Abstract
The HIV-1 genome is highly heterogeneous. This variation affords the virus a wide range of molecular properties, including the ability to infect cell types, such as macrophages and lymphocytes, expressing different chemokine receptors on the cell surface. In particular, R5 HIV-1 viruses use CCR5 as a coreceptor for viral entry, X4 viruses use CXCR4, whereas some viral strains, known as R5X4 or D-tropic, have the ability to utilize both coreceptors. X4 and R5X4 viruses are associated with rapid disease progression to AIDS. R5X4 viruses differ in that they have yet to be characterized by the examination of the genetic sequence of HIV-1 alone. In this study, a series of experiments was performed to evaluate different strategies of feature selection and neural network optimization. We demonstrate the use of artificial neural networks trained via evolutionary computation to predict viral coreceptor usage. The results indicate the identification of R5X4 viruses with a predictive accuracy of 75.5 percent.
Index Terms: Computational intelligence, evolutionary computation, artificial neural networks, HIV, AIDS, phenotype prediction, tropism, dual-tropic viruses
1 Introduction
A major challenge in the study of rapidly evolving viruses is the development of tools that can manage the unparalleled amount and complexity of genetic data. Such data contain large numbers of variables for thousands of nucleotide and/or amino acid sequences, each with their own properties and host immunological and clinical information. In addition, these data may not be independent, rather they may share an evolutionary history which should be included in scientific analysis and understanding. Accurate data mining of this information, usually based on analytic or heuristic methods, has the potential to improve medical therapies or result in the development of new theories on viral evolution.
There are many avenues for data mining. Simple statistical approaches commonly assume linearity in relating these parameters to predictions of viral phenotype. This works well when the parameters of concern are truly linear. However, features regarding biological processes are rarely linearly separable. As a result, heuristic methods based on linear models can potentially miss important relationships. Machine learning approaches that can handle both linear and nonlinear relationships are required.
Artificial neural networks (ANNs) are typically used to map input features to output decisions over a set of known examples in a database. For example, the input can be statistical features about a DNA sequence region, with the output being a decision concerning the likelihood of a particular nucleotide sequence residing in a coding or noncoding region. When example patterns are available for training, multilayer perceptrons (also sometimes described as feed-forward networks) are perhaps the most common ANN architecture used in supervised learning applications. For a given ANN architecture (that is, the type of network, the number of nodes in each layer, the connections between the nodes, and so forth) and a training set of input patterns, the collection of variable weights associated with all connections determines ANN response to each presented input pattern. The error between the actual output of the ANN and the desired target output defines a response surface over an N-dimensional hyperspace, where there are N parameters (for example, weights) to be adapted.
There are numerous approaches for ANN optimization in light of the above description. For example, back-propagation [1] implements a gradient search over the error response surface for the set of weights that minimizes the sum of the squared error between the actual and target values. Although this is a common approach in ANN optimization, it can only provide guaranteed convergence to a locally optimal solution. Even if the network’s topology provides sufficient complexity to completely solve the given pattern recognition task, the backpropagation method may be incapable of discovering an appropriate set of weights to accomplish the task.
A different approach for ANN optimization utilizes simulated evolution to discover useful models [2], [3], [4]. Natural evolution provides inspiration for algorithms that mimic random variation and selection as a means for discovering ingenious solutions to complex problems that are characterized by temporal and stochastic processes. Evolving neural networks offers not only a superior search for appropriate network parameters but can also be used to adjust ANN topology simultaneously. By mutating both the structure of the ANN and its associated parameters and coupling this variation with a process of model selection, a very fast examination of the possible model space can be made for a truly robust design. When simulated evolution has been used to train neural networks, the results have been superior to other ANN optimization methods [5], [6]. These same approaches have even been used to evolve models that meet or exceed human expert performance [7], [8].
1.1 HIV-1 Coreceptor Usage and Artificial Neural Networks in HIV Research
Models capable of examining statistics regarding viral sequences and translating these into predictions of HIV-1 coreceptor usage have many relevant biomedical applications. For example, infection of target cells by HIV-1 requires binding of the viral surface protein gp120 to the cellular surface protein CD4 and chemokine receptors CCR5 or CXCR4 [9]. Coreceptor usage can determine a virus’ behavior both in vitro and in vivo. Viruses that utilize the CCR5 coreceptor (R5 viruses) do not induce syncytia and primarily infect macrophages, but can also infect lymphocytes. On the other hand, viruses that utilize the CXCR4 coreceptor (X4 viruses) induce syncytia in transformed CD4+ cells and have the ability to infect lymphocytes and T-cells lines [10]. Additionally, there exists a subset of viruses that can utilize both the CCR5 and CXCR4 coreceptors and infect macrophages, as well as lymphocytes and T-cell lines; these viruses are usually identified as D-tropic (or otherwise called R5X4). The emergence of R5X4 and/or X4 viral variants during the course of the infection is associated with rapid progression to AIDS in about 50 percent of HIV-1 subtype B infected patients [11], in line with their accelerated replication rates in vitro [12], [13], whereas R5 viruses are associated with early infection and infection of the central nervous system [14]. Coreceptor usage also modulates viral access to various compartments within the human body due to tissue-specific cellular characteristics. For example, HIV infection in the brain is only associated with R5 viruses, whereas the thymus is mainly populated by X4 (T-tropic) variants [15], [16]. The majority of HIV-positive individuals are initially infected with R5 strains, suggesting that there may be a selective advantage of such variants with respect to transmission [17]. However, the recent report of an individual infected with a multidrug resistant, D-tropic, virus followed by rapid advancement to AIDS and death is alarming [18] and underscores the necessity for models that can successfully predict viral phenotype from genotype.
Neural networks have been applied recently to HIV coreceptor usage prediction with success limited largely by the manner in which these methods have been employed [19], [20], [21], [22], [23]. For example, Resch et al. [19] utilized neural networks to predict HIV-1 coreceptor usage from envelope V3 loop sequences. Bayesian regulation modification of backpropagation was used for neural network training with an empirically determined threshold for prediction of X4 viral strains. As mentioned previously, the use of backpropagation may be limiting the discovery of the most useful neural network models. Loannidis et al. [20] utilized ANNs optimized via backpropagation for HIV lipodystrophy prediction. Results were compared against logistic regression models using the same information that was presented to the ANN. In this case, the ANNs demonstrated improved performance over logistic regression models when using a receiver-operator characteristic (ROC) curve area as a performance metric, but only slightly. Brumme et al. [22] utilized neural networks, although the methods of neural network training were not well described. Other studies have used neural networks to predict HIV resistance to drugs such as lopinavir [21] and these are not reviewed herein as they are not directly related to prediction of coreceptor usage.
