Abstract
Acquired resistance to selective FLT3 inhibitors, is an emerging clinical problem in the treatment of FLT3-ITD+ acute myeloid leukaemia (AML). The paucity of valid pre-clinical models has limited investigations to determine the mechanism of acquired therapeutic resistance, thereby limiting the development of effective treatments. We generated selective FLT3 inhibitor-resistant cells by treating the FLT3-ITD+ human AML cell line MOLM-13 in vitro with the FLT3-selective inhibitor MLN518, and validated the resistant phenotype in vivo and in vitro. The resistant cells, MOLM-13-RES, harboured a new D835Y tyrosine kinase domain (TKD) mutation on the FLT3-ITD+ allele. Acquired TKD mutations, including D835Y, have recently been identified in FLT3-ITD+ patients relapsing after treatment with the novel FLT3 inhibitor, AC220. Consistent with this clinical pattern of resistance, MOLM-13- RES cells displayed high relative resistance to AC220 and Sorafenib. Furthermore, treatment of MOLM-13-RES cells with AC220 lead to loss of the FLT3 wild type allele and duplication of the FLT3-ITD-D835Y allele. Our FLT3-Aurora kinase inhibitor, CCT137690, successfully inhibited growth of FLT3-ITD-D835Y cells in vitro and in vivo, suggesting that dual FLT3-Aurora inhibition may overcome selective FLT3 inhibitor resistance, in part due to inhibition of Aurora kinase, and may benefit patients with FLT3-mutated AML.
Keywords: FLT3, Aurora, Kinase, AML, Resistance, Inhibitor
Introduction
Acute myeloid leukaemia (AML) is a heterogeneous class of leukaemia with prognosis predicted by a number of cytogenetic and molecular abnormalities.1 Mutation of the fms-like tyrosine kinase 3 (FLT3) gene is a frequent event in AML and usually involves internal tandem duplication (ITD) of the juxtamembrane domain coding region or point mutations of the tyrosine kinase domain (TKD).2 Both FLT3-ITD and FLT3-TKD mutations result in ligand-independent proliferation due to constitutive dimerisation and activation of the FLT3 receptor.2 High mutant-to-wild type allelic ratios of FLT3-ITD are associated with a very poor prognosis in both adults3 and children,4 but there is conflicting evidence regarding the prognostic impact of FLT3-TKD.5, 6
There has been intense interest in inhibition of FLT3 kinase in recent years, but the clinical impact of FLT3 inhibitors has thus far been limited by transient responses when used as single agents and the emergence of acquired resistance following treatment.7 Newer FLT3 inhibitors with improved selectivity and pharmacokinetic (PK) / pharmacodynamic (PD) properties may have improved single-agent efficacy,8 but clinical resistance to compounds such as AC220 is emerging. One particular mechanism of resistance is acquired secondary mutations in the FLT3-TKD. All 9 patients analysed from the current phase II study of AC220 who relapsed after achieving complete bone marrow responses, had secondary FLT3-TKD mutations on the FLT3-ITD+ allele, either F691or at D835.9 Furthermore, F691 or D835 mutations were identified in 10 of 30 patients on the AC220 trial who discontinued drug for any reason.9
Aurora kinases are a family of highly conserved serine-threonine protein kinases that play a key role in several stages of mitosis.10 Although expression analyses in leukaemia have been limited to cell lines and small patient cohorts, over-expression of Aurora A has been consistently demonstrated.11 Aurora A plays an important role in centrosome maturation, spindle assembly and metaphase I spindle orientation.10 Selective inhibition of Aurora A results in G2/M arrest, cytokinesis failure and cell death.12, 13 Aurora B kinase forms a chromosomal passenger complex (CPC) with inner centromere protein (INCENP), Survivin and Borealin.14 Selective inhibition of Aurora B classically results in cytokinesis failure and endoreduplication, leading to polyploidy and cell death.12, 13
Aurora kinase inhibitors are emerging as promising new agents in the treatment of AML, particularly when associated with FLT3-ITD.11 Recent single-agent phase I-II clinical trials of the Aurora B inhibitor AZD1152 and the Aurora A inhibitor MLN8237 in AML have shown response rates of 25% and 17% respectively.15, 16 As part of an in-house FLT3-Aurora kinase inhibitor programme, we aimed to develop a model of selective FLT3 inhibitor resistance and hypothesized that such resistance could be overcome with dual FLT3-Aurora kinase inhibitors. CCT137690 is a novel dual FLT3-Aurora kinase inhibitor based on the imidazo[4,5-b]pyridine scaffold.17-19 Here we demonstrate that a human FLT3-ITD+ AML cell line harbouring a secondary D835Y mutation has high relative resistance to the FLT3 inhibitors AC220, MLN518 and Sorafenib but not to CCT137690.
Materials and methods
Cell culture
The human AML cell lines MOLM-13 and MV4-11 were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). The DSMZ authenticates all human cell lines by DNA-typing and confirms species of origin by PCR-analysis. Working stocks for the experiments described in this study were prepared immediately upon receipt of cells from DSMZ. Both cell lines are FLT3-ITD+ 20 and were maintained in antibiotic-free RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories Ltd, Somerset, UK). MOLM-13 cells with resistance to MLN518 (henceforth referred to as “MOLM-13-RES”) were developed by culturing MOLM-13 cells in the presence of increasing concentrations of MLN518 (with ≤ 0.1% DMSO) until confluent growth was sustained in 5 μM MLN518. Experiments using MOLM-13-RES cells were carried out after at least overnight incubation in MLN518-free, RPMI 1640 medium with 10% FBS. The same method was used to generate the MOLM-13-RES-AC cell line, using increasing concentrations of AC220, up to approximately 1 μM.
Compounds
CCT137690 was discovered and synthesized at our Institute17-19 MLN518 and Sorafenib were purchased from LC Laboratories (Woburn, MA). AC220 was purchased from Activate Scientific GmbH (Prien, Germany). Nocodazole and cytarabine were purchased from Sigma-Aldrich (St Louis, MO). All compounds were dissolved in DMSO and stored at −20°C.
In vitro kinase assays
The concentration of compound that inhibited FLT3 and FLT3 (D835Y) kinase activity by 50% of normal (IC50) was determined by Z’-LYTE® assay using Invitrogen’s SelectScreen® Biochemical Kinase Profiling Service (Invitrogen, Paisley, UK). The ATP concentration used for these assays was equal to Km apparent.
