non-LJP families. A significant association was found between the detection of variants that had a deletion in the leukotoxin promoter region, indicative of a high level expression leukotoxin genotype, and conversion from a healthy periodontal status to disease. Subjects harboring A. actinomycetemcomitans of this genotype were more likely to convert to LJP than those subjects who had variants containing the full length leukotoxin promoter region (odds ratio = 22.50, 95% C.I.). These findings support the concept that highly virulent strains or clonal types of periodontal pathogens play a major role in the initiation of periodontal disease in susceptible hosts.
Keywords: Periodontitis, juvenile/epidemiology, Actinobacillus actinomycetemcomitans, leukotoxin, epidemiology, polymerase chain reaction, polymorphism, restriction fragment length
Localized juvenile Periodontitis (LJP) is a particularly aggressive form of periodontal disease that causes rapid periodontal destruction in juveniles and young adults. The facultative Gram-negative bacterium Actinobacillus actinomycetemcomitans is considered to be the primary pathogen in this disease.1–3 Clonal analysis of this species has resulted in the identification of six major lineages or electrophoretic type (ET) divisions.4 One of the major virulence determinants produced by this bacterium is a leukotoxin which is specifically cytotoxic for neutrophils and macrophages.5–7 The leukotoxin is part of an operon that is highly conserved among the 6 ET divisions.8,9 A deletion in the promoter region of the leukotoxin gene operon, found in a relatively small percentage of strains, results in a significant increase in leukotoxin expression.10 These “high leukotoxin-producing” strains appear to cluster in a single ET division.4
The world-wide distribution of the high leukotoxin-producing strains has been a subject of great interest. Haubek et al.11 screened 148 isolates of A. actinomycetemcomitans from Scandinavian and northern European patients and failed to identify high leukotoxin-producing strains. Brogan and coworkers10 found that only 3 of 17 strains examined produced high levels of leukotoxin. Zambon and coworkers12 found that the high leukotoxin-producing strain was detected in just over half of 21 LJP patients from a North American population. Haubek et al.13 found that 11 of 17 subjects of African ancestry had the high leukotoxin-producing strains and proposed that strains of this genotype may cluster in people of African origin.
It is likely that the reason for the differences in these studies is related to differences in the distribution of the high leukotoxin-producing strains in various regions of the world. Most studies to date have been cross-sectional associations of the virulent genotype with disease. To investigate the hypothesis that the virulent genotypes of A. actinomycetemcomitans are causal, a longitudinal study of a well characterized geographically homogeneous population of LJP-susceptible children was undertaken. Using restriction fragment length polymorphism (RFLP) analysis of A. actinomycetemcomitans isolated from members of these families, we showed that a single genetic variant, designated RFLP group II, correlated with the conversion of LJP-susceptible children from a healthy to diseased periodontal status.14 In addition, we found that isolates containing the deletion in the leukotoxin promoter region were present in some of these subjects.15 Since the subjects in our longitudinal study were young children from predominantly African-American families, this population was ideally structured to address the question of a possible correlation between the presence of high leukotoxin-producing strains of A. actinomycetemcomitans and LJP. In the present study we have used DNA hybridization and PCR to characterize the leukotoxin promoter region in A. actinomycetemcomitans isolated from the longitudinal study subjects. We provide clinical evidence that strains of A. actinomycetemcomitans that exhibit the high leukotoxin-producing genotype prevail in young members of LJP families who convert from a healthy periodontal status to disease.
MATERIALS AND METHODS
Subject Population
The patient population examined in this study consisted of 21 families that contained at least one member, child or adult, with clinical signs of LJP or a documented history of the disease and one or more siblings, less than 13 years of age, having no clinical evidence of LJP. Twenty-seven siblings (mean age 10.2 years at the time of enrollment) who had no evidence of LJP when accessioned into the study were defined as the LJP-susceptible subjects during the time that they were between the ages of 10 and 16 years. These subjects were deemed at risk for LJP because the disease appears to be subject to familial inheritance.16,17 Thirty-five subjects (mean age 10.3 years at the time of enrollment), matched by age, gender, and race to the LJP-susceptible children, were recruited from families with no prior history of LJP and served as the control group in the longitudinal study.
The change in the periodontal status, from health to disease, that occurred during the course of the study was defined as a conversion. The subject in each family who had active LJP at accession or a documented history of the disease was defined as the proband. The clinical evaluation of the subjects, description of the criteria used to classify the conversion of a subject from a healthy to diseased periodontal status, microbiological sampling methods, and results of RFLP analyses were described previously.14 Briefly, the plaque and gingival indexes; eruption status; decayed, missing and filled teeth; probing depth, attachment level from cemento-enamel junction (CEJ); and percentage of sites which bled on probing were collected as each subject entered the study and every 3 months thereafter from the LJP-susceptible and control children. Adults were monitored once a year. Radiographic films were taken at the time of accession and at yearly intervals. Bone loss was assessed by measuring the greatest distance between the most coronal level of the alveolar bone crest and the deepest part of the bony defect in over-laid tracings taken from the baseline and subsequent radiographs.18 Interim radiographs were taken during active periodontal inflammation. Conversions were defined as an increase of ≥ 2 mm above the baseline value, in the distance from the CEJ to the bottom of the periodontal pocket or an increase of at least 2 mm in probing depth. This change, occurring over a 3-month interval, had to coincide with a loss of attachment ≥ 1 mm above the baseline value. Bone loss was also checked at the time of surgery as supporting evidence of periodontal destruction. Over the course of the 5-year longitudinal study 9 LJP-susceptible children (mean age 12.8 years) and 3 control children (mean age 14.7 years) converted from a healthy to a diseased periodontal state.14
Subgingival microbiological samples were obtained every 3 months from 24 and 34 of the LJP-susceptible and control children, respectively. These samples were obtained from the mesial surfaces of the 4 permanent first molars, the maxillary first premolars, and any additional sites that converted from a healthy to diseased state during the study. Supragingival plaque was removed with sterile cotton pellets and subgingingival plaque was collected by inserting 3 sterile paper points to the depth of the periodontal pocket. The subgingival samples were placed in prereduced VMGA III anaerobic transport medium.19Actinobacillus actinomycetemcomitans was isolated on trypticase soy-serum-bacitracin-vancomycin (TSBV) agar.20 The isolates were identified according to conventional colony morphology and biochemical criteria and by DNA hybridization.
