Abstract
The compound eye of Drosophila melanogaster consists of about 750 ommatidia (unit eyes). Each ommatidium is composed of about 20 cells, including lens-secreting cone cells, pigment cells, a bristle cell and eight photoreceptors (PRs) R1-R8 2. The PRs have specialized microvillar structures, the rhabdomeres, which contain light-sensitive pigments, the Rhodopsins (Rhs). The rhabdomeres of six PRs (R1-R6) form a trapezoid and contain Rh1 3 4. The rhabdomeres of R7 and R8 are positioned in tandem in the center of the trapezoid and share the same path of light. R7 and R8 PRs stochastically express different combinations of Rhs in two main subtypes5: In the 'p' subtype, Rh3 in pR7s is coupled with Rh5 in pR8s, whereas in the 'y' subtype, Rh4 in yR7s is associated with Rh6 in yR8s 6 7 8.
Early specification of PRs and development of ommatidia begins in the larval eye-antennal imaginal disc, a monolayer of epithelial cells. A wave of differentiation sweeps across the disc9 and initiates the assembly of undifferentiated cells into ommatidia10-11. The 'founder cell' R8 is specified first and recruits R1-6 and then R7 12-14. Subsequently, during pupal development, PR differentiation leads to extensive morphological changes 15, including rhabdomere formation, synaptogenesis and eventually rh expression.
In this protocol, we describe methods for retinal dissections and immunohistochemistry at three defined periods of retina development, which can be applied to address a variety of questions concerning retinal formation and developmental pathways. Here, we use these methods to visualize the stepwise PR differentiation at the single-cell level in whole mount larval, midpupal and adult retinas (Figure 1).
Keywords: Neuroscience, Issue 69, Anatomy, Physiology, Immunology, Developmental Biology, Drosophila, retina, photoreceptor, imaginal disc, larva, pupa, confocal microscopy, immunohistochemistry
Protocol
1. Introduction
In this video, we describe methods for retinal dissections and immunohistochemistry at three defined developmental periods: the third instar larval, the midpupal and the adult stage. Although our protocol also works for other pupal stages (for details on earlier stages, see 16), we chose the midpupal stage, as it is optimal for imaging all PRs in one focal plane and their nuclei are easily identifiable, which facilitates the visualization of transcription factor expression patterns. The dissection of late pupal retinas is very similar to the dissection of adult retinas.
First, we demonstrate how to collect larvae and pupae at the desired stages. Then, we explain how to obtain optimal dissections of larval 17-18, midpupal and adult retinas. Second, we demonstrate how to visualize PR development at these stages using immunohistochemistry and confocal microscopy.
2. Staging and Preparation of Specimen
The developmental period of insects is temperature-dependent. To facilitate reproducible staging of larvae and pupae, we recommend raising all fly stocks at 25 °C under a 12 hr/12 hr light/dark cycle.
Third-instar larvae: About 96 hr after egg laying (at 25 °C), late third instar larvae stop feeding and wander on the walls of the food vial, where they eventually pupate. Use a soft brush or a forceps to gently remove a wandering third-instar larva from the wall of the food vial.
50% pupae ('midpupae') are observed about 48 hr after pupation. For exact timing, you can circle just-formed white pupae with a marker on the food vial. Alternatively, move all white pupae to a new vial to obtain a vial of uniform staging (for staging of pupae using morphological criteria, see 19). After 48 hr, carefully remove the midpupae from the vial using a forceps.
Adult flies: Anesthetize young adult flies (2-5 days after eclosion) on a CO2 pad. Remove heads using a forceps and store them in a glass well dish containing cold 1x PBS.
3. Dissection Procedure (About 1-2 hr per Experiment)
After collecting the specimen, place them in a drop of cold PBS on a Sylgard dissection dish. Dissect about 10-15 retinas per genotype.