FLT3 mutational analysis
Simultaneous detection of genomic DNA FLT3-ITD and FLT3-TKD (at D835) mutations was achieved by multiplex PCR and enzymatic digestion followed by fragment analysis on an automated genetic analyser, as described by Murphy et al.21 To assess the allelic distribution of FLT3 ITD and D835Y mutations in the resistant cell lines, total mRNA was extracted with the RNeasy kit (Qiagen, UK) followed by cDNA synthesis using the High-capacity cDNA kit (Life Technologies, UK). The area of the FLT3 coding region encompassing both ITD and D835Y mutations was amplified by PCR using the following primers: 5′ TCC CTT GGC ACA TCT TGT GA 3′and 5′ GGA ATG CCA GGG TAA GGA T 3′. The PCR products were cloned using the pGEM®-T vector system (Promega, UK) and at least 10 colonies containing products from each cell line were sequenced using BigDye v3.1 terminators (Life Technologies, UK).
FLT3 ploidy analysis
Cells from the suspension cultures were fixed with 3:1 methanol-acetic acid and fluorescence in-situ hybridisation (FISH) was performed using standard techniques on the slides containing each of the three cell lines. The probes used in this study were part of the Vysis (Abbott) CLL panel, with a SpectrumOrange™ signal on chromosome 13 at 13q14 and a SpectrumGreen™ signal at the centromere of chromosome 12.
Small tandem repeat (STR) analysis was performed in 1.5 ng of genomic DNA using the Powerplex16 kit (Promega, UK) as per manufacturer’s instructions and run in a 3130xl genetic analyser (Life Technologies, UK). Analysis of the STR patterns was performed using GeneMapper v4.1 (Life Technologies, UK).
Measurement of cell viability and cellular assays
To assess cell viability in vitro, all cell lines were seeded into 96-well plates at a density of 2 × 105 cells per 100 mL, then treated with 0.2% DMSO, or varying concentrations of CCT137690 or MLN518 for 72 hours. Cell viability was measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS; Promega, Madison, WI) with absorbance measured at 490 nm using a Wallac Victor2 1420 multilabel counter (PerkinElmer Life Sciences, Cambridgeshire, UK). For cellular assays to assess inhibition of kinase signaling pathways and apoptosis, cell lines were seeded at varying concentrations in 12-24-well plates or 25 cm2 flasks respectively.
Immunoblotting
Protein extraction was performed using two methods. For assays involving incubation periods up to 2 hours, whole cell lysis was performed in 2X LDS sample buffer (Invitrogen, Carlsbad, CA) with 200 mM dithiothreitol (DTT) followed by sonication for 20 seconds at 15 micron amplitude then boiling at 100°C for 10 minutes. For longer assays cells were lysed in lysis buffer: 120 mM NaCl, 50 mM Tris HCl pH 7.4, 1% NP40, 1% phosphatase inhibitor cocktail B (Santa Cruz Biotechnology, Santa Cruz, CA), 1% phosphatase inhibitor cocktail C (Santa Cruz Biotechnology, Santa Cruz, CA), 10 mM NaF, 1 “Complete Mini” protease inhibitor tablet per 10 mL (Roche Diagnostics, West Sussex, UK) with quantification of the soluble protein fraction by Bradford assay (Bio-Rad Laboratories, Cheshire, UK). Proteins were resolved by electrophoresis using 4-12% Bis-TRIS gels (Invitrogen, Carlsbad, CA), transferred to nitrocellulose membranes (Whatman, Kent, UK) then immunoblotted with the following antibodies: phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198), Aurora A, phospho-FLT3 (Tyr 842), phospho-STAT5 (Tyr694), STAT5, phospho-p44/42 MAPK (Thr202/Tyr204), p44/42 MAPK, PARP, (all from Cell Signaling Technology, Danvers, MA); FLT3 (C-20, Santa Cruz Biotechnology, Santa Cruz, CA); Aurora B, Histone H3 (both from Abcam); phospho-histone H3 (Ser10), GAPDH (both from Millipore, Billerica, MA). For detection of phosphorylated FLT3 in xenograft samples, cell lysates were immunoprecipitated with FLT3 antibody and protein A sepharose (GE Healthcare, Buckinghamshire, UK), resolved by electrophoresis, immunoblotted onto PVDF membranes (Millipore, Billerica, MA) and probed with phospho-tyrosine 4G10 antibody (Millipore, Billerica, MA). After exposure, membranes were stripped with 5% acetic acid then reprobed for total FLT3 antibody. All membranes were incubated with secondary anti-rabbit or anti-mouse mouse antibodies conjugated to horseradish peroxidase (Dako, Glostrup, Denmark) then immunoreactive bands were detected using ECL substrate (Thermo Fisher Scientific, Rockford, IL) and exposed on Amersham Hyperfilm ECL (GE Healthcare, Buckinghamshire, UK).
Measurement of apoptosis
Apoptosis was measured by flow cytometry using the Annexin V-FITC Apoptosis Detection Kit (Enzo Life Sciences, Exeter, UK) as per the manufacturer’s instructions. Apoptosis was further confirmed by detection of PARP cleavage using immunoblotting.
Cell cycle analysis
Cells were fixed overnight at 4°C in 70% ethanol, washed in PBS (with 1% FBS) then incubated for 30 minutes in PBS with 1% FBS, 0.04% propidium iodide (PI) and 0.25% RNase. Cells were analysed using a BD LSRII flow cytometer (BD Biosciences).
Human tumour xenograft studies in athymic mice
All animal studies were approved by the local research ethics committee and carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 and national guidelines.22 Athymic (CrTac:NCr-Fox1(nu) mice were bred in house. Female mice 6-8 weeks of age were injected subcutaneously in the right flank with 2 × 106 MOLM-13 or MOLM-13-RES cells. When mean tumour diameter was 6 mm (approximately day 5), mice were assigned to treatment or control cohorts (8 mice each) and dosing began twice daily orally at 12 hour intervals with vehicle, 75 mg/kg/dose CCT137690 or 160 mg/kg/dose MLN518. Tumours were routinely measured across two perpendicular diameters and volumes calculated using the formula V = 4/3π [(d1 +d2)/4]3. Cohorts of mice were culled at specified times after the final dose, with tumours excised, weighed, measured and processed for PK and PD analyses. For survival analysis, animals were culled when subcutaneous tumours approached UK Home Office license limits (maximum mean diameter 1.2 cm).