1.2 Evolved Neural Networks for Coreceptor Usage Prediction
HIV-1 coreceptor usage is determined either experimentally in the laboratory or inferred by examining the charged positions and overall charge of the V3 loop region in the envelope protein [23]. Although prediction of CCR5 or CXCR4 coreceptor usage based on charge for V3 loop residues may be > 80 percent accurate [24], [25], [26], [27], [28], it leaves room for 20 percent improvement in terms of predictive accuracy, with insufficient confidence in what other factors are involved in natural coreceptor usage determination. More importantly, a prediction algorithm able to identify R5X4 strains has not yet been described. ANNs optimized by backpropagation have been employed to predict HIV coreceptor usage [19]. However, a mean reliability of predicting X4 sequences of 69 percent is considered insufficient for use in a clinical setting despite the 80 percent sensitivity and 89 percent specificity of the best neural network on the testing examples for the decision of R5 versus X4. In this paper, we demonstrate the use of evolved neural networks to predict HIV-1 coreceptor usage, including R5X4 viral strains. As shown below, such a method not only allows us to increase the prediction of R5 and X4 viruses to a mean predictive accuracy of 88.9 percent, but it is also able, for the first time, to predict R5X4 HIV-1 variants with 75.5 percent accuracy.
2 Materials and Methods
2.1 Data Collection and Feature Generation
There were 149 sequence isolates for the HIV V3 loop representing three experimentally determined viral tropisms (77 R5, 31 R5X4, and 41 X4 sequences) from a variety of HIV subtypes identified from the Los Alamos National Laboratory HIV Sequence Database (www.hiv.lanl.gov/content/hiv-db/main-page.html) and downloaded to a local computer. The accession numbers for these sequences are provided in Table 1. Sampling bias was minimized by ensuring that the sequences did not originate from similar sources or studies. V3-loop sequence contained 35 amino acid positions. Alignments were performed by hand to maximize homology between sequences [28]. Gaps in the alignment were treated as positions where no information was available and were assigned a quantitative value of 0. Amino acid positions 1, 3, 26, and 38 were removed from the alignment because they were invariant and, thus, uninformative for the purpose of classification.
TABLE 1.
Accession Numbers for Sequences of Different Tropisms
HIVbase software [29] was used to calculate nine statistics per position (for example, amino acid type, Chou-Fasman helix index, Chou-Fasman sheet index, pKa value for free amino acid carboxylate, pKa value for free amino acid amine, volume, polarizability index, charge, and surface index; see Table 2), in addition to two V3-domain level features (isoelectric point and V3-domain total charge) for a total of 317 features. Amino acids can be further grouped into several classes based on overall features, as in Table 3. Given that positional information is known to be important for coreceptor usage [23], we employed a direct encoding method for these statistical features. The above features were chosen because of their relevance and/or statistical correlation with HIV ability to use different coreceptors. The data set was exported to a Microsoft Excel spreadsheet via the HIVbase query engine.
TABLE 2.
Amino Acid Properties
| Amino acid residues | Chemical property | Charge | Volume (A3) | Mass (daltons) | HP Scale | Surface Area | 2D structure propensity
|
||
|---|---|---|---|---|---|---|---|---|---|
| alpha helix | B-strand | Turn | |||||||
| Alanine (A) | aliphatic | 0 | 67 | 71.09 | 1.8 | 0.74 | 1.41 | 0.72 | 0.82 |
| Arginine I | basic | +1 | 148 | 156.19 | −4.5 | 0.64 | 1.21 | 0.84 | 0.90 |
| Asparagine (N) | amide | 0 | 96 | 114.11 | −3.5 | 0.63 | 0.76 | 0.48 | 1.34 |
| Aspartic Acid (D) | acidic | −1 | 91 | 115.09 | −3.5 | 0.62 | 0.99 | 0.39 | 1.24 |
| Cysteine(C) | reactive | 0 | 86 | 103.15 | 2.5 | 0.91 | 0.66 | 1.40 | 0.54 |
| Glutamine (Q) | amide | 0 | 114 | 128.14 | −3.5 | 0.62 | 1.27 | 0.98 | 0.84 |
| Glutamic Acid (E) | acidic | −1 | 109 | 129.12 | −3.5 | 0.62 | 1.59 | 0.52 | 1.01 |
| Glycine (G) | small | 0 | 48 | 57.05 | −0.4 | 0.72 | 0.43 | 0.58 | 1.77 |
| Histidine (H) | aromatic | 0 | 118 | 137.14 | −3.2 | 0.78 | 1.05 | 0.80 | 0.81 |
| Isoleucine (I) | aliphatic | 0 | 124 | 113.16 | 4.5 | 0.88 | 1.09 | 1.67 | 0.47 |
| Leucine (L) | aliphatic | 0 | 124 | 113.16 | 3.8 | 0.85 | 1.34 | 1.22 | 0.57 |
| Lysine (K) | basic | +1 | 135 | 128.17 | −3.9 | 0.52 | 1.23 | 0.69 | 1.07 |
| Methionine (M) | aliphatic | 0 | 124 | 131.19 | 1.9 | 0.85 | 1.30 | 1.14 | 0.52 |
| Phenylalanine (F) | aromatic | 0 | 135 | 147.18 | 2.8 | 0.88 | 1.16 | 1.33 | 0.59 |
| Proline (P) | cyclic imino | 0 | 90 | 97.12 | −1.6 | 0.64 | 0.34 | 0.31 | 1.32 |
| Serine (S) | hydroxyl | 0 | 73 | 87.08 | −0.8 | 0.66 | 0.57 | 0.96 | 1.22 |
| Threonine (T) | hydroxyl | 0 | 93 | 101.11 | −0.7 | 0.70 | 0.76 | 1.17 | 0.90 |
| Tryptophane (W) | aromatic | 0 | 163 | 186.21 | −0.9 | 0.85 | 1.02 | 1.35 | 0.65 |
| Tyrosine (Y) | aromatic | 0 | 141 | 163.18 | −1.3 | 0.76 | 0.74 | 1.45 | 0.76 |
| Valine (V) | aliphatic | 0 | 105 | 99.14 | 4.2 | 0.86 | 0.90 | 1.87 | 0.41 |
Volume = volume enclosed by van der Waals radii. Mass = molecular weight of nonionized amino acid minus that of water, both adopted from [28]. HP scale = degree of hydrophobicity of amino acid side chains, based on [29]. Surface area = mean fraction buried, based on [30]. Secondary structure propensity = normalized frequencies for each conformation, adopted from [28], is the fraction of residues of each amino acid that occurred in that conformation, divided by this fraction for all residues.
TABLE 3.