Please consult the Material Safety Data Sheets as well as your institution's Environmental Health and Safety Office for proper handling of the reagents and equipment (see tables at the end of this protocol).
1. Larval Eye Imaginal Discs
Locate the anterior end of the larva, which contains the mouth hooks. Use a forceps to press the middle part of the larva gently against the base of the Sylgard dish. Use another forceps to grab the mouth hooks and pull gently and slowly away from the body. Discard the main part of the body and use the mouthparts to hold the anterior part in order to remove extraneous tissue from the imaginal discs that remain attached to the brain.
Transfer brain with imaginal discs by grabbing the mouth hooks with a forceps to 1x PBS in a three-well glass dish. Perform fixation step (step 4.1.).
After fixation, remove salivary glands, fat tissue and pull the brain away from the eye-antennal imaginal disc. To improve accessibility of 'difficult' antibodies, the peripodial membrane, which is located on the surface of the epithelium, can be removed by holding down the antennal part of the disc and peeling the membrane away with a dissecting pin. Next, perform immunohistochemistry (step 5.1.).
2. 50% Pupation/Midpupal Retinas
Hold posterior end of the pupal case with a forceps. Gently remove the anterior surface of the pupal case by grabbing the cuticle with another forceps and slowly pull away from the puparium. Peel away residual cuticle to expose the head.
Use the forceps' tips to pierce the underlying, soft sack in the dorsal anterior region. This provides access to the head. Gently separate the head from the thorax by slowly sucking up the head with a P20 pipette. Expel head from pipette tip into fresh cold 1x PBS and remove excess tissue; grab the head cuticle and remove proboscis. Proceed to fixation step (step 4.1.).
Pierce the optic lobe with one dissecting pin to steady the tissue. Insert the other dissecting pin between the lamina and retina to separate the retina from the lamina/optic lobe. Perform immunohistochemistry (step 5.1.).
3. Adult Eyes
Grab the center of the posterior head capsule with a forceps and remove mouth parts (proboscis), fat tissue and tracheae.
Grab the dorsal head cuticle with one forceps and use the other to separate eyes from brain tissue by gently pulling sideways. In most cases, the lamina, a thin and transparent sheath of brain neuropil, will stay attached to the retina. It is easier to remove the lamina after fixation.
Remove cuticle at the eye margin, but leave a small piece behind to facilitate the transfer of retinas to different solutions. Perform fixation (step 4.1.).
Remove the lamina by grabbing it at one side with a forceps and gently pulling sideways with the other forceps without impairing the retina. Remove residual cuticle, as it could fold onto the retina during mounting. These two steps are critical for visualizing photoreceptors, which would otherwise be obscured by the lamina and remains of cuticle during the imaging process. Proceed to immunohistochemistry (step 5.1.).
4. Fixation and Washing (About 1 hr)
Store the dissected retinas in 1x PBS in a three-well glass dish placed on ice while freshly preparing the fixative (dilute 40 microliters of 37% formaldehyde solution with 360 μl of 1x PBS, vortex and store on ice). We recommend to proceed with the fixation protocol and to avoid long storage times on ice (>15 min).
Replace 1x PBS with 200 μl of 3.7% formaldehyde solution. Make sure the tissue is submerged in fixative. This is critical, as it may otherwise not be fixed properly. If necessary, remove air bubbles and tracheae with the forceps.
Incubate on shaker for 15 min at room temperature.
Replace fixative with 1x PBS and rinse twice with 1x PBS. Continue washing in PBS for 30 min (on shaker, at room temperature).
Replace PBS with PBST. Rinse twice with PBST.
Proceed with removing the peripodial membrane from larval eye discs (3.1.3.) and the laminas from midpupal/adult retinas (3.2.3., 3.3.4.).
5. Immunohistochemistry
Blocking: Replace PBS with 200 μl of 5% normal serum in PBST and incubate retinas for 20 min on shaker at room temperature.