Compound measurement from in vivo studies
CCT137690 and MLN518 were quantified in extracted mouse plasma and tissue samples by high performance liquid chromatography (HPLC) with tandem mass spectrometry using reverse phase gradient elution chromatography and multiple reaction monitoring.
Statistics
All statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software Inc, La Jolla, CA). In vitro log dose-response curves were calculated using non-linear regression with variable slope after normalizing absorbance to untreated and cellular controls with the concentration required to inhibit the MTS response by 50% reported as the viability IC50. For in vivo studies, survival was calculated using the Kaplan-Meier method.
Results
Long-term exposure of MOLM-13 cells to the selective FLT3 inhibitor MLN518 results in selection of a secondary FLT3-TKD mutation (D835Y) on the FLT3-ITD+ allele and resistance
To generate a human AML cell line with resistance to selective FLT3 inhibition, MOLM-13 cells were cultured for approximately 7 weeks in the presence of progressively increasing concentrations of MLN518, the most selective FLT3 inhibitor commercially available at the time the work was carried out. To control for the possibility of spontaneous FLT3 mutations occurring during prolonged culture, parental MOLM-13 cells were cultured in parallel. Once confluent growth was sustainable in concentrations of 5 μM MLN518, aliquots of the MLN518-resistant cells, termed “MOLM-13-RES”, and the parental MOLM-13 cells (in parallel prolonged culture) were analysed for FLT3 mutations as described and compared to freshly-thawed MOLM-13 cells. We used the multiplex PCR assay with enzymatic digestion and fragment analysis to simultaneously detect both FLT3-ITD mutations and any point mutation of the FLT3-TKD at residue D835 (ref. 22). By this assay, the FLT3 status of parental MOLM-13 cells after prolonged culture was the same as freshly-thawed cells, indicating that prolonged culture had not lead to a change in the FLT3-ITD or selection of a secondary FLT3-TKD mutation at D835. The MOLM-13-RES cells, however, had a new point mutation p.D835Y, in addition to the original FLT3-ITD.
Subsequent to our development of the MOLM-13-RES cell line, it was reported that secondary D835Y mutations were present on the FLT3-ITD+ allele of patients relapsing after treatment with AC220,9 and murine BaF3 cells transfected with a doubly-mutated FLT3-ITD-D835Y gene were resistant in vitro to AC220, as well as Sorafenib.23 We therefore tested the in vitro sensitivity of MOLM-13-RES cells to AC220 and Sorafenib. Whilst the parental MOLM-13 cells were highly sensitive to AC220 and Sorafenib, MOLM-13-RES cells displayed marked relative resistance to both compounds. AC220 was approximately 23-fold less potent against MOLM-13-RES, whilst Sorafenib was approximately 60-fold less potent. To further assess the potential mechanism underlying clinical relapse following treatment with AC220, we cultured MOLM-13-RES cells in the presence of increasing concentrations of AC220 (up to approximately 1 μM). This population of cells was termed MOLM-13-RES-AC.
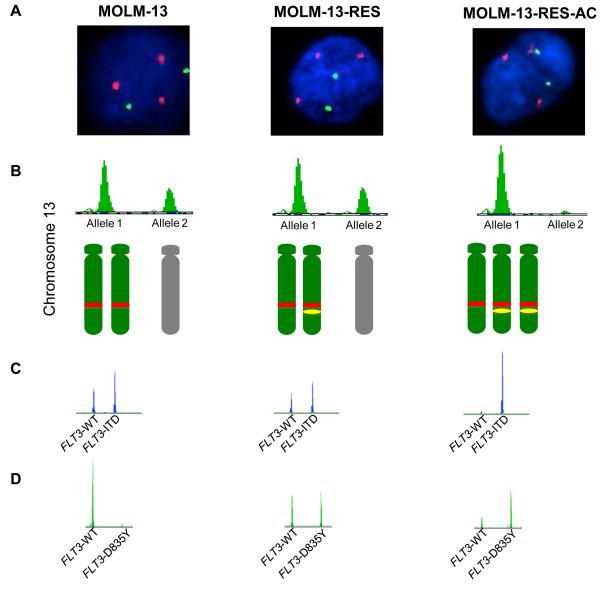
MOLM-13 cells are known to have three copies of chromosome 13q and to harbour a FLT3- ITD mutation, but mutations at D835 have not been described.20, 24 The FLT3 gene resides at 13q12 (ref. 25), therefore, we first assessed the ploidy status of this region by FISH and STR analysis (Figure 1A). STR analysis of D13S317 showed the parental MOLM-13 and MOLM-13-RES cell lines contained 3 copies of the marker (i.e., 2 copies of one allele and 1 copy of the remaining allele) while the MOLM-13-RES-AC cell line has undergone LOH and contains only copies of one allele (Figure 1B). Given that FISH analyses showed that all cell lines contained 3 copies of 13q, this likely reflects the loss of the allele with double wild-type FLT3 (FLT3-WT with no ITD and no D835Y) and gain of the allele with both ITD and D853Y mutations (Figure 1A-D). To confirm that the acquired D835Y mutation occurred on the same allele as the FLT3-ITD, the tyrosine kinase domain of FLT3 was sequenced in individual colonies (Supplementary Figure 1). The results showed that the acquired D835Y mutation occurred on one of the FLT3-ITD+ alleles in MOLM-13-RES, and this allele was then duplicated in MOLM-13-RES-AC whilst the FLT3-WT allele was lost.
Figure 1.
Clonal evolution of the FLT3 alleles in MOLM-13 cell lines treated with FLT3 inhibitors. The figure shows the genetic evolution of the FLT3 alleles in the parental MOLM-13 cell line (MOLM-13), MOLM-13-RES and MOLM-13-RES-AC. (A) FISH analysis show how all three cell lines have 3 copies of 13q14 (red) and 2 copies of chromosome 12 centromere (green). (B) STR analysis of DS13317 (upper panel) shows the presence of 2 copies of the shorter marker allele (allele 1) and 1 copy of the larger marker allele (allele 2) in MOLM-13 and MOLM-13-RES, whilst MOLM13-RES-AC cell line shows loss of heterozygosity (LOH) with only 1 marker allele present (allele 1). Depiction of the copy number and ploidy status of chromosome 13q according to the data from FISH, STR and FLT3 mutation analyses; parental and maternal copies of 13q are represented in green and grey, the red rectangle represents a FLT3-ITD mutation and the yellow oval represents a FLT3-D835Y mutation (lower panel). (C) FLT3-ITD results on the DNA of the 3 cell lines showing heterozygous mutations in MOLM-13 and MOLM-13-RES (mutant / wild type ratio = 2:1) and homozygous FLT3-ITD mutation in MOLM-13-RES-AC. (D) FLT3-D835Y results on DNA showing no mutation in the parental MOLM-13 cell line, heterozygous mutation in the MOLM-13-RES and heterozygous mutation with increased mutant ratio in the MOLM-13-RES-AC.
