Abstract
The method described here consists in redesigning E. coli adherence properties by assembling the minimum number of curli genes under the control of a strong and metal-overinducible promoter, and in visualizing and quantifying the resulting gain of bacterial adherence. This method applies appropriate engineering principles of abstraction and standardization of synthetic biology, and results in the BBa_K540000 Biobrick (Best new Biobrick device, engineered, iGEM 2011).
The first step consists in the design of the synthetic operon devoted to curli overproduction in response to metal, and therefore in increasing the adherence abilities of the wild type strain. The original curli operon was modified in silico in order to optimize transcriptional and translational signals and escape the "natural" regulation of curli. This approach allowed to test with success our current understanding of curli production. Moreover, simplifying the curli regulation by switching the endogenous complex promoter (more than 10 transcriptional regulators identified) to a simple metal-regulated promoter makes adherence much easier to control.
The second step includes qualitative and quantitative assessment of adherence abilities by implementation of simple methods. These methods are applicable to a large range of adherent bacteria regardless of biological structures involved in biofilm formation. Adherence test in 24-well polystyrene plates provides a quick preliminary visualization of the bacterial biofilm after crystal violet staining. This qualitative test can be sharpened by the quantification of the percentage of adherence. Such a method is very simple but more accurate than only crystal violet staining as described previously 1 with both a good repeatability and reproducibility. Visualization of GFP-tagged bacteria on glass slides by fluorescence or laser confocal microscopy allows to strengthen the results obtained with the 24-well plate test by direct observation of the phenomenon.
Keywords: Bioengineering, Issue 69, Microbiology, Molecular Biology, curli, cobalt, biofilm, Escherichia coli, synthetic operon, synthetic biology, adherence assay, biofilm quantification, microscopy
Introduction
Bacterial adherence to abiotic support plays a major role in bioremediation, biocatalysis or microbial fuel cells. Bioremediation processes use the capacities of microorganisms to degrade organic substances, or to modify the metal distribution (immobilization, volatilization) or speciation. These beneficial activities are observed in aquatic and terrestrial ecosystems, but also in the artificial systems developed to treat polluted water of industrial and domestic wastes. The intensity and the quality of the microbial activity depend on physico-chemical factors, but also on the lifestyle of microorganisms (free-floating or embedded into biofilm). The biofilm formation is associated with a metabolism promoting resistance to biocides by diverse mechanisms. This phenomenon will therefore be encouraged in most bioremediation processes. Moreover, engineering Escherichia coli cells to control the biofilm formation has been successfully applied to immobilize whole-cell sensors on biochips2-3.
Adaptation of microorganisms to high concentration of metals occurs via diverse mechanisms such as adsorption to extracellular matrix components, activation of efflux pump or specific carriers able to concentrate the metal into the cell. Boosting these bacterial activities via genetic engineering allows efficient and cheap treatment of metal pollution at the laboratory scale, especially in the case of highly toxic metals in weak quantity as described by Raghu et al. 2008 4. Bacterial remediation represents in this case a competitive and cost saving method compared to classical chemical processes using ion exchange resins. The authors described an E. coli chassis genetically engineered for cobalt uptake and retention first by knocking out the efflux pump encoding gene rcnA, and then by transformation with a multi copy plasmid allowing overproduction of a transporter with preferential uptake for cobalt. Such a strain appears as an efficient alternative to ion exchange resins to treat radioactive effluent, but a key unresolved issue is the recovery of contaminated bacteria at the end of the process 4. The objective of our work was therefore to engineer a custom-designed strain able to stick to abiotic supports such as glass or plastic.