Amino Acid Membership Classes
| Name | Size | Feature | Membership classes | Numeric Class Conversion |
|---|---|---|---|---|
| Exchange Group | 6 | Conservative substitution | {HRK} {DENQ} {C} {STPAG} {MILV} {FYW} | 1,2,3,4,5,6 |
| Charge Polarity | 4 | Charge and Polarity | {HRK} {DE} {CTSGNQY} {APMLIVFW} | 1,2,3,4 |
| Hydrophobicity | 3 | Hydrophobicity | {DENQRK} {CSTPGHY} {AMILVFW} | 1,2,3 |
| Mass | 3 | Mass | {GASPVTC} {NDQEHILKM} {RFWY} | 1,2,3 |
| Structural | 3 | Surface Exposure | {DENQHRK} {CSTPAGWY} {MILVF} | 1,2,3 |
| 2D Propensity | 3 | 2D Propensity | {AEQHKMLR} {CTIVFYW} {SGPDN} | 1,2,3 |
2.2 Preprocessing of Features
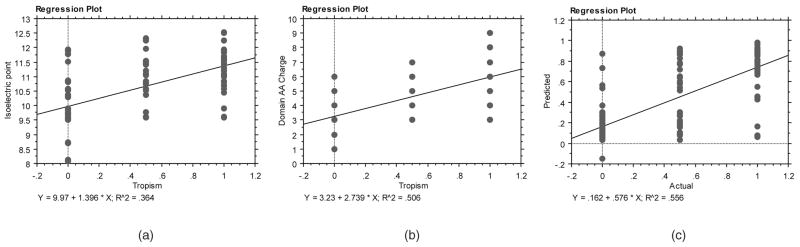
Initial screening of the data indicated that many of the statistical features were invariant across the three coreceptor usage classes (due to invariant amino acid positions in the alignment) and could be removed from further analysis as noninformative in discriminating coreceptor usage. This reduced the total number of features from 317 to 248. Further statistical analysis of the features demonstrated that the two domain-level features (isoelectric point and domain charge) were only poorly correlated with coreceptor usage when taken individually (see Figs. 1a and 1b; R2 values of 0.364 and 0.506, respectively). Despite this, these domain-level features were still incorporated in model development. For the experiments that follow, first the two domain-level features were used as input, followed by a combination of these domain-level features with 28 charge features, followed by affording the evolutionary process itself, determine the appropriate features to use over a prespecified range from the entire set of 250 possible features. This latter method allows for simultaneous weight and topology optimization of the neural network models.
Fig. 1.
Correlation of domain-level features (a) isoelectric point and (b) domain charge with coreceptor usage (x-axis values for presentation purposes only: 0 = R5, 0.5 = R5X4, 1 = X4). (c) Predictions from the best evolved neural networks developed with just these two input features using leave-one-out cross validation over all samples.
2.3 Evolved Neural Networks
Evolved neural networks were used to map the input vectors to coreceptor usage predictions using increasingly more complex feature sets. For additional information on evolved neural networks, the reader is directed to [4], which provides a thorough review of the approach. Unless specified otherwise, for the purpose of developing ANN models, fully connected, feed-forward architectures were used with input, hidden, and output nodes. Leave-one-out cross validation was used over all samples, with an initial step size of 0.1, tournament selection with four opponents, and a population size of 50 parent and 50 offspring neural networks. Evolutionary optimization was employed over a prespecified number of generations. All hidden nodes used a sigmoid activation function. Fitness was measured by taking the mean squared error (MSE) of the prediction from the neural network for all training examples, relative to the actual value for each sample, using the equation
| (1) |
where P is the predicted activity for the kth sample, O is the observed activity for the kth sample, and N is the number of patterns in the training set. MSE was to be minimized over evolutionary optimization.
For the purpose of investigating the utility of using only domain-level features for coreceptor usage prediction, a simple ANN with two inputs, two hidden nodes, and one output node was used for 5,000 generations of evolution. The choice of two hidden nodes was arbitrary but felt to be sufficient in light of only two input nodes and for the purpose of this preliminary investigation. For the purpose of combining these domain-level features with charge features from the V3 loop to discriminate R5X4 sequences from either R5 or X4 sequences, a random selection of 30 inputs, two hidden nodes, and one output node was used over 1,000 generations of evolution. R5X4 sequences were assigned a value of 0, whereas both R5 and X4 sequences were assigned a target value of 1 over all samples (n = 149). Although it is possible that additional hidden nodes would increase the ability of the ANN to discriminate tropism classes, we wished to determine a minimal nonlinear representation capable of doing this task to determine how well such a parsimonious model compared to prior results in the literature.
When using evolved neural networks it is possible to select a subset of features as input over a range of possible features and make the selection of which inputs to use subject to evolutionary variation simultaneously with weight optimization. This approach leads to simultaneous feature reduction and optimal model development. To discriminate R5 and X4 sequences (n = 118), 30 features were allowed as possible input. A subsequent series of seven experiments was conducted, forcing the number of inputs to the ANN to be 2, 5, 10, 15, 20, 25, or 30 features out of the space of possible features chosen at random. For each of these experiments, the choice of precisely which features to use as input was itself subject to evolutionary optimization concurrent with weight assignment optimization.
As a final test of the process, eight new sequences were added to the data and the data was divided randomly into 127 training samples and 30 testing samples. The number of inputs used by each neural network was subject to evolutionary variation in addition to the weight of importance assigned to all connections. Given that the best results with leave-one-out cross validation were when using 10 inputs, for this experiment, the number of inputs was also set to a random choice of 10 features from the available 248 total (the domain-level features were not included). The evolutionary process was allowed to continue for 1,000 generations and was repeated 30 times with random initial settings.
3 Results and Discussion
To our knowledge, prediction of R5X4 viral variants, separately from R5 or X4 strains, remains entirely novel. We chose to break down the problem of coreceptor usage classification into a two-step process of 1) classifying R5X4 sequences from sequences previously classified as R5 or X4 followed by 2) classification of R5 sequences from X4 sequences. This two-tiered approach to classification provided a reasonable preliminary test of evolved neural networks.
Multiple linear regression using the domain-level features of isoelectric point and domain charge combined yielded a correlation to coreceptor usage of R2 = 0.517, which was slightly better than either of these domain-level features in isolation (Figs. 1a and 1b). As shown in Fig. 1c, the predictions of the best resulting neural networks using these two inputs had slightly improved correlation with coreceptor usage (R2 = 0.556) versus multiple linear regression.
Table 4 presents the results when combining these two domain features with the charges for 28 amino acid positions in the V3 loop when discriminating R5X4 from R5 or X4 variants and then selecting different combinations of features as input to the neural network from this set of 30 features.