Characterisation of CCT137690: a dual inhibitor of FLT3 and Aurora kinases
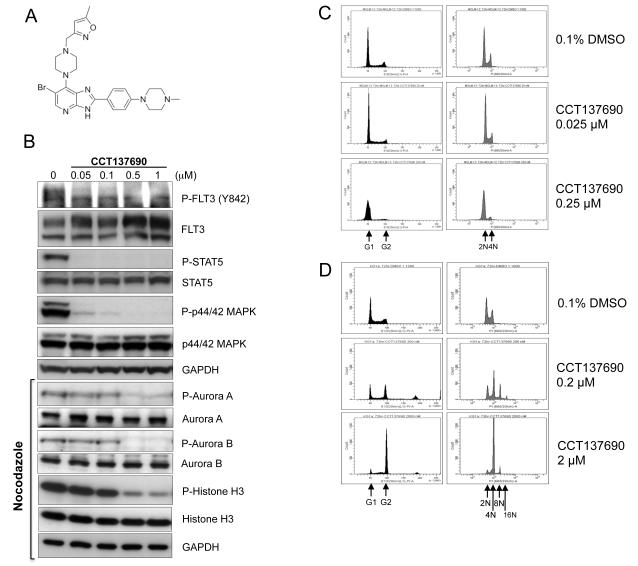
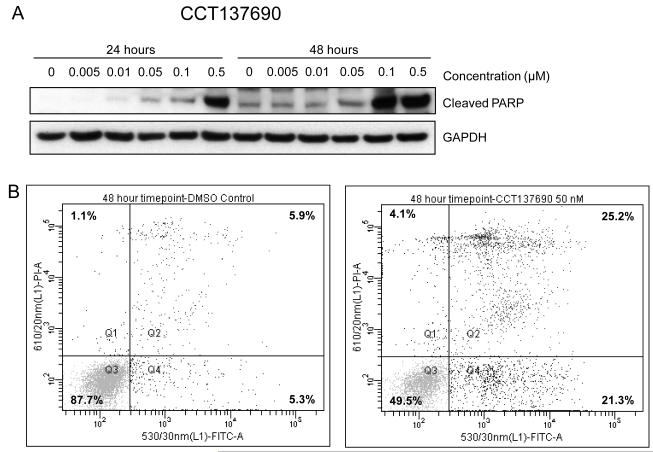
CCT137690 is an orally bioavailable imidazo[4,5-b]pyridine derivative (Figure 2A), which selectively inhibits Aurora (Aurora A, B and C IC50 0.015 μM, 0.025 μM and 0.019 μM respectively)17 and FLT3 kinases (FLT3-WT IC50 0.0025 μM, FLT3-D835Y IC50 0.0033 μM). Viability of the FLT3-ITD+ human AML cell lines MOLM-13 and MV4-11 was potently inhibited in vitro by CCT137690. Using MTS assays, the viability IC50 for these cell lines was 0.023 and 0.062 μM respectively. Consistent with the kinase inhibitory profile in biochemical assays, cellular assays of CCT137690 in MOLM-13 cells show that inhibition of FLT3 signaling occurs at lower concentrations than those inhibiting Aurora kinases (Figure 2B). Furthermore, CCT137690 causes G1/S arrest in MOLM-13 cells, similar to the profile seen with MLN518 (Figure 2C and supplementary Figure 2). By contrast, the classic pan-Aurora inhibition phenotype of polyploidy is seen in the FLT3-WT cell line KG1a (Figure 2D). Taken together, these data suggest that in FLT3-ITD+ AML, where FLT3 kinase is constitutively activated, the FLT3-inhibitory effects of CCT137690 predominate, whilst in FLT3-WT AML, the Aurora inhibitory effects predominate. Finally, cell death of MOLM-13 cells in response to CCT137690 occurs via apoptosis, with concentration- and time-dependent increases in PARP cleavage and Annexin V positivity (Figure 3A-B).
Figure 2.
In vitro activity of CCT137690. (A) Chemical structure of CCT137690. (B) MOLM-13 cells were incubated with CCT137690 for 2 hours at the concentrations indicated. Cell lysates were prepared and analysed for the expression of the indicated proteins by immunoblotting. Immunoblots marked “nocodazole” were obtained from MOLM-13 cells incubated overnight with 50 ng/mL nocodazole followed by inhibitor treatment for 2 hours at the concentrations indicated. GAPDH was used as loading control. Cell cycle profile of AML cells treated with CCT137690 at concentrations approximating viability IC50 and 10x IC50. FLT3-ITD+ MOLM-13 (C) and FLT3-WT KG-1a (D) cells were treated for 72 hours at concentrations shown or 0.1% DMSO, fixed in 70% ethanol, then stained with PI (x-axes) and analysed by FACS. Y-axes represent FACS event counts (same scale for all histograms). Black histograms are on linear-scaled x-axes to demonstrate cell cycle. Grey histograms are on log-scaled x-axes to demonstrate DNA content.
Figure 3.
CCT137690 induces apoptosis in MOLM-13 cells. (A) Immunoblots of MOLM-13 lysates showing increases in cleaved PARP following exposure to CCT137690 at the indicated concentrations for 24 and 48 hours. GAPDH was used as a loading control. (B) Representative flow cytometry dot-plot of Annexin V positivity after MOLM-13 cells were exposed to 0.05 μM CCT137690 for 48 hours. X-axes represent Annexin V positivity, Y-axes represent propidium iodide positivity.