Amongst the whole set of adhesins and adherent fimbriae identified in Gram- bacteria, we chose to design a system allowing curli production. Curli are thin (2-5 nm diameter) and highly aggregative amyloid fibers that protrude from the E. coli and Salmonella surface as a non-crystalline and insoluble matrix 5-7. Curli are also involved in the colonization of abiotic surfaces and the development of biofilms 8. Curli were recently shown to bind mercury ions 9. Amyloids are indeed known to possess high affinity for metals ions such as Cu2+, Zn2+ and Fe3+ 10. This property might further improve the decontamination of metal polluted effluents. The csg cluster is responsible for the production of curli fibers and is constituted of two divergently transcribed operons (Figure 1). The csgB, csgA and csgC genes constitute the sense operon, encoding the two curli subunits, CsgA and CsgB. CsgC seems to be involved in redox activity within the curli biogenesis system and to affect CsgG pore behavior 11. However, the absence of csgC in the majority of curli-producing bacteria indicates that the corresponding protein provides only a secundary level of control over the curli biogenesis. To simplify the system, we have been chosen to work with the minimum number of genes.
The csgDEFG operonencodes proteins essential in the regulation and transportation of CsgA and CsgB to the cell surface. CsgD is a transcriptional activator of the csgBAC operon and plays a key role in the control of biofilm formation by controlling the production of curli fimbriae and other biofilm components such as cellulose 12 and by inhibiting the flagellum production 13. CsgE, CsgF and CsgG constitute a curli-specific secretory apparatus in the outer-membrane through which the major curli subunit protein CsgA is secreted as a soluble protein. The polymerization of CsgA is dependent in vivo on the membrane-bound nucleator protein CsgB (reviewed in 14). Complex regulatory pathways involving several two-component systems have been shown to control curli gene expression 15-16. These complex regulations allow bacteria to form thick biofilms via the curli production in response to environmental cues, but are difficult to control for industrial applications. To facilitate the recovery of the metal-stuffed bacteria during an industrial process, bacterial fixation to a solid support indeed needs to be controlled by well defined parameter(s). The adherent properties of curli are linked to their amyloid nature 17 and could be used to improve bioremediation processes, but a simpler and easily controlled device has to be created.
Amongst these 7 genes 18, a set of 5 absolutely required genes for curli synthesis (csgB and csgA encoding fiber monomers) and export (csgE csgF and csgG, encoding the curli secretion complex) were selected to construct the synthetic operon. To escape the "natural" regulation of curli, a synthetic operon comprising these 5 csg genes under the control of a strong and cobalt-overinducible promoter (Figure 2) was designed and synthesized. The step-by-step analysis of the curli-encoding region and the design procedure for a functional synthetic operon are described. Two methods to visualize and quantify bacterial adherence to polystyrene and glass are explained.
Protocol
1. Biobrick Design and Synthesis of the Curli Operon
Determine the genetic organization and localize the endogenous transcriptional and translational signals of the curli genes. These informations are gathered in specialized databases such as RegulonDBa or EcoGeneb and completed by a careful reading of pertinent publications. Data management and in silico preanalysis were performed with Clone Manager softwarec.
Select the pertinent coding sequences. A set of five absolutely required genes for curli synthesis (csgB and csgA encoding fiber monomers) and export (csgE csgF and csgG, encoding the curlin secretion complex) were selected to construct the synthetic operon. Extract the chosen sequences from the data base in FASTA format. As the csgGFE cluster is antisense in E. coli genome, convert this sequence into its reverse complement counterpart by using the Clone Manager tools Operations> Process molecule>Invert molecule. Paste the csgEFG sequence behind csgBA by using the function "Ligate" (Clone> Ligate).
Add the appropriate promoter. By placing the five selected curli genes under control of the promoter Prcn, curli are predicted to be over-produced in presence of cobalt and nickel. The rcn locus encodes an efflux pump responsible for Ni and Co detoxification (rcnA) and its cognate metallo-regulator (rcnR). RcnR controls the expression of rcnA and its own gene in response to Ni and Co 19. The rcn sequence comprising rcnR CDS, the whole rcn intergenic region plus the 41 first nucleotides of rcnA was placedin front of the csgBAEFG chimerical sequence (Figure 1, the sequence of the whole construct is provided as supplementary data).
Optimize the transcriptional signals. A perfect ribosome binding site (or perfect RBS= AAGGAGGTATATA) was added in front of the first ATG of the csgBA DNA sequence. A second perfect RBS was added in front of the csgEFG sequence. Endogenous RBS for the csgB and csgF and csgG genes were conserved.