Human use protocols were approved by the University of Pennsylvania Committee on Studies Involving Human Beings (assurance M1025).
Leukotoxin Promoter Deletion Assays
Total DNA was prepared from A. actinomycetemcomitans isolates according to standard methods.21 A representative isolate of each RFLP type, that was found in each family member that had A. actinomycetemcomitans, was used. Microbial analysis was limited in this manner because we have previously found that multiple isolates exhibiting the same RFLP type were often replicates of the same genetic variant or genotype.14,15 The RFLP types were previously identified based on the hybridization of a 4.7 kb EcoRI chromosomal DNA fragment, cloned from strain FDC Y4, to total DNA.15
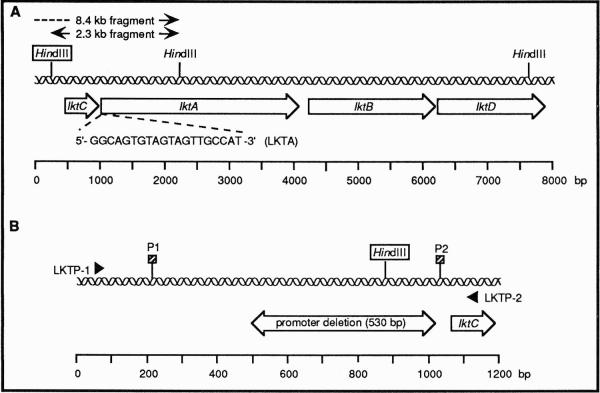
To identify the leukotoxin promoter deletion by DNA hybridization, approximately 1 μg of total DNA from each isolate was digested to completion with HindIII, according to the manufacturer's instructions, and the restriction fragments were separated and analyzed on Southern blots. Hybridizations were performed with an oligonucleotide 5′-GGCAGTGTAGTAGTTGCCAT-3′ (LKTA), homologous to the 5′ end of the lktA gene (Fig. 1A), to detect restriction site polymorphism in the upstream non-coding region of the leukotoxin promoter.15 The probe was end labeled with [γ-32P]ATP (>5,000 Ci/mmol) as previously described.22 To obtain molecular size markers on the autoradiograms, bacteriophage λ DNA was digested to completion with HindIII and run on the Southern blots. A portion of the λ DNA sample was labeled by nick translation and used as a hybridization probe (5 × 106 cpm/blot). Hybridizations were performed as described previously.15 A single hybridization-positive fragment of approximately 8.4 or 2.3 kb was observed if the A. actinomycetemcomitans isolate contained or lacked, respectively, the leukotoxin promoter deletion (Fig. 1A). This heterogeneity was due to a HindIII recognition site that resided within the sequence marked by the deletion.
Figure 1.
DNA hybridization probe and PCR primers used in this study. (A) Physical map of the leukotoxin operon showing the location of the hybridization probe sequence (LKTA). The boxed HindIII site is present (2.3 kb hybridization-positive fragment) or absent (8.4 kb hybridization-positive fragment) in low or high leukotoxin-producing strains, respectively. Information for the construction of the map was obtained from sequences deposited in GenBank (accession numbers X16829, X53955, and X53956). (B) Physical map of the leukotoxin promoter region. The portion of the sequence missing in high leukotoxin-producing strains is indicated by the double-headed arrow. The binding sites of the PCR primers, LKTP-1 and LKTP-2, used to detect the deleted region, are shown. The boxed HindIII site is the same one designated in panel A. The proposed transcriptional start sites are labeled P1 and P2. Information for the construction of the map was obtained from reference 10.
The PCR was also used to identify the leukotoxin promoter deletion. Approximately 100 pg of total DNA and 100 pmol of the primers 5′-GGAGTCGACTTGAGAAATATGACAGT-3′ (LKTP-1) and 5′-GGCGAATTCTCTATGCAAAGGAGAAT-3′ (LKTP-2) (Nucleic Acid Facility, University of Pennsylvania) were used. The primers flank the promoter region and are homologous to positions −1002 to −1018 and +56 to +72, respectively, in the leukotoxin operon (position +1 is the first base of the lktC start codon) (Fig. 1B). Recognition sites (underlined bases) for the restriction endonucleases SalI and EcoRI were incorporated into LKTP-1 and LKTP-2, respectively, to facilitate future cloning experiments. PCR reactions were performed in 50 or 100 μl volumes using a PCR reagent kit† and DNA Polymerase.‡ Reagent concentrations used were the same as recommended in the manufacturer's instructions. Reactions were run in a DNA thermal cycler.§ The samples were heated at 97°C for 4 minutes and amplifications were performed at 95°C for 1 minute, 55°C for 1 minute and 72°C for 1 minute for 25 cycles. The amplified products were extended at 72°C for 10 minutes. Twenty μl of the amplified products were analyzed on 1% agarose gels in 0.5× TBE buffer [1×x TBE buffer contains 89 mM Tris base, 89 mM boric acid and 2.5 mM EDTA (pH 8.3)] and were detected by staining with 0.5 μg/ml of ethidium bromide. Lambda DNA digested with HindIII and HindIII/EcoRI was run on the gels as molecular size standards. A single PCR product of approximately 560 or 1050 bp was observed if the isolate contained or lacked, respectively, the leukotoxin promoter deletion (Fig. 1B).