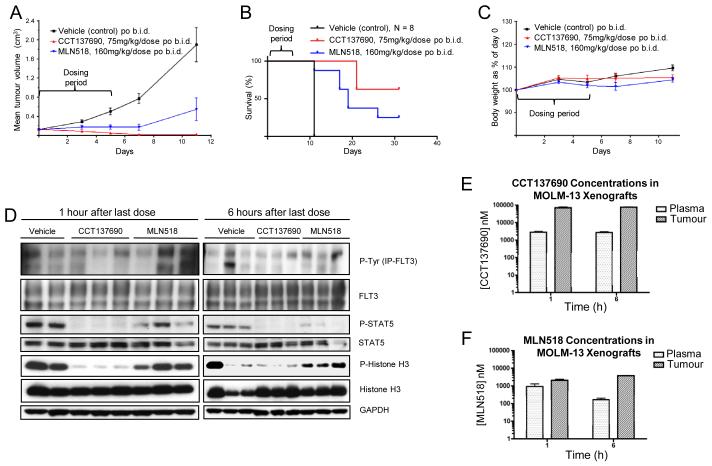
Based on the in vitro activity, human tumour xenograft experiments in athymic mice were undertaken to confirm the efficacy of CCT137690 in vivo. We used a MOLM-13 subcutaneous xenograft model with MLN518 as a positive control. The efficacy of MLN518 when administered orally at a maximal tolerated dose of 160 mg/kg twice daily had been previously reported using this model.26 As demonstrated in Figure 4A, both MLN518 and CCT137690 showed reduction of tumour growth compared with the vehicle-treated mice. More pronounced reduction of tumour growth was seen with the dual FLT3-Aurora inhibitor CCT137690 than the selective FLT3 inhibitor MLN518. Half of the mice treated with CCT137690 achieved complete remission with disappearance of their subcutaneous tumours compared to only 2 of 8 (25%) mice treated with MLN518 (Figure 4B). Furthermore, tumour regrowth was more pronounced in the MLN518-treated mice and CCT137690 appeared to confer a survival advantage (log-rank P = 0.056). Both CCT137690 and MLN518 were well tolerated, with all mice appearing to be in good health and maintaining body weight (Figure 4C).
Figure 4.
Dual inhibition of Aurora and FLT3 kinases by CCT137690 inhibits growth of FLT3-ITD+ AML xenografts. (A) Athymic mice (8 per cohort) were injected subcutaneously with 2 × 106 MOLM-13 cells. Five days after implantation mean tumour diameter was 6 mm (day 0 on graphs) and dosing began twice daily orally for 5 days with vehicle (black), 75 mg/kg/dose CCT137690 (red) or 160 mg/kg/dose MLN518 (blue). Mean tumour volumes ± s.e.m. are shown. (B) Mouse survival. Mice were culled when tumour size reached licence limits. Five CCT137690 treated mice (63%) and two MLN518 treated mice (25%) were still alive at day 31 when the study was terminated (log-rank P = 0.056). (C) CCT137690 and MLN518 were both well tolerated with no loss of body weight. (D) Tumour lysates from the 3-day PK/PD xenograft study were analysed by immunoblotting for the expression of the indicated proteins. GAPDH used as loading control. Total plasma and tumour concentrations from the 3-day PK/PD xenograft study of CCT137690 (E) and MLN518 (F) 1 and 6 hours after the final dose.
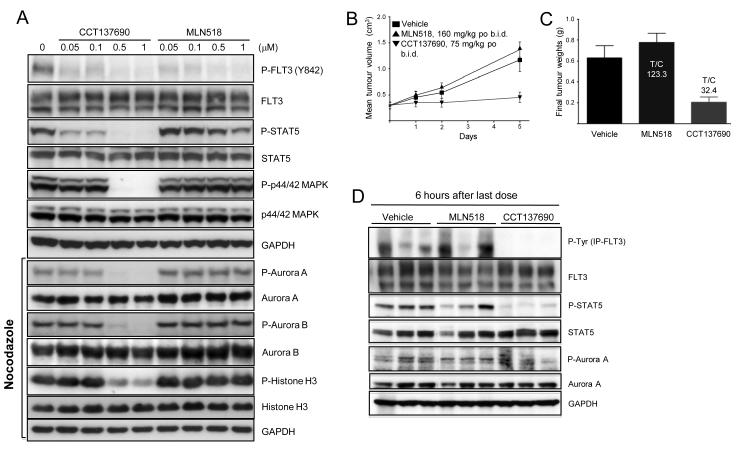
A repeat in vivo study was undertaken to assess PK and PD biomarkers in MOLM-13 tumours. In this experiment, mice were culled after 3 days of twice daily dosing (75 mg/kg/dose CCT137690 and 160 mg/kg/dose MLN518). Tumours were collected at 1 and 6 hrs after the last dose and the lysates were analysed by immunoblotting. Phospho-FLT3 and the downstream target phospho-STAT5 were reduced in comparison to the total FLT3 and STAT5 levels respectively. It should be noted that due to technical challenges using the haemorrhagic subcutaneous tumours, the quality of the phospho-FLT3 blots (Figure 4D) was suboptimal with either IP or the specific phospho-FLT3 (Y842) antibody. However, since STAT5 is a direct downstream substrate of FLT3-ITD,27 the robust inhibition of STAT5 phosphorylation, consistent with in vitro signaling, can be used as a surrogate marker of FLT3 inhibition in vivo. Reduction of phospho-histone H3 indicated pan-Aurora inhibition. In summary, these data suggest that although the FLT3 inhibitory effects on MOLM-13 cells appear to predominate in vitro, CCT137690 causes inhibition of both FLT3 and Aurora kinases at well-tolerated doses in vivo. Consistent with its kinase selectivity profile, MLN518 only inhibited FLT3 signaling, as evidenced by inhibition of phospho-STAT5 (Figure 4D). Total plasma and tumour concentrations of both CCT137690 and MLN518 were significantly above the kinase IC50 for their relevant targets (FLT3 0.0025 μM and 0.003 μM for CCT137690 and MLN518 respectively; Aurora A 0.015 μM and Aurora B 0.025 μM for CCT137690; Figure 4E, F). Moreover, the free plasma concentration of CCT137690 at 1 and 6 hours after the last dose was approximately 0.06 μM, higher than the IC50 for Aurora A, Aurora B, FLT3 kinases. Based on the reported plasma protein binding of MLN518 (80% in mice),28 the free plasma concentrations of MLN518 at 1 and 6 hours after the last dose (approximately 0.2 μM and 0.04 μM) were also higher than the FLT3 IC50. These data are also in accordance with biomarker modulation as shown in Figure 4D indicating prolonged target modulation in vivo.