Replace blocking solution with primary antibody solution. Seal the three-well dish with parafilm and incubate overnight on shaker at room temperature.
Replace solution with PBST and rinse 3 times. Continue washing with PBST for a minimum of 1 hr on shaker at room temperature.
Transfer in secondary antibody solution and incubate in a parafilm-sealed glass well dish on shaker overnight at room temperature.
Replace antibody solution with PBST and rinse three times. Continue washing with PBST for a minimum of one hr on shaker at room temperature. The samples can be kept in PBST for several days, but it is critical to add fresh PBST at least once per day to prevent them from drying out.
6. Mounting
For mounting larval and midpupal retinas, use the P20 to transfer retinas to the center of a clean glass slide. Remove excess PBST. Use a forceps to align retinas in order to facilitate imaging.
Add 10 μl of mounting medium on top of the retinas. Cover them gently with a 24x40-1 cover slip and seal with clear nail polish.
For mounting adult retinas, prepare 'bridges' 20: Add two 5 μl droplets of 50% glycerol on both ends of a glass side and cover them with 22x22-1 cover slips (0.13 to 0.17 mm thick).
Remove the PBST with a P20 pipette; make sure that the retinas do not dry out. Add 20 μl of mounting medium to the well. Use mounting medium to transfer retinas with the P20 to the 'bridge' glass slide.
Use a forceps to orient (lenses should face the glass slide) and align retinas in order to facilitate imaging. If necessary, remove air bubbles with the P20.
Slowly place 24x40-1 cover slip (0.13 to 0.17 mm thick) from one side on top and seal edges with clear nail polish or Scotch tape. Store samples at 4 °C in a microscope slide saver box (keep in the dark).
7. Representative Results
As an example for the application of this protocol, we show representative results for visualizing photoreceptor differentiation at the three developmental stages in the video. A larval eye-antennal disc and a midpupal retina were stained with antibodies that label different photoreceptor types during their development (Figure 1A, B) and an adult retina was stained with two Rhodopsin antibodies to visualize two photoreceptor subtypes (Figure 1C).
Discussion
1. Troubleshooting
In our experience, dissections require practice (up to several weeks) and are facilitated by achieving a comfortable hand position 21 by resting the elbows and forearms on the table and with the fingers making contact with the dissection dish. That way, only the thumbs, index and middle fingers perform subtle movements.
Removing the lamina without damaging the photoreceptors is probably the most challenging step. Practice with red-eyed wildtype flies first before dissecting white-eyed mutants, as the lamina is much easier to see against the red eye pigmentation. Also, use lighting from behind and a dark background below the dissection dish to achieve optimal contrast.
However, the red eye pigment is fluorescent and can cause high background during the imaging process. This can weaken the signals from the secondary antibodies. A good strategy is to keep washing the adult retinas for four to five days and to replace the washing solution with fresh PBST at least once per day. The pigment will be washed out; be sure to wash experimental and control genotypes for the same duration.
2. Applications
Eye-specific loss- and gain-of-function approaches 22 23 make the Drosophila eye a particularly powerful system for genetic manipulations and screens. Moreover, recently developed computational methods for single-cell analysis in 3D reconstructions of entire retinas allow the analysis of mutant phenotypes at high resolution 24. This protocol can therefore be applied to a variety of questions concerning eye development and the underlying regulatory mechanisms 16,25-28 .
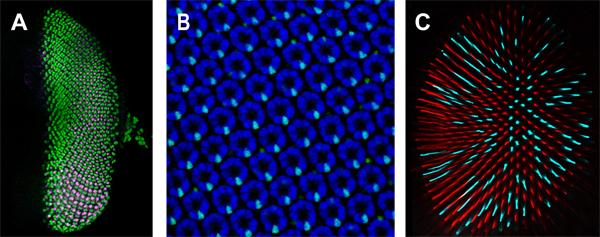 Figure 1. Retinal whole mounts at three different developmental stages. A) Larval eye imaginal disc stained with antibodies against Seven-up (green) and Runt (magenta). B) Midpupal retina. Two antibodies were used to visualize all photoreceptors R1-R8 (Elav, blue) and R7 photoreceptors (Prospero, cyan). C) Adult retina. Stochastic and mutually exclusive expression of photosensitive pigments Rh5 (cyan) and Rh6 (red) in two subtypes of R8 photoreceptors.