Statistical Methods
A total of 58 subjects, 24 LJP-susceptible and 34 control, were analyzed. The Fisher's exact test was used to evaluate differences in distributions between LJP-susceptible subjects and control subjects who converted to LJP and those who remained healthy. This test was also used to assess differences in distribution between the families that had LJP-susceptible children convert to LJP during the course of the study and those families that did not. Odds ratios were calculated to evaluate the risk that subjects containing the high leukotoxin-producing variant would convert from a healthy to diseased periodontal status.23
RESULTS
The first objective was to establish if an association was evident between the detection of A. actinomycetemcomitans and conversion from a healthy to diseased periodontal status in a geographically homogeneous LJP family population. To more clearly present the data from these analyses, the 21 LJP families used in the study were divided into two categories. The first category, listed in Table 1, contained members of the 7 LJP families that had at least one child convert from a healthy to diseased periodontal state. The second category, listed in Table 2, contained all members of the 14 LJP families in which no child converted from health to disease. A. actinomycetemcomitans was detected in at least one member of each of the conversion subject families included in Table 1. The bacterium was not detected in members of 3 of the families shown in Table 2. Differences between families that had at least one new case of LJP (Table 1) and those that did not (Table 2), with respect to the detection of A. actinomycetemcomitans, were not statistically significant.
Table 1.
Leukotoxin Promoter Deletion Analysis of Isolates of A. actinomycetemcomitans From LJP Families Having 1 or More Conversion Subjects
| LJP Family (N = 7) | Subject (N = 27) | Familial Relation | Age at Baseline* | Race† | Clinical Status | RFLP of Isolate‡ | Hybridization-positive lkt DNA Fragment (kb)§ | lkt PCR Product (bp)∥ |
|---|---|---|---|---|---|---|---|---|
| 6 | 060025 | Father | 34 | AA | NA | XI | 2.3 | 1050 |
| 6 | 060026 | Mother | 34 | AA | Proband | IV | ND | 1050 |
| 6 | 060027¶ | Son | 13 | AA | Conversion | IV | 2.3 | 1050 |
| 6 | 060028¶ | Son | 9 | AA | Healthy | IV | ND | 1050 |
| 6 | 060029 | Son | 1 | AA | Healthy | IV | ND | 1050 |
| 7 | 070030 | Mother | 29 | AA | Proband | None | NA | NA |
| 7 | 070031¶ | Daughter | 12 | AA | Conversion | II | 8.4 | 560 |
| 7 | 070032¶ | Daughter | 6 | AA | Healthy | None | NA | NA |
| 7 | 070084¶ | Son | <1 | AA | Healthy | None | NA | NA |
| 10 | 100039 | Mother | 25 | AA | Proband | II | 8.4 | 560 |
| 10 | 100040¶ | Son | 11 | AA | Conversion | II | 8.4 | 560 |
| V | 2.3 | 1050 | ||||||
| 10 | 100054 | Father | 40 | AA | NA | VI | 2.3 | 1050 |
| 17 | 170070 | Mother | 34 | AA | NA | ND | NA | NA |
| 17 | 170072 | Daughter | 14 | AA | Proband | None | NA | NA |
| 17 | 170073¶ | Son | 11 | AA | Conversion | None | NA | NA |
| 17 | 170074¶ | Son | 9 | AA | Conversion | II | 8.4 | 560 |
| 18 | 180075 | Mother | 27 | AA | Proband | V | ND | 1050 |
| 18 | 180076¶ | Son | 10 | AA | Conversion | V | ND | 1050 |
| 21 | 210090 | Mother | 38 | AA | NA | None | NA | NA |
| 21 | 210091 | Son | 16 | AA | Proband | II | 8.4 | 560 |
| 21 | 210092¶ | Son | 12 | AA | Conversion | II | 8.4 | 560 |
| ND | NA | NA | ||||||
| 23 | 230125 | Mother | 33 | AA | NA | IX | ND | 1050 |
| 23 | 230124 | Daughter | 9 | AA | Proband | II | 8.4 | 560 |
| 23 | 230126¶ | Son | 9 | AA | Conversion | VIII | 2.3 | 1050 |
| II | 8.4 | 560 | ||||||
| 23 | 230127¶ | Daughter | 11 | AA | Conversion | IX | ND | 1050 |
| VI | ND | 1050 | ||||||
| 23 | 230128 | Father | 35 | AA | NA | ND | NA | NA |
| 23 | 230129 | Daughter | 18 | AA | Healthy | None | NA | NA |
The time at which the subject entered the study is defined as the baseline.
A, Asian; AA, African-American; C, Caucasian; O, other.
Determined on Southern blots using a 4.7 kb EcoRl DNA fragment cloned from strain Y4.15 ND, not determined; these subjects were not included in the statistical analysis due to the lack of microbiological data.
Determined by RFLP analysis using HindIII-digested DNA and an oligonucleotide (LKTA) homologous to the 5′-end of lktA gene. ND, not determined. NA, not applicable.
Amplified using primers LKTP-1 and LKTP-2.
LJP-susceptible subjects used in the statistical analysis (see Table 4).
Table 2.