TABLE 4.
Performance of Evolved Neural Networks for the Discrimination of D-Tropic Sequences from R5 and X4 Sequences Following Leave-One-Out Cross Validation in Terms of Area under the ROC Curve (A(z))
| Number of Input Features | Number of Hidden Nodes | Number of Output Nodes | ROC curve area (A(z)) |
|---|---|---|---|
| 2 | 2 | 1 | 0.689 |
| 5 | 2 | 1 | 0.731 |
| 10 | 2 | 1 | 0.760 |
| 15 | 2 | 1 | 0.713 |
| 20 | 2 | 1 | 0.761 |
| 25 | 2 | 1 | 0.665 |
| 30 | 2 | 1 | 0.707 |
A maximum of 30 possible input features was available (2 domain charge features and 28 amino acid features). Forcing neural networks to use fewer than 30 inputs can still produce neural network models with reasonable A(z) values.
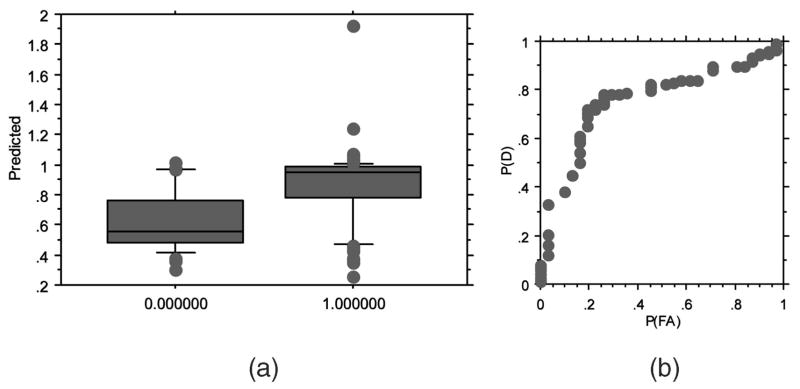
The coupling of domain-level features and amino acid position features results in a neural network that can discriminate R5X4 sequences from R5 and X4 sequences. Fig. 2 presents the results for the most parsimonious evolved ANN model, which utilized 10 inputs (Table 4). In this case, the neural network with 20 features has an ROC area of 0.761, whereas the neural network with 10 features has a nearly equivalent ROC area of 0.760. Given that the model with 20 features as input has twice the number of features with essentially an identical ROC area, the model utilizing 10 features was considered parsimonious. The box plot in Fig. 2a suggests that further separation may be possible with large population sizes or additional generations of evolution. The mean output prediction for the 10-2-1 neural network was 0.807 (Fig. 2a). Using this mean prediction as a decision threshold, a confusion matrix can be generated (Table 5). These results suggest that R5X4 HIV-1 strains can be predicted with 77.4 percent accuracy and R5 and X4 strains with 73.7 percent accuracy. The mean predictive accuracy for these two categories is 75.5 percent. The neural network has a slightly higher error in predicting R5X4 sequences as R5 and X4 phenotype versus predicting R5 and X4 as R5X4 sequences.
Fig. 2.
Results of leave-one-out cross validation on R5X4 versus R5 or X4 corecptor usage prediction. (a) Ability of the best neural networks to discriminate between D-tropic samples (0) and R5 or X4 samples (1). (b) ROC curve showing predictive performance on the held-out samples. The best probability of detection (P(D)) of 0.80 is achieved with probability of false alarm (P(FA)) of slightly > 0.2 (area under the ROC curve A(z) = 0.760).
TABLE 5.
Performance of the Best Evolved Neural Network on the Discrimination of R5X4 HIV Sequences from R5 and X4 Sequences
| R5X4Pred | (R5 & X4)Pred | |
|---|---|---|
| R5X4ACT | 24/31 | 7/31 |
| (R5 & X4)ACT | 31/118 | 87/118 |
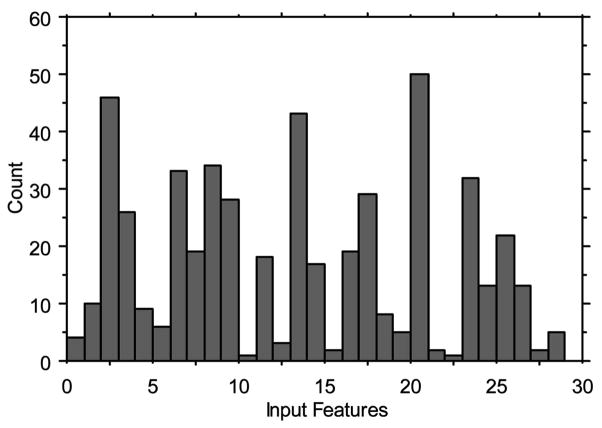
From the best evolved neural network, it is possible to identify which of the 30 possible features were used most often over the leave-one-out samples. Fig. 3 presents these data for the first 50 leave-one-out cross validation models. Features 3 (charge for V3 loop amino acid position 2), 14 (charge for V3 loop amino acid position 21), and 21 (charge for V3 loop amino acid position 29) were used most often in the resulting models. Surprisingly, the two domain-level features (features 1 and 2) were rarely used in the best resulting leave-one-out models.
Fig. 3.
Input features used most often over the first 50 leave-one-out trials for the discrimination of R5X4 from R5 and X4 strains.
Table 6 presents the results when combining the two domain-level features with the charges for 28 amino acid positions in the V3 loop when discriminating R5 from X4 sequences and then selecting different subsets of features to be used as input to the neural network over the range [2–30].
TABLE 6.
Performance of Evolved Neural Networks for the Discrimination of R5 and X4 Sequences Following Leave-One-Out Cross Validation in Terms of Area under the ROC Curve (A(z))
| Number of Input Features | Number of Hidden Nodes | Number of Output Nodes | ROC curve area (A(z)) |
|---|---|---|---|
| 2 | 2 | 1 | 0.884 |
| 5 | 2 | 1 | 0.912 |
| 10 | 2 | 1 | 0.908 |
| 15 | 2 | 1 | 0.842 |
| 20 | 2 | 1 | 0.904 |
| 25 | 2 | 1 | 0.906 |
| 30 | 2 | 1 | 0.927 |
A maximum of 30 possible input features was available (2 domain charge features and 28 amino acid features). Forcing neural networks to use fewer than 30 inputs can still produce neural network models with reasonable A(z) values.