MOLM-13-RES cells are resistant to MLN518 but sensitive to CCT137690 in vitro and in vivo
MOLM-13-RES cells remained sensitive to CCT137690, with an in vitro viability IC50 of 0.08 μM, reflecting a 3.5-fold difference in sensitivity compared with parental MOLM-13 cells. In contrast, the relative resistance of MOLM-13-RES cells to MLN518, Sorafenib and AC220 was approximately 105-, 60- and 23-fold respectively. Cellular assays of MOLM-13-RES cells with CCT137690 showed a similar pattern of FLT3 and Aurora inhibition as in MOLM-13 cells, although inhibition of STAT5 and p44/42 MAPK phosphorylation occurred at slightly higher concentrations in MOLM-13-RES cells, possibly due to increased downstream signaling by the doubly-mutated FLT3 kinase (Figure 5A).
Figure 5.
Dual inhibition of Aurora and FLT3 kinases with CCT137690 overcomes resistance to the selective FLT3 inhibitor MLN518 in vitro and in vivo. (A) MOLM-13-RES cells were incubated with CCT137690 for 2 hours at the concentrations indicated. Cell lysates were prepared and analysed for the expression of the indicated proteins by immunoblotting. Immunoblots marked “nocodazole” were obtained from MOLM-13-RES cells incubated overnight with 50 ng/mL nocodazole followed by inhibitor treatment for 2 hours at the concentrations indicated. GAPDH was used as loading control. (B) Athymic mice (6 per cohort) were injected subcutaneously with 2 × 106 MOLM-13-RES cells. Thirteen days after implantation mean tumour diameter was 6 mm (day 0 on graphs) and dosing began twice daily orally for 5 days with solvent vehicle (■), 75 mg/kg/dose CCT137690 (▼) or 160 mg/kg/dose MLN518 (▲). Mice were culled at 1 and 6 hours following the final dose. Mean tumour volumes ± s.e.m. are shown. (C) Final tumour weights. T/C calculated from the mean weight of treated tumours/control tumours (%). CCT137690 and MLN518 were both well tolerated with all mice appearing to be in good health and maintaining body weight (not shown). (D) Tumour lysates were analysed by immunoblotting for the expression of the indicated proteins. GAPDH was used as loading control.
To confirm the in vitro activity of CCT137690 against the MOLM-13-RES model of selective FLT3 inhibitor resistance, in vivo studies were performed using subcutaneous human tumour xenografts of MOLM-13-RES cells in athymic mice. As illustrated in Figure 5B and 5C, CCT137690 caused significant in vivo growth inhibition. As in the parental MOLM-13 in vivo studies, biomarker modulation was consistent with inhibition of both FLT3 and Aurora kinases at well-tolerated doses (Figure 5D). In contrast, MLN518 caused no growth inhibition of MOLM-13-RES xenografts (Figure 5B, C). Importantly, the drug concentrations measured in this experiment confirmed that MLN518 concentrations in plasma and MOLM-13-RES tumour tissue were similar to efficacious concentrations in studies of parental MOLM-13 xenografts (not shown). These data indicate that the therapeutic failure of MLN518 in vivo was due to inherent MOLM-13-RES resistance, rather than inadequate drug exposure.
Discussion
FLT3 remains an attractive target for AML therapy, although the success of FLT3 inhibitors when used as single agents has thus far been limited by transient responses and the emergence of resistance.7 Although newer more potent FLT3 inhibitors such as AC220 and Sorafenib have held promise of improved efficacy, resistance to these drugs may also become a significant clinical issue.
It has previously been shown that cells harbouring isolated D835Y and D835V mutations are resistant to MLN518.29 These experiments, however, were based upon transfection of mutated constructs into murine BaF3 cells. Transfection studies in FLT3-ITD+ BaF3 cells have also been used to demonstrate non-overlapping and variable resistance to the FLT3 inhibitors PKC412, Sorafenib and SU5614.30 Point mutations associated with FLT3 inhibitor resistance included, but were not limited to, those at N676, F691 and Y842. Similarly, in vitro studies using BaF3 cells transfected with FLT3 harbouring both ITD and TKD mutations (at F691L, D835V, D835Y, Y842C and Y842H), have shown variable resistance to AC220, Sorafenib, Ponatinib, PLX3397 and DCC2036.23, 31-33 In the present study, a spontaneously occurring D835Y mutation was detected after long-term treatment of human FLT3-ITD+ AML cells with MLN518. To our knowledge, this is the first report of a human AML cell line harbouring both FLT3-ITD and FLT3-TKD mutations. Unlike transfected murine BaF3 cells, our model more likely reflects the clinical scenario whereby treatment with FLT3 inhibitors may result in clonal selection pressure and emergence of acquired resistance. Indeed, it was recently reported that of 9 patients analysed who relapsed after therapy with AC220, all had secondary TKD mutations in the FLT3-ITD+ allele, as did one third of patients who had ceased AC220 for any reason.9 Heidel et al reported the case of a patient with FLT3-ITD+ AML who developed resistance to the FLT3 inhibitor PKC412.34 Sequencing of the FLT3 gene in blasts obtained at relapse revealed the presence of an N676K point mutation in the TKD of the FLT3-ITD+ allele, but not the FLT3 wild-type allele. Furthermore, this TKD mutation was not present in blasts obtained prior to PKC412 treatment.34
Loss of the FLT3-WT allele and an increased FLT3-ITD:FLT3-WT allelic ratio are often seen at relapse in AML,35 and blasts may be more “addicted” to FLT3 signaling.36 Acquired uniparental disomy of chromosome 13q with subsequent homozygosity of FLT3-ITD and FLT3-D835Y has also been described in patients at relapse.37 Furthermore, loss of the FLT3- WT allele in FLT3-ITD+ mouse models has recently been shown to induce a more aggressive myeloproliferative phenotype,38 suggesting that the process of MOLM-13 first acquiring a D835Y mutation (MOLM-13-RES), then increasing the FLT3-ITD-D835Y allelic ratio with further exposure to AC220 through LOH (MOLM-13-RES-AC), may have contributed to clinical relapses following AC220 therapy.