Leukotoxin Promoter Deletion Analysis of Isolates of A. actinomycetemcomitans from LJP Families Having No Conversion Subjects
| LJP Family (N = 14) | Subject (N = 56) | Familial Relation | Age at Baseline* | Race† | Clinical Status | RFLP of Isolate‡ | Hybridization-positive lkt DNA Fragment (kb)§ | lkt PCR Product (bp)∥ |
|---|---|---|---|---|---|---|---|---|
| 1 | 010001 | Father | 41 | A | NA | V | ND | 1050 |
| 1 | 010002 | Mother | 41 | A | NA | III | ND | 1050 |
| V | ND | 1050 | ||||||
| 1 | 010003 | Daughter | 16 | A | Proband | III | 2.3 | 1050 |
| V | 2.3 | 1050 | ||||||
| 1 | 010004¶ | Son | 13 | A | Healthy | VII | 2.3 | 1050 |
| 2 | 020005 | Grandmother | 31 | AA | NA | None | NA | NA |
| 2 | 020006 | Daughter | 16 | AA | Proband | X | 2.3 | 1050 |
| 2 | 020007 | Son | 15 | AA | Proband | II | 8.4 | 560 |
| 2 | 020008 | Daughter | 11 | AA | Healthy | ND | NA | NA |
| 2 | 020050 | Son | <1 | AA | Healthy | None | NA | NA |
| 3 | 030009 | Father | 39 | C | Proband | III | ND | 1050 |
| 3 | 030010 | Mother | 36 | C | NA | III | ND | 1050 |
| 3 | 030011 | Son | 16 | C | Healthy | None | NA | NA |
| 3 | 030012 | Son | 12 | C | Healthy | ND | NA | NA |
| 4 | 040013 | Grandmother | 42 | AA | NA | XI | ND | 1050 |
| 4 | 040014 | Daughter | 25 | AA | NA | None | NA | NA |
| 4 | 040015 | Granddaughter | 3 | AA | Healthy | IV | ND | 1050 |
| 4 | 040016¶ | Grandson | 6 | AA | Healthy | IV | ND | 1050 |
| IX | ND | 1050 | ||||||
| 4 | 040017 | Daughter | 25 | AA | Proband | III | ND | 1050 |
| XI | ND | 1050 | ||||||
| 4 | 040018 | Granddaughter | 2 | AA | Healthy | IV | ND | 1050 |
| 4 | 040019 | Grandson | <1 | AA | Healthy | None | NA | NA |
| 4 | 040020 | Daughter | 16 | AA | Healthy | None | NA | NA |
| 4 | 040021¶ | Daughter | 13 | AA | Healthy | XI | ND | 1050 |
| 5 | 050022 | Mother | 30 | C | Proband | None | NA | NA |
| 5 | 050023 | Father | 29 | C | NA | None | NA | NA |
| 5 | 050024¶ | Daughter | 8 | C | Healthy | None | NA | NA |
| 8 | 080033 | Father | 28 | AA | Proband | XII | ND | 1050 |
| 8 | 080034¶ | Daughter | 7 | AA | Healthy | IV | ND | 1050 |
| V | ND | 1050 | ||||||
| 8 | 080056 | Mother | 30 | AA | NA | IV | ND | 1050 |
| 9 | 090035 | Mother | 47 | AA | NA | None | NA | NA |
| 9 | 090036¶ | Daughter | 15 | AA | Healthy | None | NA | NA |
| 9 | 090037¶ | Son | 12 | AA | Healthy | None | NA | NA |
| 9 | 090038 | Son | 18 | AA | Proband | II | 8.4 | 560 |
| 11 | 110041 | Mother | 36 | AA | NA | None | NA | NA |
| 11 | 110042 | Daughter | 16 | AA | Proband | None | NA | NA |
| 11 | 110043¶ | Son | 8 | AA | Healthy | IV | ND | 1050 |
| 12 | 120044 | Mother | 36 | AA | NA | None | NA | NA |
| 12 | 120045 | Son | 15 | AA | Proband | II | 8.4 | 560 |
| 12 | 120046 | Son | 8 | AA | Healthy | ND | NA | NA |
| 12 | 120047 | Son | 3 | AA | Healthy | IV | ND | ND |
| 13 | 130048 | Mother | 29 | AA | Proband | None | NA | NA |
| 13 | 130049¶ | Son | 8 | AA | Healthy | None | NA | NA |
| 13 | 130055 | Aunt | 27 | AA | NA | None | NA | NA |
| 14 | 140052 | Son | 14 | AA | Proband | II | 8.4 | 560 |
| 14 | 140053¶ | Son | 12 | AA | Healthy | II | 8.4 | 560 |
| 16 | 160066 | Mother | 39 | C | Proband | None | NA | NA |
| 16 | 160067 | Father | 37 | C | NA | None | NA | NA |
| 16 | 160068¶ | Son | 8 | C | Healthy | None | NA | NA |
| 16 | 160069 | Son | 3 | C | Healthy | None | NA | NA |
| 19 | 190077 | Mother | 31 | AA | NA | XII | ND | 1050 |
| 19 | 190078¶ | Son | 10 | AA | Healthy | II | 8.4 | 560 |
| XII | 2.3 | 1050 | ||||||
| XII | 2.3 | 1050 | ||||||
| 19 | 190079 | Daughter | 14 | AA | Proband | II | 8.4 | 560 |
| XII | ND | 1050 | ||||||
| 20 | 200085¶ | Daughter | 12 | AA | Healthy | II | 2.3 | 1050 |
| 20 | 200086 | Grandmother | 33 | AA | NA | ND | NA | NA |
| 20 | 200087 | Daughter | 3 | AA | Healthy | II | 2.3 | 1050 |
| 20 | 200088 | Daughter | 16 | AA | Proband | II | 2.3 | 1050 |
| 20 | 200089 | Grandson | <1 | AA | Healthy | IX | 2.3 | 1050 |
The time at which the subject entered the study is defined as the baseline.