Figure 1. Retinal whole mounts at three different developmental stages. A) Larval eye imaginal disc stained with antibodies against Seven-up (green) and Runt (magenta). B) Midpupal retina. Two antibodies were used to visualize all photoreceptors R1-R8 (Elav, blue) and R7 photoreceptors (Prospero, cyan). C) Adult retina. Stochastic and mutually exclusive expression of photosensitive pigments Rh5 (cyan) and Rh6 (red) in two subtypes of R8 photoreceptors.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by an Ehrman fellowship to H-Y. H., a Jane Coffin Childs Memorial Fund for Medical Research postdoctoral fellowship to R.J.J., NIH Grant F32EY016309 to D.V., a New York University Dean's Dissertation Fellowship to D.J., NIH GrantR01 EY13010 to C.D. and a DFG fellowship to J.R. (RI 2208/1-1). We thank Nina Vogt and Pamela Boodram for comments on the manuscript.
References
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Hardie RC. Functional organization of the fly retina. In: D Ottoson., editor. Sensory Physiology. Vol. 5. Springer-Verlag; 1985. pp. 1–79. [Google Scholar]
- O'Tousa JE. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 2011;71:1212–1226. doi: 10.1002/dneu.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WH. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Chou WH. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int. J. Dev. Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsachaki M, Sprecher SG. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Dev. Dyn. 2012;241:40–56. doi: 10.1002/dvdy.22738. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in Drosophila ommatidium. Dev. Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Zipursky SL. Molecular and genetic analysis of Drosophila eye development: sevenless, bride of sevenless and rough. Trends Neurosci. 1989;12:183–189. doi: 10.1016/0166-2236(89)90069-6. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Specification of cell fate in the developing eye of Drosophila. Bioessays. 1991;13:621–631. doi: 10.1002/bies.950131202. [DOI] [PubMed] [Google Scholar]
- Charlton-Perkins M, Cook TA. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr. Top Dev. Biol. 2010;93:129–173. doi: 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther RF, Pichaud F. Immunofluorescent staining and imaging of the pupal and adult Drosophila visual system. Nat. Protoc. 2006;1:2635–2642. doi: 10.1038/nprot.2006.379. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological Techniques for the Drosophila Eye Part I: Larva and Pupa. In: Sullivan Wea., editor. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Wolff T. Dissection techniques for pupal and larval Drosophila eyes. CSH Protoc. 2007;2007:pdb prot4715. doi: 10.1101/pdb.prot4715. [DOI] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- Morante J, Desplan C. Dissection and staining of Drosophila optic lobes at different stages of development. Cold Spring Harb Protoc. 2011. pp. 652–656. [DOI] [PMC free article] [PubMed]
- Williamson WR, Hiesinger PR. Preparation of Developing and Adult Drosophila Brains and Retinae for Live Imaging. J. Vis. Exp. 2010. p. e1936. [DOI] [PMC free article] [PubMed]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Sood P, Johnston RJ, Kussell E. Stochastic De-repression of Rhodopsins in Single Photoreceptors of the Fly Retina. PLoS Comput. Biol. 2012;8:e1002357. doi: 10.1371/journal.pcbi.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Desplan C. Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev. Cell. 2011;21:874–887. doi: 10.1016/j.devcel.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskas D. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature. 2011;479:108–112. doi: 10.1038/nature10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. Building an ommatidium one cell at a time. Dev. Dyn. 2012;241:136–149. doi: 10.1002/dvdy.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]