Although usually mutually exclusive at diagnosis, it has been reported that double FLT3-ITD and FLT3-TKD mutations occur spontaneously in approximately 1-2% of all patients and may occur in the same or opposite allele.39-41 Thiede et al reported an incidence of 1.7% (n = 17) with the majority (88.2%) occurring in patients with cytogenetically normal AML, but the small number of patients precluded the detection of a survival difference compared with those harbouring FLT3-ITD mutations alone.39 The type of TKD in the patients with FLT3-ITD was not reported. Chen et al characterized 21 patients (1.3%) with dual FLT3-ITD-TKD mutations.40 A number of these patients had dual mutations present in the first sample analysed, albeit at low levels, whilst others had gains or losses of FLT3-TKD mutations over time.40 From the experiences with AC220 and PKC412, it appears that dual ITD and TKD mutations of FLT3 may therefore represent not only a natural clonal evolution of FLT3- mutated AML, but an important cause of acquired resistance to FLT3 inhibitors.
The dual FLT3-Aurora kinase inhibitor CCT137690 overcame resistance to selective FLT3 inhibition both in vitro and in vivo. The extent to which Aurora kinase inhibition by CCT137690 contributes to efficacy is unknown. CCT137690 inhibits FLT3-D835Y kinase with similar potency to FLT3-ITD kinase, but MLN518 is much less active against FLT3-D835Y in biochemical assays (IC50 1.8 μM). However, differences in potency against individual FLT3 tyrosine kinase variants alone cannot account for the sensitivity to CCT137690. AC220 and Sorafenib also display relatively similar affinity toward FLT3-ITD and FLT3-D835Y kinases individually,28 yet are dramatically less potent against both MOLM-13-RES cells and the doubly mutated BaF3 models reported by Smith et al.24 It is therefore possible that the additional inhibition of Aurora kinases by CCT137690, based on the observed Aurora kinase-specific biomarker modulation in vitro and in vivo, is important in maintaining efficacy against AML with doubly-mutated FLT3.
In summary, we have developed a clinically-relevant model of selective FLT3 inhibitor resistance and hypothesize that dual FLT3-Aurora inhibitors may overcome such resistance in the clinic. However, CCT137690 has a narrow in vitro safety margin against the hERG channel which may limit its pre-clinical development.17 Nevertheless, CCT137690 represents a useful tool compound for pre-clinical studies. Orally bioavailable, dual FLT3-Aurora kinase inhibitors with improved properties are currently under development.
Supplementary Material
Acknowledgements
We acknowledge NHS funding to the NIHR Biomedical Research Centre. This work, and all authors from the Cancer Research UK Cancer Therapeutics Unit, are supported by core funding from Cancer Research UK (grant numbers C309/A8274, C309/A11566). ASM is supported by Cancer Research UK (grant number C1178/A10294) and in part by a New Investigator Scholarship awarded by the Haematology Society of Australia and New Zealand. ADJP is supported by Cancer Research UK (programme grant C1178/A10294). PW is a Cancer Research UK Life Fellow. SL is also supported by Breakthrough Breast Cancer.
Funding: We acknowledge NHS funding to the NIHR Biomedical Research Centre. This work, and all authors from the Cancer Research UK Cancer Therapeutics Unit, are supported by core funding from Cancer Research UK (grant numbers C309/A8274, C309/A11566). ASM is supported by Cancer Research UK (grant number C1178/A10294) and in part by a New Investigator Scholarship awarded by the Haematology Society of Australia and New Zealand. ADJP is supported by Cancer Research UK (programme grant C1178/A10294). PW is a Cancer Research UK Life Fellow. SL is also supported by Breakthrough Breast Cancer.
Footnotes
Conflicts of interest All authors are employees of The Institute of Cancer Research which has a commercial interest in drug development programmes (see www.icr.ac.uk). Please note that all authors who are, or have been, employed by The Institute of Cancer Research are subject to a ‘Rewards to Inventors Scheme’ which may reward contributors to a programme that is subsequently licensed.
Supplementary information is available at Leukemia’s website.
REFERENCES
- 1.Smith ML, Hills RK, Grimwade D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2010;25:39–51. doi: 10.1016/j.blre.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Meshinchi S, Appelbaum FR. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res. 2009;15:4263–4269. doi: 10.1158/1078-0432.CCR-08-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 4.Meshinchi S, Alonzo TA, Stirewalt DL, Zwaan M, Zimmerman M, Reinhardt D, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–3661. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mead AJ, Linch DC, Hills RK, Wheatley K, Burnett AK, Gale RE. FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia. Blood. 2007;110:1262–1270. doi: 10.1182/blood-2006-04-015826. [DOI] [PubMed] [Google Scholar]
- 6.Whitman SP, Ruppert AS, Radmacher MD, Mrozek K, Paschka P, Langer C, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knapper S. The clinical development of FLT3 inhibitors in acute myeloid leukemia. Expert Opin Investig Drugs. 2011;20:1377–1395. doi: 10.1517/13543784.2011.611802. [DOI] [PubMed] [Google Scholar]
- 8.Levis MJ. Will newer tyrosine kinase inhibitors have an impact in AML? Best Pract Res Clin Haematol. 2010;23:489–494. doi: 10.1016/j.beha.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CC, Chin J, Wang Q, Salerno S, Damon LE, Hunt JP, et al. Validation of FLT3-ITD as a therapeutic target in human acute myeloid leukemia. Blood (ASH Annual Meeting Abstracts) 2011;118 abstract 937. [Google Scholar]
- 10.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 11.Moore AS, Blagg J, Linardopoulos S, Pearson AD. Aurora kinase inhibitors: novel small molecules with promising activity in acute myeloid and Philadelphia-positive leukemias. Leukemia. 2010;24:671–678. doi: 10.1038/leu.2010.15. [DOI] [PubMed] [Google Scholar]