A, Asian; AA, African-American; C, Caucasian; O, other.
Determined on Southern blots using a 4.7 kb EcoRl DNA fragment cloned from strain Y4.15 ND, not determined; these subjects were not included in the statistical analysis due to the lack of microbiological data.
Determined by RFLP analysis using HindIII-digested DNA and an oligonucleotide (LKTA) homologous to the 5′-end of lktA gene. ND, not determined. NA, not applicable.
Amplified using primers LKTP-1 and LKTP-2.
LJP-susceptible subjects used in the statistical analysis (see Table 4).
The 35 children who served as control subjects from non-LJP families are listed in Table 3. The presence or absence of A. actinomycetemcomitans could not be confirmed in one of the control subjects. Likewise, 3 of the LJP-susceptible subjects were not available for microbiological sampling. Therefore, among the 24 LJP-susceptible subjects (Tables 1 and 2) and 34 control subjects (Table 3) available for study, 12 subjects developed disease (9 LJP-susceptible and 3 control children). Actinobacillus actinomycetemcomitans was detected in 30 of the 58 children (Table 4). There were no statistically significant differences between the proportion of subjects who developed LJP and those that did not, with respect to the detection of the bacterium either in LJP-susceptible subjects, the control group or the total sample (Table 5).
Table 3.
Leukotoxin Promoter Deletion Analysis of Isolates of A. actinomycetemcomitans From Control Subjects
| Subject (N = 35) | Gender | Age at Baseline* | Race† | Clinical Status | RFLP of Isolate‡ | Hybridization-positive Ikt DNA Fragment (kb)§ | lkt PCR Product (bp)∥ |
|---|---|---|---|---|---|---|---|
| 1020064 | Female | 11 | AA | Healthy | XIII | 2.3 | 1050 |
| 1020102 | Female | 11 | AA | Healthy | XIII | 2.3 | 1050 |
| 1030093 | Male | 11 | C | Healthy | None | NA | NA |
| 1030094 | Male | 11 | C | Healthy | XIII | 2.3 | 1050 |
| 1040133 | Male | 7 | AA | Healthy | XIII | 2.3 | 1050 |
| 1040123 | Male | 7 | AA | Healthy | None | NA | NA |
| 1060097 | Male | 13 | O | Conversion | None | NA | NA |
| 1060096 | Male | 8 | O | Healthy | None | NA | NA |
| 1070117 | Female | 12 | AA | Healthy | None | NA | NA |
| 1070099 | Female | 7 | AA | Healthy | V | ND | 1050 |
| XIII | 2.3 | 1050 | |||||
| 1070101 | Female | 6 | AA | Healthy | None | NA | NA |
| 1080081 | Female | 7 | AA | Healthy | XIII | 2.3 | 1050 |
| 1080113 | Female | 9 | AA | Healthy | VI | ND | 1050 |
| IX | ND | 1050 | |||||
| 1090063 | Female | 13 | AA | Healthy | XII | ND | 1050 |
| XIII | 2.3 | 1050 | |||||
| XIV | 2.3 | 1050 | |||||
| 1090100 | Female | 14 | AA | Conversion | III | ND | 1050 |
| 1090109 | Female | 14 | AA | Healthy | None | NA | NA |
| 1090111 | Male | 12 | AA | Healthy | None | NA | NA |
| 1100062 | Male | 11 | AA | Healthy | None | NA | NA |
| 1100119 | Male | 11 | AA | Healthy | XII | ND | 1050 |
| 1110112 | Male | 8 | AA | Healthy | XII | ND | 1050 |
| XIII | 2.3 | 1050 | |||||
| 1120114 | Male | 8 | AA | Healthy | ND | NA | NA |
| 1130107 | Male | 9 | AA | Healthy | XIII | 2.3 | 1050 |
| 1140098 | Male | 14 | O | Conversion | None | NA | NA |
| 1140108 | Male | 12 | AA | Healthy | XIV | 2.3 | 1050 |
| 1160095 | Male | 7 | C | Healthy | None | NA | NA |
| 1160122 | Male | 9 | C | Healthy | None | NA | NA |
| 1170110 | Male | 11 | AA | Healthy | None | NA | NA |
| 1170120 | Male | 9 | O | Healthy | None | NA | NA |
| 1170134 | Male | 10 | AA | Healthy | None | NA | NA |
| 1180115 | Male | ND | AA | Healthy | None | NA | NA |
| 1190121 | Male | 11 | AA | Healthy | None | NA | NA |
| 1200131 | Female | 12 | AA | Healthy | None | NA | NA |
| 1210118 | Male | 13 | AA | Healthy | None | NA | NA |
| 1230132 | Male | 10 | AA | Healthy | None | NA | NA |
| 1230130 | Female | 11 | O | Healthy | None | NA | NA |
The time at which the subject entered the study is defined as the baseline.
A, Asian; AA, African-American; C, Caucasian; O, other.
Determined on Southern blots using a 4.7 kb EcoRI DNA fragment cloned from strain Y4.15 ND, not determined; these subjects were not included in the statistical analysis due to the lack of microbiological data.
Determined by RFLP analysis using HindIII-digested DNA and an oligonucleotide (LKTA) homologous to the 5′ -end of lktA gene. ND, not determined. NA, not applicable.