To fit iGEM standards, the device must be flanked by a standard BioBrick prefix and suffix, containing restriction sites for EcoRI, PstI (prefix) and SpeI and XbaI (suffix). Paste the corresponding sequences at each end of the deviced.
Eliminate any EcoRI, PstI, SpeI and XbaI recognition site in the device. To facilitate further assembly process, the BioBrick part itself may not contain any of these restriction sites. In silico restriction analysis of the device revealed one PstI site in the csgA sequence, one PstI site in the rcnR sequence and one EcoRI site in the csgE gene. These sites are respectively located in position 80 (Mut1), 1340 (Mut2) and 1830 (Mut3) of the device sequence (supplementary data) and were modified as follow by silent mutations. Mut1 PstI site CTGCAG changed in CGTCTG Mut2 PstI site CTGCAG changed in CAGCAG Mut3 EcoRI site GAATTC changed in GAATT
Run through the translation simulation using the Clone Manager software (Operation>Process molecules>Translate molecules). Use the sequence alignment program BLAST to verify the perfect homology between the wild type curli protein and the proteins encoded by the synthetic operon.
Order the synthetic operon. Commercial gene synthesis services are available from numerous companies worldwide, our partner was Genecust (Luxembourg). 4 μg of the artificial operon (3165 bp) were received few weeks later and inserted into pUC57 (pIG2).
2. Visualize and Quantify Adherent Bacteria on Polystyrene
Fill each well of a 24-well polystyrene plate with 2 ml of M63 minimal medium (glucose 0.2%) and inoculate each well with 106 cells of an overnight culture. Grow bacteria at 30 °C for 18 to 48 hr without shaking. Each column (4 wells) represents a modality. Add the proper amount of cobalt (25 to 100 mM) and antibiotic (ampicillin 100 μg.L-1) when needed. The 3 first rows of the 24-well plate are used to quantify the adherent bacteria by averaging the 3 repetitions, the last row left is used to visualize the biofilm.
For the 3 first rows of the 24-well plate: for each well, recover the supernatant containing the planktonic cells.
Carefully rinse each well with 1 ml of M63 and pool the 1 ml wash with the initial supernatant obtained in 2.2). This pool is referred to as swimming cells (=S).
Recover the biofilm in 1 ml of M63 by scraping and pipetting up and down (=B). Vortex 15 sec. Estimate the number of surface-attached and swimming bacteria from the optical density at 600 nm (OD600) to give the adherence percentage corresponding to each modality.
The percentage of adherence is calculated by using the formula: Bx100/(3xS+B).
Only for the last row of the 24-well plate: discard planktonic cells.
Rinse with 1 ml of M63 the biofilm which had developed on the bottom of the plate.
Dry the open plate for 1 hr at 80 °C.
Add 100 μl of 20% crystal violet for 2 min in each well followed by extensive washes with water to visualize the surface-attached bacteria. Three independent assays (plates) have to be performed to ensure reproducibility.
3. Visualize Adherent Bacteria on Glass by Microscopy
Get a fluorescent chassis. The E. coli SCC1e strain which constitutively expresses green fluorescent protein (GFP) with no observable difference from the parent strain MG1655 20 is a suitable host for the plasmid bearing the synthetic curli operon.
Transform SCC1 with the pIG2 plasmid (=synthetic curli operon inserted at the EcoRI/PstI site of the pUC57 plasmid) to obtain the S23 strain. Transform SCC1 with a control plasmid (pUC18) to obtain the control strain S24 21.
Grow overnight precultures of the S23 and of the control (S24) strains at 30 °C in M63 medium supplemented with glucose (0.2 %) and ampicillin (100 μg.L-1).
Inoculate 15 ml of the same medium in Petri dishes with 100 μl of the precultures. Add the appropriate concentration of cobalt (i.e. 25 μM) and don't forget the negative control without cobalt. Introduce 3 rectangular glass coverslip in each Petri dish. Incubate overnight at 30 °C without shaking.
Remove the coverslip from the Petri dish and carefully drain it. Carefully clean the lower face with a small cotton stuff impregnated with 70 ° ethanol by wiping off every bacterial residue. For confocal observation, clean both upper ends on a few millimeters to allow further fixation of the coverslip.