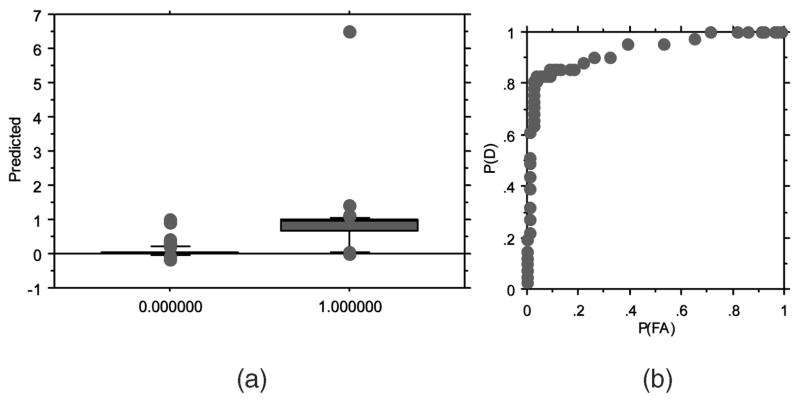
Fig. 4 presents the results for the most parsimonious evolved ANN model that utilized all 30 inputs (Table 6). The box plot in Fig. 4a suggests that the best evolved ANN models can easily discriminate the held-out samples in leave-one-out cross validation. The mean output prediction for the 30-2-1 neural network was 0.361 (Fig. 4a). Using this mean prediction as a decision threshold, a confusion matrix can be generated (Table 7). These data suggest that R5 strains can be predicted with 94.8 percent accuracy and X4 with 82.9 percent accuracy, with a mean predictive accuracy of 88.9 percent. The neural network has a slightly higher error in predicting X4 sequences as R5 versus predicting R5 sequences as X4.
Fig. 4.
Results of leave-one-out cross validation on R5 versus X4 strains prediction. (a) Ability of the best neural networks to discriminate between R5 samples (0) and non-X4 samples (1). (b) The best probability of detection (P(D)) of 0.81 is achieved with a probability of false alarm (P(FA)) of 0.05 (area under the ROC curve A(z) = 0.927).
TABLE 7.
Performance of the Best Evolved Neural Network on the Discrimination of R5 HIV Sequences from X4 Sequences
| R5Pred | X4Pred | |
|---|---|---|
| R5ACT | 73/77 | 4/77 |
| X4ACT | 7/41 | 34/41 |
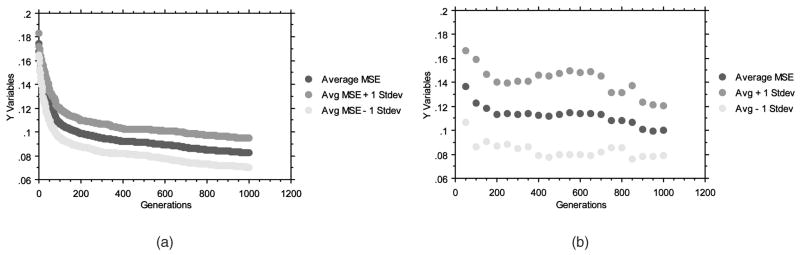
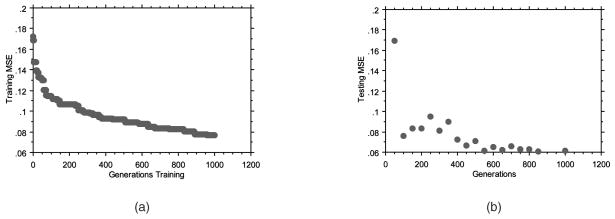
Fig. 5a provides the average convergence over the number of generations over all 30 trials when using a division of training and testing data rather than leave-one-out cross validation. As anticipated, MSE (y-axis) is minimized over time. At every 50 generations, the best evolved neural network was evaluated over the testing examples and an MSE was calculated (Fig. 5b). The MSE on the testing examples decreased over all 1,000 generations, indicating that no overtraining had occurred. Over all 30 trials, the best evolved neural network (the neural network resulting from trial number 22) had the lowest resulting MSE on the testing examples (MSE = 0.0612). The convergence plots over the training and testing examples for this single evolutionary optimization (Figs. 6a and 6b) demonstrate how rapidly useful ANNs can be discovered.
Fig. 5.
Evolutionary optimization of neural networks. (a) Reduction of average MSE over all 30 trials on the training examples. (b) Average performance of the best evolved neural networks on the testing examples sampled every 50 generations during training. No overtraining is observed.
Fig. 6.
Evolutionary optimization of neural networks. (a) Reduction of average MSE over all 30 trials on the training examples. (b) Average performance of the best evolved neural networks on the testing examples sampled every 50 generations during training. No overtraining is observed.
When evaluating performance over all 127 examples in the training set, the best ANN from trial 22 had a predictive accuracy of 75 percent, 40 percent, and 79.3 percent (Table 8a) when predicting all three patterns of coreceptor usage, R5, R5X4, and X4, using decision thresholds of R5 = x < 0.2108, R5X4 = 0.2108 < x < 0.5728, and X4 = x > 0.5728, where x was the prediction made by the single output node of the ANN. When using these same decision thresholds for the testing examples, this same best network had a predictive accuracy of 79 percent, 50 percent, and 70 percent for all three classes (Table 8b). When evaluating the number and type of errors made, it is interesting to note that, during training, R5 sequences were never misclassified as X4 sequences, while some X4 sequences were misclassified as R5. The difficulty in predicting R5X4 strains, a category that by definition shares characteristics of both R5 and X4 sequences, may not only be inherent to this problem but may also be the result of different numbers of training examples or insufficient training length (given that no overtraining was yet observed) for each of the three classes in the training data. Further research and development will help to resolve these issues, including the use of neural networks with two output nodes that may discriminate these three classes with better resolution.
TABLE 8.
(a) Predictive Accuracy on the 127 Training Examples by Coreceptor Usage. (b) Predictive Accuracy on the 30 Testing Examples by Coreceptor Usage
| R5PRED | R5X4PRED | X4PRED | |
|---|---|---|---|
| R5ACT | 51/68 | 17/68 | 0/68 |
| R5X4ACT | 8/30 | 12/30 | 10/30 |
| X4ACT | 2/29 | 4/29 | 23/29 |
| R5PRED | R5X4PRED | X4PRED | |
|---|---|---|---|
| R5ACT | 11/14 | 3/14 | 0/14 |
| R5X4ACT | 2/6 | 3/6 | 1/6 |
| X4ACT | 3/10 | 0/10 | 7/10 |
Actual (ACT) versus predicted (PRED) outcomes are provided for each coreceptor usage. On-diagonals are correct predictions. Off-diagonals are incorrect predictions.
The best evolved neural network from trial 22 utilized the following input features: Chou-Fasman helix index for positions 17, 22, and 26, Chou-Fasman sheet index for position 7, pKa value for free amino acid amine for positions 4, 5, and 8, polarizability index for position 21, volume for position 34, and charge at position 28. The amino acid positions related to these features are shown in Fig. 7.