Amplified using primers LKTP-1 and LKTP-2.
LJP-susceptible subjects used in the statistical analysis (see Table 4).
Table 4.
Percentage of Subjects Containng A. actinomycetemcomitans and Variants With the Leukotoxin (lkt) Promoter Deletion
| Category | A. actinomycetemcomitans | lkt Promoter Region Deletion |
|---|---|---|
| LJP-susceptible subjects from families with conversion subjects* | 81.8(9/11) | 54.5(6/11) |
| LJP-susceptible subjects from families without conversion subjects† | 61.5 (8/13) | 15.4 (2/13) |
| Probands‡ | 72.7 (16/22) | 31.8 (7/22) |
| Control subjects§ | 38.2(13/34) | 0 (0/34) |
Table 5.
Distribution of LJP-Susceptible and Control Subjects According to Detection of A. actinomycetemcomitans and Conversion From a Healthy to Diseased Periodontal Status
Since there was no association between the presence of A. actinomycetemcomitans and the conversion of a subject from a healthy to LJP status, at either the family or subject level, we proceeded to determine if an association existed between specific genotypes of the bacterium and the development of disease. DNA hybridization and PCR were used to distinguish between A. actinomycetemcomitans isolates containing and lacking a deletion in the promoter region of the leukotoxin operon representative of high and low leukotoxin-producing genotypes, respectively. The results are shown in Tables 1 through 3. Our early experiments employed the DNA hybridization method since a strain variable HindIII site in the leukotoxin locus had been reported.3 Later investigations showed that this HindIII site was variable because it resided within the region encompassed by the leukotoxin promoter deletion. This meant that PCR could be used as the primary method for the analysis of high leukotoxin-producing strains.
The genetic variant containing the leukotoxin promoter region deletion was found in members of 10 of 21 LJP families as evidenced by a 8.4 kb hybridization-positive DNA fragment and a 560 bp PCR product (Tables 1 and 2). Five of these families had at least one LJP-susceptible child convert to disease. Two of 11 LJP families that did not have members infected with A. actinomycetemcomitans containing the leukotoxin promoter deletion had a member who converted to LJP.
Eight of 24 LJP-susceptible subjects and none of the 34 control subjects had A. actinomycetemcomitans with the leukotoxin promoter deletion genotype. Six of these 8 subjects converted to disease during the study period. Three LJP-susceptible and 3 control children who did not have genetic variants containing the leukotoxin promoter region deletion converted from a healthy to periodontally diseased status. There was a statistically significant association between the detection of genetic variants with the promoter deletion and conversion to LJP among the LJP-susceptible subjects as well as in the total sample (Table 6). Those subjects that had A. actinomycetemcomitans with the leukotoxin promoter deletion were at higher risk to develop LJP (odds ratio = 22.5).
Table 6.
Distribution of LJP-Susceptible and Control Subjects According to Detection of the Leu-kotoxin (lkt) Promoter Deletion Variant and Conversion From a Healthy to Diseased Periodontal Status
Examination of A. actinomycetemcomitans isolates from the probands of the LJP families revealed that among 22 probands (2 probands in family 2), 16 had the bacterium and 7 of these contained the genotype with the leukotoxin promoter deletion (Table 4). Two of these 7 probands resided in LJP families that had an LJP-susceptible child convert from a healthy to diseased periodontal status. No significant association between the presence of the high leukotoxin-producing variant in the proband and conversion to disease of a LJP-susceptible sibling was observed (P > 0.05).
Seventeen of the 21 LJP families studied were African-American. This high proportion of African-American subjects in our study population precluded a statistical determination of an association between the presence of the high leukotoxin-producing genotype and disease on the basis of race. All 7 LJP families that had a conversion subject were African-American (Table 1). Eight of 46 African-American subjects harbored the promoter deletion variant and 6 of these 8 subjects developed LJP. In addition, 4 of 37 subjects who did not contain the promoter deletion variant, also converted to a diseased status. Two of the control subjects who converted to LJP did not have cultivable A. actinomycetemcomitans and were of undetermined ancestry (Table 3). The third control subject who converted to disease contained a genetic variant with the full length promoter region and was African-American.
Without exception, the A. actinomycetemcomitans isolates that exhibited the leukotoxin promoter deletion were members of RFLP group II (Tables 1 and 2). This genetic variant was not found in any of the control subjects (Table 3). Fifteen of 18 of the RFLP group II isolates contained the promoter deletion. The remaining 3 isolates that had the full length promoter region were obtained from the proband and each of two LJP-susceptible children in the same non-conversion LJP family (family 20).
DISCUSSION
We have shown that isolates of A. actinomycetemcomitans that have a deletion in the promoter region of the leukotoxin operon are prevalent in young LJP-susceptible children who are members of LJP families. In total, 15 subjects in 10 African-American families harbored this specific genotype and 6 of these were children who converted from a healthy to diseased periodontal status. Eight of the 15 subjects were probands infected with the specific promoter deletion genotype. This demonstrates a relationship between the leukotoxin promoter deletion and disease even in the non-conversion LJP families.
Actinobacillus actinomycetemcomitans was detected in 8 of 9 LJP-susceptible subjects who converted to LJP during the study (89%) and in 1 of 3 control subjects who exhibited attachment loss. However, these conversions in the control subjects appeared to be atypical since they occurred at a later age than those in the LJP-susceptible subjects.14 Despite the association between A. actinomycetemcomitans and LJP reported by others, and as shown here, there was no statistical association between conversion to disease and the presence of the bacterium.3,24,25 This outcome was most likely due to the frequent isolation of A. actinomycetemcomitans from the healthy subjects. Nine of 15 healthy subjects belonging to LJP families (60%) and 12 of 31 healthy control subjects (38.7%) harbored the bacterium. The percentage of healthy subjects positive for A. actinomycetemcomitans in this study was higher than that reported previously.3,24,26 This may have been the result of higher sampling frequencies which increased the chances of detecting the bacterium. Several hypotheses, such as increased host resistance, presence of a neutralizing microflora,26 or presence of a less virulent strain, could explain the presence of A. actinomycetemcomitans in healthy subjects.