Adherent bacteria can be visualized directly but quickly under fluorescence microscope, but carefully avoid drying of the sample.
For confocal observation, deposit the coverslip on a glass slide with the upper face covered by the biofilm placed face up. The biofilm is then covered with a larger coverslip, which is fixed to the glass slide with varnish. Invert the setup so that the biofilm will now be on the lower face.
Place the setup under the confocal laser microscope. A right Axioplan2 LSM510 (Zeiss) confocal laser microscope was used at the Platim platformf.
Setting. Excite GFP at 488 nm, and collect the bacterial fluorescence in the range 500 to 600 nm. Use 40x oil immersion objective for acquiring images in laser scanning confocal mode. Scan the overall three-dimensional structures of the biofilms from the solid surface to the interface with the growth medium, using a step of 1 μm.
Perform three-dimensional projections with IMARIS software (Bitplane, Zürich, Switzerland). Determine biofilm thickness by the analysis of the average z value along statistical longitudinal sections. Quantification of biofilm biovolumes can be extracted from confocal z-stacks as described elsewhere 22.
a RegulonDB provides mechanistic information about operon organization and their decomposition into transcription units, promoters and their sigma type, genes and their ribosome binding sites, terminators, binding sites of specific transcriptional regulators,as well as their organization into regulatory phrases. http://regulondb.cs.purdue.edu/index.jsp
bThe EcoGene database contains updated information about the E. coli K-12 genome and proteome sequences, including extensive gene bibliographies. A major EcoGene focus has been the re-evaluation of translation start sites. http://ecogene.org/
c http://www.scied.com/dl_cmp9d.htm
d http://partsregistry.org/wiki/index.php?title=Help:BioBrick_Prefix_and_Suffix
eGift of Chun Chau Sze, Nan Yang Technical University, Singapore.
fPlatim microscopic platform UMS3444 BioSciences Gerland - Lyon Sud.
Representative Results
In silico annotation of the wild type csg sequence of E. coli K12 associated with the optimization of transcriptional and translational signals has allowed to design the single synthetic curli operon Prcn-csg shown in Figure 3 (full sequence in supplementary data). Protocol 2 and protocol 3 were used to visualize and quantify adherence associated with curli production. Using crystal violet staining on 24-well polystyrene plates (protocole 2) biofilm formation at the bottom of each well can be rapidly visualized and it appears that the wild type strain is less adherent than the engineered strain as shown by the difference in purple color intensity (Figure 4A). This qualitative approach is strengthened by quantitative measurement which reveals that the percentage of adherence is 1.5 fold higher in the engineered strain (Figure 4B). Moreover, accuracy of the quantitative approach allows measurement of a significant reinforcement of adherence in presence of increasing concentrations of cobalt. Indeed, in media with cobalt concentrations of 0 μM, 25 μM or 50 μM, percentages of adherent cells reach 25%, 30% and 40% respectively (Figure 4B). Adherence abilities can also be compared using microscopy with GFP-tagged bacteria grown on glass slides. Epifluorescence microscopy observations (green bacteria on black background) reveal that the engineered strain forms a several layers large aggregate considered as a biofilm whereas the wild type strain forms only micro-colonies (Figure 5). Confocal microscopy analysis provides an overall view of the biofilm three-dimensional structure and confirms that the engineered strain is more adherent, forming a denser and thicker biofilm (Figure 6).