Fig. 7.
Consensus sequence from alignment of X5, X4, and R5X4 sequences. Invariable positions 1, 3, 26, and 38 were removed. Lowercase letters indicate that the final alignment contained greater than 51 percent identity, whereas a question mark indicates less than 51 percent identity. Highlighted positions indicate a column in the alignment containing informative features for the best evolved neural network.
4 Conclusion
Accurate assessment of coreceptor usage is critical for many aspects of HIV research, including viral transmission, evolution, the study of reservoirs and other in vivo and in vitro studies. Typically, coreceptor usage is determined in the laboratory or predicted by overall charge of the V3 loop and/or the appearance of charged residues at certain positions. This preliminary study has demonstrated that evolved ANNs can also be used to determine coreceptor usage and, furthermore, can distinguish R5X4 viruses, which can use either CCR5 or CXCR4 coreceptor, from pure R5 or X4 viral sequences.
In the second set of experiments, R5 and X4 sequences were identified with a probability of correct classification of 0.906, which is comparable to or exceeds the performance of previous methods used to identify HIV-1 coreceptor usage; however, R5X4 sequences were discriminated from R5 and X4 sequences with a probability of correct classification of 0.755.
R5X4 HIV viruses are important to monitor within individuals and populations because of their association to rapid disease progression [18], [31]. Although R5 viruses appear to be more transmissible, infection with a dual tropic strain has been recently noted within a high-risk individual [18]. It is unclear if the viral phenotype found in this patient was from the transmission of such a virus or if it emerged rapidly. In either case, the possibility of such a virus infecting a population should be of great concern.
To date, the possibility of identifying R5X4 sequences without time consuming experimental assays has been unattainable because these viruses share genotypic characteristics with both R5 and X4 viral variants; overall charge analysis or positional information is not as clear cut as when identifying R5 from X4 strains. The system developed here is interesting because it used a collection of parameters in combination to identify dual tropic variants. These combinations of features and their relative importance would not be immediately apparent after examining databases of genetic sequence information alone. Considering the impact that the R5X4 virus may have on clinical progression, the ability to accurately identify these viruses within individuals or populations could lead to more aggressive clinical treatments for individuals infected with or quickly progressing with dual tropic strains. Furthermore, regions of the HIV genome outside of the V3 domain influence viral coreceptor usage. As the genetic databases grow and more viral sequences with experimentally determined coreceptor usage are generated, new tools, such as evolved ANNs, will catalyze new insight as to how various properties of amino acid sequences may influence HIV-1 ability to employ different coreceptors and infect specific cells.
Acknowledgments
The authors would like to thank the US National Science Foundation (NSF), particularly the Small Business Innovative Research Program. This material is based on the work supported by NSF Grants DMI-0349669 and OII-0610688. G.F. Fogel is supported through NSF SBIR Grants DMI-0522270 and OII-0610688. S.L. Lamers is supported through NSF SBIR Grant DMI-0349669 and is a consultant on NSF Grant OII-0610688. M.S. McGrath is supported through Grants NIH R01 MH073510-01 and West Coast AIDS and Cancer Specimen Resource Consortium U01 CA066529-12. M. Salemi is supported by Grants AI065265 and HD32259 and the Department of Pediatrics at the University of Florida, Gainesville, and is a consultant on NSF Grant OII-0610688. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the US National Science Foundation.
Biographies
 Susanna L. Lamers received the BS degree from the University of Texas in 1987. She has worked in the field of HIV molecular evolution for 18 years. She is currently the scientific director for BioInfoExperts, Inc., and is developing software to assist in the management and analysis of HIV and HCV specific data. She collaborates with researchers at the University of California, San Francisco, the University of Florida, Gainesville, the South African National Bioinformatics Institute, Cape Town, and the US National Institutes of Health, Bethesda, Maryland, with a focus on the evolutionary aspects of the HIV virus in different tissues and cell types, viral markers that may be linked to HIV-associated illnesses, and the epidemiology of AIDS in Africa.
Susanna L. Lamers received the BS degree from the University of Texas in 1987. She has worked in the field of HIV molecular evolution for 18 years. She is currently the scientific director for BioInfoExperts, Inc., and is developing software to assist in the management and analysis of HIV and HCV specific data. She collaborates with researchers at the University of California, San Francisco, the University of Florida, Gainesville, the South African National Bioinformatics Institute, Cape Town, and the US National Institutes of Health, Bethesda, Maryland, with a focus on the evolutionary aspects of the HIV virus in different tissues and cell types, viral markers that may be linked to HIV-associated illnesses, and the epidemiology of AIDS in Africa.
 Marco Salemi received the BA degree in chemistry from the University of Pavia, Italy, in 1991, a specialization in biotechnology from the University of Milano, Italy, in 1994, and the PhD degree in molecular evolution from the Catholic University of Leuven, Belgium, in 1999. He is currently an assistant professor in the Pathology Immunology and Laboratory Medicine Department of the College of Medicine at the University of Florida, Gainesville. He has been working on HIV/HTLV research since 1991 and, for the last 10 years, his main interests have been molecular evolution and molecular phylogenetics of human and simian retroviruses. He is a coeditor of the Phylogenetic Handbook, one of the major reference textbooks in the field of phylogenetic analysis, and the author of more than 50 publications in peer-reviewed journals. He was named a Distinguished International Educator in 2004 for his efforts in organizing and teaching several molecular evolution and bioinformatics workshops at universities in the US, Latin America, Europe, and East Asia.
Marco Salemi received the BA degree in chemistry from the University of Pavia, Italy, in 1991, a specialization in biotechnology from the University of Milano, Italy, in 1994, and the PhD degree in molecular evolution from the Catholic University of Leuven, Belgium, in 1999. He is currently an assistant professor in the Pathology Immunology and Laboratory Medicine Department of the College of Medicine at the University of Florida, Gainesville. He has been working on HIV/HTLV research since 1991 and, for the last 10 years, his main interests have been molecular evolution and molecular phylogenetics of human and simian retroviruses. He is a coeditor of the Phylogenetic Handbook, one of the major reference textbooks in the field of phylogenetic analysis, and the author of more than 50 publications in peer-reviewed journals. He was named a Distinguished International Educator in 2004 for his efforts in organizing and teaching several molecular evolution and bioinformatics workshops at universities in the US, Latin America, Europe, and East Asia.