- 12.Girdler F, Gascoigne KE, Eyers PA, Hartmuth S, Crafter C, Foote KM, et al. Validating Aurora B as an anti-cancer drug target. J Cell Sci. 2006;119:3664–3675. doi: 10.1242/jcs.03145. [DOI] [PubMed] [Google Scholar]
- 13.Carpinelli P, Moll J. Aurora kinase inhibitors: identification and preclinical validation of their biomarkers. Expert Opin Ther Targets. 2008;12:69–80. doi: 10.1517/14728222.12.1.69. [DOI] [PubMed] [Google Scholar]
- 14.Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowenberg B, Muus P, Ossenkoppele G, Rousselot P, Cahn JY, Ifrah N, et al. Phase I/II study to assess the safety, efficacy, and pharmacokinetics of barasertib (AZD1152) in patients with advanced acute myeloid leukemia. Blood. 2011;118:6030–6036. doi: 10.1182/blood-2011-07-366930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg SL, Fenaux P, Craig MD, Gyan E, Lister J, Kassis J, et al. Phase 2 Study of MLN8237, An investigational Aurora A kinase (AAK) inhibitor in patients with acute myelogenous leukemia (AML) or myelodysplastic syndromes (MDS) Blood (ASH Annual Meeting Abstracts) 2010;116 abstract 3273. [Google Scholar]
- 17.Bavetsias V, Large JM, Sun C, Bouloc N, Kosmopoulou M, Matteucci M, et al. Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate. J Med Chem. 2010;53:5213–5228. doi: 10.1021/jm100262j. [DOI] [PubMed] [Google Scholar]
- 18.Bavetsias V, Sun C, Bouloc N, Reynisson J, Workman P, Linardopoulos S, et al. Hit generation and exploration: imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases. Bioorg Med Chem Lett. 2007;17:6567–6571. doi: 10.1016/j.bmcl.2007.09.076. [DOI] [PubMed] [Google Scholar]
- 19.Chan F, Sun C, Perumal M, Nguyen QD, Bavetsias V, McDonald E, et al. Mechanism of action of the Aurora kinase inhibitor CCT129202 and in vivo quantification of biological activity. Mol Cancer Ther. 2007;6:3147–3157. doi: 10.1158/1535-7163.MCT-07-2156. [DOI] [PubMed] [Google Scholar]
- 20.Drexler HG. Guide to Leukemia-Lymphoma Cell Lines. 2 ed Braunschweig; 2010. [Google Scholar]
- 21.Murphy KM, Levis M, Hafez MJ, Geiger T, Cooper LC, Smith BD, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CC, Damon L, Salerno S, Shah NP. Saturation mutagenesis of FLT3-ITD: AC220-resistance-conferring kinase domain mutations are restricted to a limited number of residues and are cross-resistant to sorafenib in vitro. American Association for Cancer Research (AACR) Meeting Abstracts. 2011 abstract 4737. [Google Scholar]
- 24.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003;17:120–124. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 25.Rosnet O, Stephenson D, Mattei MG, Marchetto S, Shibuya M, Chapman VM, et al. Close physical linkage of the FLT1 and FLT3 genes on chromosome 13 in man and chromosome 5 in mouse. Oncogene. 1993;8:173–179. [PubMed] [Google Scholar]
- 26.Kiyoi H, Shiotsu Y, Ozeki K, Yamaji S, Kosugi H, Umehara H, et al. A novel FLT3 inhibitor FI-700 selectively suppresses the growth of leukemia cells with FLT3 mutations. Clin Cancer Res. 2007;13:4575–4582. doi: 10.1158/1078-0432.CCR-07-0225. [DOI] [PubMed] [Google Scholar]
- 27.Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Müller C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 28.Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML) Blood. 2009;114:2984–2992. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark JJ, Cools J, Curley DP, Yu JC, Lokker NA, Giese NA, et al. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood. 2004;104:2867–2872. doi: 10.1182/blood-2003-12-4446. [DOI] [PubMed] [Google Scholar]
- 30.von Bubnoff N, Engh RA, Aberg E, Sanger J, Peschel C, Duyster J. FMS-like tyrosine kinase 3-internal tandem duplication tyrosine kinase inhibitors display a nonoverlapping profile of resistance mutations in vitro. Cancer Res. 2009;69:3032–3041. doi: 10.1158/0008-5472.CAN-08-2923. [DOI] [PubMed] [Google Scholar]
- 31.Smith CC, Damon LE, Zhu X, Salerno S, Shah N. Analysis of in vitro activity of the clinically-active ABL/FLT3 inhibitor Ponatinib (AP24534) against AC220-resistant FLT3-ITD mutants. Blood (ASH Annual Meeting Abstracts) 2011;118 abstract 930. [Google Scholar]
- 32.Smith CC, Perl AE, Lasater E, Zhang C, Jeschke GR, Damon LE, et al. PLX3397 is an investigational selective FLT3 inhibitor that retains activity against the clinically-relevant FLT3-ITD/F691L “gatekeeper” mutation in vitro. Blood (ASH Annual Meeting Abstracts) 2011;118 abstract 764. [Google Scholar]
- 33.Fiskus W, Smith CC, Smith J, Wise SC, Lasater E, Damon LE, et al. Activity of allosteric, switch-pocket, ABL/FLT3 kinase inhibitor DCC2036 against cultured and primary aml progenitors with FLT-ITD or FLT3 kinase domain mutations. Blood (ASH Annual Meeting Abstracts) 2011;118 abstract 2611. [Google Scholar]
- 34.Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 35.Shih LY, Huang CF, Wu JH, Lin TL, Dunn P, Wang PN, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: a comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2002;100:2387–2392. doi: 10.1182/blood-2002-01-0195. [DOI] [PubMed] [Google Scholar]
- 36.Pratz KW, Sato T, Murphy KM, Stine A, Rajkhowa T, Levis M. FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood. 2010;115:1425–1432. doi: 10.1182/blood-2009-09-242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghavan M, Smith LL, Lillington DM, Chaplin T, Kakkas I, Molloy G, et al. Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood. 2008;112:814–821. doi: 10.1182/blood-2008-01-132431. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Bailey E, Greenblatt S, Huso D, Small D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood. 2011;118:4935–4945. doi: 10.1182/blood-2011-01-328096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 40.Chen W, Jones D, Medeiros LJ, Luthra R, Lin P. Acute myeloid leukaemia with FLT3 gene mutations of both internal tandem duplication and point mutation type. Br J Haematol. 2005;130:726–728. doi: 10.1111/j.1365-2141.2005.05666.x. [DOI] [PubMed] [Google Scholar]
- 41.Carnicer MJ, Nomdedeu JF, Lasa A, Estivill C, Brunet S, Aventin A, et al. FLT3 mutations are associated with other molecular lesions in AML. Leuk Res. 2004;28:19–23. doi: 10.1016/s0145-2126(03)00125-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.