There was a statistically significant association between the detection of the high leukotoxin-producing genotype of A. actinomycetemcomitans and the conversion of an LJP-susceptible subject from health to disease. These data indicate that the high leukotoxin-producing genotype of the species has the potential to contribute to the virulence of this genetic variant. It was also observed that the genetic variant with the leukotoxin promoter deletion was detected in only 2 of the probands in the 7 LJP families that had a child convert to disease (families 10 and 21). However, 6 of the 9 conversion subjects from the LJP families harbored this genotype. This difference may have been due to the sampling frequency. Probands were sampled only once a year while the LJP-susceptible subjects were sampled quarterly.14 Also, some of the probands were adults and had been treated for LJP prior to their entry into the study. Treatment may have reduced numbers of A. actinomycetemcomitans in these subjects. The risk of acquiring this bacterium may be even higher in subjects belonging to LJP families in which no member has been treated for the disease.
The virulence potential of the high leukotoxin-producing variant of A. actinomycetemcomitans may not be due solely to the enhanced expression of the leukotoxin genes. We have shown previously that variants of a single specific RFLP group (II), out of 14 different groups, have a statistically significant correlation with the development of LJP in the same subject population.14,15 While it is clear that the high leukotoxin-producing variants are unique members of RFLP group II, it is likely that additional genotypic differences account for the correlation between the members of this RFLP group and LJP. Increased expression of other potential vimlence genes, in addition to the leukotoxin, by the RFLP group II variants may contribute to the association between this variant and conversion from health to disease.
Our results are consistent with those of Zambon et al.12 who found that 57% of LJP subjects contained the high leukotoxin-producing variant while adult Periodontitis and periodontally healthy subjects lacked this variant. Also, there was a significantly higher prevalence of the high leukotoxin-producing strains in our study population than in those examined by Haubek et al.11 These investigators reported that strains that have the leukotoxin promoter deletion are rare. Their conclusion was based on the analysis of a collection of 88 isolates from Finnish dental patients and over 60 strains obtained from northern European patients. However, the strains they examined originated from subjects of varied clinical status including a significant number of healthy individuals and adult Periodontitis patients. Brogan and coworkers10 showed that two strains obtained from the Haubek et al.11 group were highly leukotoxic. Presumably, these strains were of northern European origin. More recently, Haubek et al.13 reported a high prevalence of leukotoxin-producing strains in subjects originating from the Cape Verde Islands, Morocco, and Algeria. Our data are consistent with these findings and support epidemiological studies promoting a higher prevalence of LJP in people of African ancestry than in peoples of other origins.27–29 Since our study population was composed of predominantly African-American families, we cannot show a statistical correlation between the high leukotoxin-producing genotype and LJP based on race. All other races were drastically underrepresented in the study population.
Studies reporting the relatively high prevalence of LJP in peoples of African origins, the data presented by Haubek et al.13 and the results of our investigation suggest that the detection of the leukotoxin promoter deletion by PCR may be a useful diagnostic test for the identification of disease risk subjects in specific highly homogeneous patient populations.
Acknowledgments
We thank Terry McKay for providing able technical assistance during the early stages of the study and Fred Correia for critically reading the manuscript. Dr. Marcia Mayer was supported by grant 1996-3328-0 from the Fundação de Auxilio a Pesquisa de Estado de São Paulo, Brazil. This study was supported by a University of Pennsylvania Research Foundation award and National Institutes of Health grant DE/OD10891.
Footnotes
Department of Microbiology, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA.
GeneAmp PCR Reagent Kit, Perkin Elmer Cetus, Norwalk, CT.
Amplitaq, Perkin Elmer Cetus.
Perkin Elmer Cetus.