 Michael S. McGrath received the MD and PhD degrees from Stanford University in 1980 and 1985, respectively. He is currently a professor of laboratory medicine, medicine, and pathology at the University of California San Francisco. He has been the director of the San Francisco General Hospital AIDS Immunobiology Research Laboratory for the past 20 years. He has been the director of the largest AIDS and cancer specimen bank (ACSR) in the US for the past 12 years. His research interests include evaluating the role pathogenic macrophages play in AIDS and chronic diseases, as well as macrophage targeted drug development. He has published more than 100 papers on cancer and AIDS.
Michael S. McGrath received the MD and PhD degrees from Stanford University in 1980 and 1985, respectively. He is currently a professor of laboratory medicine, medicine, and pathology at the University of California San Francisco. He has been the director of the San Francisco General Hospital AIDS Immunobiology Research Laboratory for the past 20 years. He has been the director of the largest AIDS and cancer specimen bank (ACSR) in the US for the past 12 years. His research interests include evaluating the role pathogenic macrophages play in AIDS and chronic diseases, as well as macrophage targeted drug development. He has published more than 100 papers on cancer and AIDS.
 Gary B. Fogel received the BA degree in biology from the University of California, Santa Cruz, in 1991 and the PhD degree in biology from the University of California, Los Angeles, in 1998. He is currently the vice president of Natural Selection, Inc., La Jolla, California. His experience includes more than 14 years of applying computational intelligence methods to bioinformatics problems. He has more than 40 publications in the technical literature, the majority treating the science and application of evolutionary computation, and he is a coeditor of the book Evolutionary Computation in Bioinformatics (Morgan Kauffman, 2003). He served as a coeditor for a recent special issue on computational intelligence approaches in computational biology and bioinformatics for the IEEE/ACM Transactions on Computational Biology and Bioinformatics. He serves as an associate editor for IEEE/ACM Transactions on Computational Biology and Bioinformatics, IEEE Computational Intelligence Magazine, and the IEEE Transactions on Evolutionary Computation. He is on the editorial board of three other technical journals. He served as the general chairman for the 2005 and 2006 IEEE Computational Intelligence in Bioinformatics and Computational Biology Symposia, as cotechnical chair for the 2001 and 2006 IEEE Congress on Evolutionary Computation, and as program chair for the 2004 IEEE Congress on Evolutionary Computation. He was the chair of the IEEE Computational Intelligence Society Bioinformatics and Bioengineering Technical Committee from 2004 to 2005. He is a senior member of the IEEE.
Gary B. Fogel received the BA degree in biology from the University of California, Santa Cruz, in 1991 and the PhD degree in biology from the University of California, Los Angeles, in 1998. He is currently the vice president of Natural Selection, Inc., La Jolla, California. His experience includes more than 14 years of applying computational intelligence methods to bioinformatics problems. He has more than 40 publications in the technical literature, the majority treating the science and application of evolutionary computation, and he is a coeditor of the book Evolutionary Computation in Bioinformatics (Morgan Kauffman, 2003). He served as a coeditor for a recent special issue on computational intelligence approaches in computational biology and bioinformatics for the IEEE/ACM Transactions on Computational Biology and Bioinformatics. He serves as an associate editor for IEEE/ACM Transactions on Computational Biology and Bioinformatics, IEEE Computational Intelligence Magazine, and the IEEE Transactions on Evolutionary Computation. He is on the editorial board of three other technical journals. He served as the general chairman for the 2005 and 2006 IEEE Computational Intelligence in Bioinformatics and Computational Biology Symposia, as cotechnical chair for the 2001 and 2006 IEEE Congress on Evolutionary Computation, and as program chair for the 2004 IEEE Congress on Evolutionary Computation. He was the chair of the IEEE Computational Intelligence Society Bioinformatics and Bioengineering Technical Committee from 2004 to 2005. He is a senior member of the IEEE.
Footnotes
For information on obtaining reprints of this article, please send to: tcbb@computer.org, and reference IEEECS Log Number TCBB-0136-0706.
Contributor Information
Susanna L. Lamers, Email: Susanna@HIVanalysis.net, BioInfoExperts, 4133 NW 44th Dr., Gainesville, FL 32606
Marco Salemi, Email: msalemi68@yahoo.com, Department of Pathology, Immunology, and Laboratory Medicine, University of Florida (UF-COM) Gainesville, 1600 S.W. Archer Road, Gainesville, FL 32610.
Michael S. McGrath, Email: mmcgrath@php.ucsf.edu, Department of Medicine, University of California San Francisco, San Francisco, CA 94143-0874
Gary B. Fogel, Email: gfogel@natural-selection.com, Natural Selection, Inc., 9330 Scranton Rd., Suite 150, San Diego, CA 92121
References
- 1.Werbos PJ. The Roots of Backpropagation. John Wiley & Sons; 1994. [Google Scholar]
- 2.Fogel DB, Fogel LJ, Porto VW. Evolving Neural Networks. Biological Cybernetics. 1990;63(6):487–493. [Google Scholar]
- 3.Porto VW, Fogel DB, Fogel LJ. Alternative Neural Network Training Methods. IEEE Expert. 1995;10(3):16–22. [Google Scholar]
- 4.Yao X. Evolving Artificial Neural Networks. Proc IEEE. 1999;87(9):1423–1447. [Google Scholar]
- 5.Landavazo DG, Fogel GB, Fogel DB. Quantitative Structure-Activity Relationships by Evolved Neural Networks for the Inhibition of Dihydrofolate Reductase by Pyrimidines. BioSystems. 2002;65:37–47. doi: 10.1016/s0303-2647(01)00192-7. [DOI] [PubMed] [Google Scholar]
- 6.Weekes D, Fogel GB. Evolutionary Optimization, Back-propagation, and Data Preparation Issues in QSAR Modeling of HIV Inhibition by HEPT Derivatives. BioSystems. 2003;72:149–158. doi: 10.1016/s0303-2647(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 7.Fogel DB. Blondie24: Playing at the Edge of AI. Morgan Kaufmann; 2002. [Google Scholar]