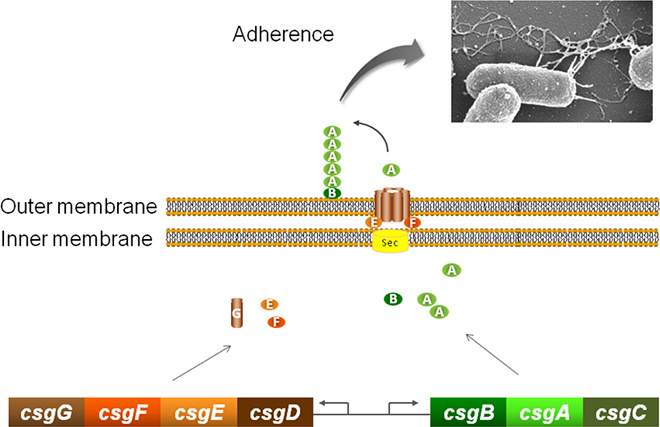 Figure 1. Curli wild-type system. The curli are made of two monomers encoded by the csgB and csgA genes. Thanks to their signal peptide, the CsgA and CsgB curli monomers are translocated across the cytoplasmic membrane via the Sec system. A specific machinery composed of 3 main components, namely CsgE, CsgF and CsgG allows the translocation of curli monomers across the outer membrane.
Figure 1. Curli wild-type system. The curli are made of two monomers encoded by the csgB and csgA genes. Thanks to their signal peptide, the CsgA and CsgB curli monomers are translocated across the cytoplasmic membrane via the Sec system. A specific machinery composed of 3 main components, namely CsgE, CsgF and CsgG allows the translocation of curli monomers across the outer membrane.
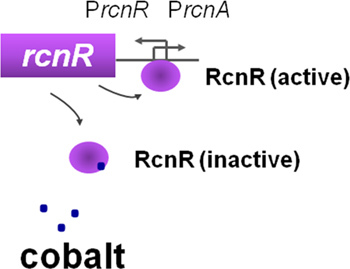 Figure 2. A strong and cobalt inducible-promoter (BBa_K540001). The promoter PrcnA is controlled by the transcriptional repressor RcnR. The rcnR gene is transcribed from its endogenous promoter PrcnR, divergently from PrcnA. The presence of the rcnR gene ensures a correct ratio of repressor copies versus PrcnA regulatory region. In absence of cobalt, RcnR binds to the RcnR box on DNA and prevents the full transcriptional activation of the downstream genes. If the intracellular concentration of cobalt rises, the binding of cobalt to the RcnR protein prevents the fixation of the repressor to the promoter and the transcriptional activity of PrcnA increases 19.
Figure 2. A strong and cobalt inducible-promoter (BBa_K540001). The promoter PrcnA is controlled by the transcriptional repressor RcnR. The rcnR gene is transcribed from its endogenous promoter PrcnR, divergently from PrcnA. The presence of the rcnR gene ensures a correct ratio of repressor copies versus PrcnA regulatory region. In absence of cobalt, RcnR binds to the RcnR box on DNA and prevents the full transcriptional activation of the downstream genes. If the intracellular concentration of cobalt rises, the binding of cobalt to the RcnR protein prevents the fixation of the repressor to the promoter and the transcriptional activity of PrcnA increases 19.
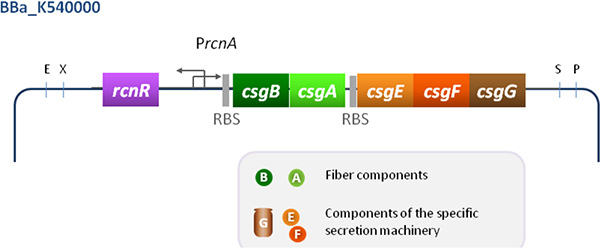 Figure 3. The synthetic curli operon. Curli genes are placed under the control of PrcnA. The rcnR gene expressed from its own promoter should provide enough repressor to control the expression of curli genes in a cobalt dependent manner. To avoid periplasmic traffic jam due to curli monomer overproduction (CsgB and CsgA), the components of the curli specific secretion apparatus (CsgE, CsgF and CsgG) have to be overproduced too. Added traductional signals are indicated in grey (RBS= Perfect RBS). E= EcoRI X= XbaI S= SphI P= PstI. This synthetic part referred to as Bba_K540000 allows the engineered strain to become adherent to glass, sand or plastics via curli overproduction.