REFERENCES
- 1.Moore WEC. Microbiology of periodontal disease. J Periodont Res. 1987;22:335–341. doi: 10.1111/j.1600-0765.1987.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 2.Slots J, Zambon JJ, Rosling BG, Reynolds HS, Christersson LA, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: Association, serology, leukotoxicity, and treatment. J Periodont Res. 1982;17:447–455. doi: 10.1111/j.1600-0765.1982.tb02022.x. [DOI] [PubMed] [Google Scholar]
- 3.Zambon JJ, Christersson LA, Slots J. Actinobacillus actinomycetemcomitans in human periodontal disease: Prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontol. 1983;54:707–111. doi: 10.1902/jop.1983.54.12.707. [DOI] [PubMed] [Google Scholar]
- 4.Poulsen K, Theilade E, Lally ET, Demuth DR, Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiol. 1994;140:2049–2060. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- 5.Taichman NS, McArthur WR, Tsai C-C, et al. Leukocidal mechanisms of Actinobacillus actinomycetemcomitans. In: Genco RJ, Mergenhagen SE, editors. Host-Parasite Interactions in Periodontal Diseases. American Society for Microbiology; Washington, DC: 1982. pp. 261–269. [Google Scholar]
- 6.Taichman NS, Simpson DL, Sakurada S, Cranfield M, DiRienzo J, Slots J. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol Immunol. 1987;2:97–104. doi: 10.1111/j.1399-302x.1987.tb00270.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsai C-C, Taichman NS. Dynamics of infection by leucotoxic strains of A. actinomycetemcomitans in juvenile Periodontitis. J Clin Periodontol. 1986;11:330–331. doi: 10.1111/j.1600-051x.1986.tb02231.x. [DOI] [PubMed] [Google Scholar]
- 8.Kraig E, Dailey Kolodrubetz D. Nucleotide sequence of the leukotoxin gene from Actinobacillus actinomycetemcomitans: homology to the alpha-hemolysin/leukotoxin gene family. Infect Immun. 1990;58:920–929. doi: 10.1128/iai.58.4.920-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lally ET, Golub EE, Kieba IR, et al. Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. J Biol Chem. 1989;264:15451–15456. [PubMed] [Google Scholar]
- 10.Brogan JM, Lally ET, Poulsen K, Kilian M, Demuth DR. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect Immun. 1994;62:501–508. doi: 10.1128/iai.62.2.501-508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubek D, Poulsen K, Asikainen S, Kilian M. Evidence for absence in northern Europe of especially virulent clonai types of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1995;33:395–401. doi: 10.1128/jcm.33.2.395-401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zambon JJ, Haraszthy VI, Harihara G, Lally ET, Demuth DR. The microbiology of early-onset Periodontitis: Association of highly toxic Actinobacillus actinomycetemcomitans strains with localized juvenile Periodontitis. J Periodontol. 1996;67(Suppl.):282–290. doi: 10.1902/jop.1996.67.3s.282. [DOI] [PubMed] [Google Scholar]
- 13.Haubek D, Poulsen K, Westergaard J, Dahlèn G, Kilian M. Highly toxic clone of A. actinomycetemcomitans in geographically widespread cases of juvenile Periodontitis in adolescents of African origin. J Clin Microbiol. 1996;34:1576–1578. doi: 10.1128/jcm.34.6.1576-1578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRienzo JM, Slots J, Sixou M, Sol M-A, Harmon R, McKay TL. Specific genetic variants of A. actinomycetemcomitans correlate with disease and health in a regional population of localized juvenile Periodontitis families. Infect Immun. 1994;62:3058–3065. doi: 10.1128/iai.62.8.3058-3065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiRienzo JM, McKay TL. Identification and characterization of genetic cluster groups of A. actinomycetemcomitans isolated from the human oral cavity. J Clin Microbiol. 1994;32:75–81. doi: 10.1128/jcm.32.1.75-81.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaty TH, Boughman JA, Yang R, Astemborski JA, Suzuki JB. Genetic analysis of juvenile Periodontitis in families ascertained through an affected proband. Am J Hum Genet. 1987;40:443–452. [PMC free article] [PubMed] [Google Scholar]
- 17.Saxén L. Juvenile Periodontitis. J Clin Periodontol. 1980;7:1–19. doi: 10.1111/j.1600-051x.1980.tb01944.x. [DOI] [PubMed] [Google Scholar]
- 18.Rosling B, Nyman S, Lindhe J. The effect of systemic plaque control on bone regeneration in infrabony pockets. J Clin Periodontol. 1976;3:38–53. doi: 10.1111/j.1600-051x.1976.tb01849.x. [DOI] [PubMed] [Google Scholar]
- 19.Möller ARJ. Microbial examination of root canals and periapical tissues of human teeth. Odontol Tidskr. 1966;74:1–38. [PubMed] [Google Scholar]
- 20.Slots J. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1982;15:606–609. doi: 10.1128/jcm.15.4.606-609.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stull TL, LiPuma JJ, Edlind TD. A broad-spectrum probe for the molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988;167:280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd ed Vol. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. pp. 11.31–11.34. [Google Scholar]
- 23.Greenberg RS, Daniels SR, Flanders WD, Eley JW, Boring JR. Medical Epidemiology. 2nd ed Appleton & Lange; Norwalk CT: 1996. [Google Scholar]
- 24.Genco RJ, Zambon JJ, Christersson LA. Use and interpretation of microbiological assays in periodontal diseases. Oral Microbiol Immunol. 1986;1:73–79. doi: 10.1111/j.1399-302x.1986.tb00324.x. [DOI] [PubMed] [Google Scholar]
- 25.Asikainen S, Jousimes-Somer H, Kanervo A, Summanen P. Certain bacterial species and morphotypes in localized juvenile Periodontitis and in matched controls. J Periodontol. 1987;58:224–230. doi: 10.1902/jop.1987.58.4.224. [DOI] [PubMed] [Google Scholar]
- 26.Asikainen S, Alaluusua S, Kleemola-Kujala E. A 2-year follow-up on the clinical and microbiological condition of periodontium in teenagers. J Clin Periodontol. 1991;18:16–19. doi: 10.1111/j.1600-051x.1991.tb01113.x. [DOI] [PubMed] [Google Scholar]
- 27.Harley AF, Floyd PD. Prevalence of juvenile Periodontitis in school-children in Lagos, Nigeria. Community Dent Oral Epidemiol. 1988;16:299–301. doi: 10.1111/j.1600-0528.1988.tb01780.x. [DOI] [PubMed] [Google Scholar]
- 28.Löe H, Brown LJ. Early-onset Periodontitis in the United States of America. J Periodontol. 1991;62:608–616. doi: 10.1902/jop.1991.62.10.608. [DOI] [PubMed] [Google Scholar]
- 29.Saxby M. Prevalence of juvenile Periodontitis in a British school population. Community Dent Oral Epidemiol. 1984;12:185–187. doi: 10.1111/j.1600-0528.1984.tb01435.x. [DOI] [PubMed] [Google Scholar]