- 8.Fogel DB, Hays TJ, Hahn SL, Quon J. A Self-Learning Evolutionary Chess Program. Proc IEEE. 2004;92:1947–1954. [Google Scholar]
- 9.Berger EA, Murphy PM, Farber JM. Chemokine Receptors as HIV-1 Coreceptors: Roles in Viral Entry, Tropism, and Disease. Ann Rev Immunology. 1999;17:675–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 10.Goodenow MM, Collman RG. HIV-1 Coreceptor Preference Is Distinct from Target Cell Tropism: A Dual-Parameter Nomenclature to Define Viral Phenotypes. J Leukocyte Biology. 2006;80(5):965–972. doi: 10.1189/jlb.0306148. [DOI] [PubMed] [Google Scholar]
- 11.Koot M, van’t Wout AB, Koostra NA, Degoede REY, Tersmette M, Schitemaker H. Prognostic Value of HIV-1 Syncytium-Inducing Phenotype for Rate of CD4+ Cell Depletion and Progression to AIDS. Annals of Internal Medicine. 118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 12.Kreisberg JF, Kwa D, Schramm B, Trautner V, Connor R, Schuitemaker H, Mullins JI, van’t Wout AB, Goldsmith MA. Cytopathicity of Human Immunodeficiency Virus Type 1 Primary Isolates Depends on Coreceptor Usage and Not Patient Disease Status. J Virology. 2001;75(18):8842–8847. doi: 10.1128/JVI.75.18.8842-8847.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuttle DL, Anders CB, Aquino-De Jesus MJ, Poole PP, Lamers SL, Briggs DR, Pomeroy SM, Alexander L, Peden KW, Andiman WA, Sleasman JW, Goodenow MM. Increased Replication of Non-Syncytium-Inducing HIV Type 1 Isolates in Monocyte-Derived Macrophages Is Linked to Advanced Disease in Infected Children. AIDS Research and Human Retroviruses. 2002;18(5):353–362. doi: 10.1089/088922202753519133. [DOI] [PubMed] [Google Scholar]
- 14.Cheng-Mayer C, Weiss C, Seto D, Levy JA. Isolates of Human Immunodeficiency Virus Type 1 from the Brain May Constitute a Special Group of the AIDS Virus. Proc Nat’l Academy of Sciences USA. 1989;86(21):8575–8579. doi: 10.1073/pnas.86.21.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitchen SG, Zack JA. CXCR4 Expression during Lymphopoiesis: Implications for Human Immunodeficiency Virus Type 1 Infection of the Thymus. J Virology. 1997;71(9):6928–6934. doi: 10.1128/jvi.71.9.6928-6934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salemi M, Lamers SL, Yu S, de Oliveira T, Fitch WM, McGrath MS. HIV-1 Phylodynamic Analysis in Distinct Brain Compartments Provides a Model for the Neuropathogenesis of AIDS. J Virology. 2005;79:11343–11352. doi: 10.1128/JVI.79.17.11343-11352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-van Lent N, Scherpbier HJ, Veenstra J, Boer K, Coutinho RA, Miedema F, Schuitemaker H. Macrophage-Tropic Variants Initiate Human Immunodeficiency Virus Type 1 Infection after Sexual, Parenteral, and Vertical Transmission. J Clinical Investigation. 1994;94(5):2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz M, Mohri H, Mehandru S, Shet A, Berry L, Kalyanaraman R, Kim A, Chung C, Jean-Pierre P, Horowitz A, La Mar M, Wrin T, Parkin N, Poles M, Petropoulos C, Mullen M, Boden D, Ho DD. Infection with Multidrug Resistant, Dual-Tropic HIV-1 and Rapid Progression to AIDS: A Case Report. Lancet. 2005;365(9464):1031–1038. doi: 10.1016/S0140-6736(05)71139-9. [DOI] [PubMed] [Google Scholar]
- 19.Resch W, Hoffman N, Swanstrom R. Improved Success of Phenotype Prediction of the Human Immunodeficiency Virus Type 1 from Envelope Variable Loop 3 Sequence Using Neural Networks. J Virology. 2001;76:3852–3864. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 20.Loannidis JA, Trikalinos TA, Law M. HIV Lipodystrophy Case Definition Using Artificial Neural Network Modeling. Antiviral Therapy. 2003;8:435–441. [PubMed] [Google Scholar]
- 21.Wang D, Larder B. Enhanced Prediction of Lopinavir Resistance from Genotype by Use of Artificial Neural Networks. J Infectious Diseases. 2003;188:653–660. doi: 10.1086/377453. [DOI] [PubMed] [Google Scholar]
- 22.Brumme ZL, Dong WWY, Yip B, Wynhoven B, Hoffman NG, Swanstrom R, Jensen MA, Mullins JI, Hogg RS, Montaner JSG, Harrigan PR. Clinical and Immunological Impact of HIV Envelope V3 Sequence Variation after Starting Initial Triple Antiretroviral Therapy. AIDS. 2004;18:F1–F9. doi: 10.1097/00002030-200403050-00001. [DOI] [PubMed] [Google Scholar]
- 23.Milich L, Margolin B, Swanstrom R. V3 Loop of the Human Immunodeficiency Virus Type 1 Env Protein: Interpreting Sequence Variability. J Virology. 1993;67(9):5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foucier RA, Brouwer M, Broersen SM, Schuitemaker H. Determination of Human Immunodeficiency Virus Type 1 Syncytium-Inducing V3 Genotype by PCR. J Clinical Microbiology. 1995;33(4):906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen MA, Li FS, van ’t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved Coreceptor Usage Prediction and Genotypic Monitoring of R5-to-X4 Transition by Motif Analysis of Human Immunodeficiency Virus Type 1 env V3 Loop Sequences. J Virology. 2003;77(24):13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable Prediction of T-Cell Epitopes Using Neural Networks with Novel Sequence Representations. Protein Science. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai SK, Good B, Pond SK, Wong JK, Strain MC, Richmand DD, Smith DM. Semen-Specific Genetic Characteristics of Human Immunodeficiency Virus Type 1 env. J Virology. 2005;39:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salemi M, Goodenow MM, Lamers SL. Inferring Correct Positional Homology in Human Immunodeficiency Virus Type 1 Envelope V1-V2 Hypervariable Domains by Motif-Based Alignment: Consequence for Phylogenetic and Selection Pressure Analyses. 2006. submitted. [Google Scholar]
- 29.Lamers S, Beason S, Dunlap L, Compton R, Salemi M. HIVbase: A PC/Windows-Based Software Offering Storage and Querying Power for Locally Held HIV-1 Genetic, Experimental and Clinical Data. Bioinformatics. 2002;20:436–438. doi: 10.1093/bioinformatics/btg445. [DOI] [PubMed] [Google Scholar]
- 30.Creighton TE. Proteins. W.H. Freeman; 1993. [Google Scholar]
- 31.Cayabyab M, Rohne D, Pollakis G, Mische C, Messele T, Abebe A, Etemad-Moghadam B, Yang P, Henson S, Axthelm M, Goudsmit J, Letvin NL, Sodroski J. Rapid CD4+ T-Lymphocyte Depletion in Rhesus Monkeys Infected with a Simian-Human Immunodeficiency Virus Expressing the Envelope Glycoproteins of a Primary Dual-Tropic Ethiopian Clade C HIV Type 1 Isolate. AIDS Research and Human Retroviruses. 2004;20(1):27–40. doi: 10.1089/088922204322749477. [DOI] [PubMed] [Google Scholar]