Figure 3. The synthetic curli operon. Curli genes are placed under the control of PrcnA. The rcnR gene expressed from its own promoter should provide enough repressor to control the expression of curli genes in a cobalt dependent manner. To avoid periplasmic traffic jam due to curli monomer overproduction (CsgB and CsgA), the components of the curli specific secretion apparatus (CsgE, CsgF and CsgG) have to be overproduced too. Added traductional signals are indicated in grey (RBS= Perfect RBS). E= EcoRI X= XbaI S= SphI P= PstI. This synthetic part referred to as Bba_K540000 allows the engineered strain to become adherent to glass, sand or plastics via curli overproduction.
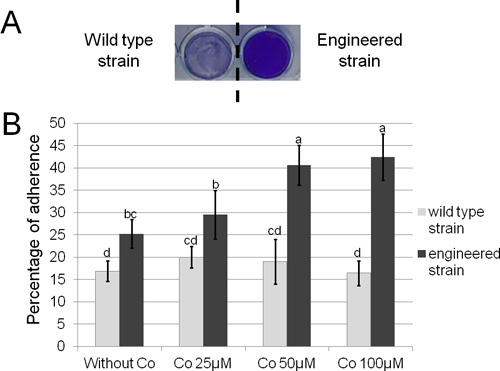 Figure 4. Visualization and quantification of the adherent bacteria on 24-well polystyrene plate. A. Crystal violet stained biofilm formed by adherent bacteria on polystyrene. B. Percentage of adherence on polystyrene of the wild type and the engineered strains with various concentrations of cobalt. Statistical differences between treatments are indicated with lowercase letters (analysis of variance and Fisher's least significant difference test; P<0.05).
Figure 4. Visualization and quantification of the adherent bacteria on 24-well polystyrene plate. A. Crystal violet stained biofilm formed by adherent bacteria on polystyrene. B. Percentage of adherence on polystyrene of the wild type and the engineered strains with various concentrations of cobalt. Statistical differences between treatments are indicated with lowercase letters (analysis of variance and Fisher's least significant difference test; P<0.05).
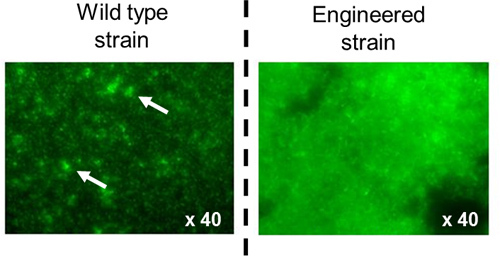 Figure 5. Epifluorescence microscopy images of adherent GFP-tagged bacteria on glass slides. Bacteria are green on a black background. Arrows point at microcolonies.
Figure 5. Epifluorescence microscopy images of adherent GFP-tagged bacteria on glass slides. Bacteria are green on a black background. Arrows point at microcolonies.
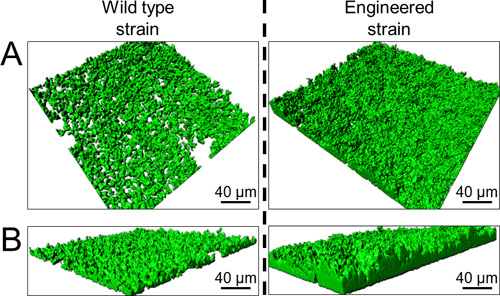 Figure 6. Confocal microscropy assisted three-dimensional reconstructions of biofilm structure on glass slides: A. Top view reconstructions B. Side view reconstructions.
Figure 6. Confocal microscropy assisted three-dimensional reconstructions of biofilm structure on glass slides: A. Top view reconstructions B. Side view reconstructions.
Discussion
Critical steps
The most critical step in this synthetic biology approach is the gene design. Synthetic gene design has to be meticulous to ensure an efficient system production. Two genes encoding the fiber monomers and three genes encoding proteins involved in their secretion system have been assembled with a strong and metal-inducible promoter to create a new functional unit for a novel application: the bio-decontamination of nuclear effluent. As planned and predicted, this device leads to an enhanced high curli production, which is reinforced by increasing amount of cobalt in the medium. Such a success results from the presence of both the appropriate transcriptional signals in the chosen promoter (PrcnA), and the efficient translational signals added in front of each set of curli genes (csgBA and csgEFG) (see Figure 3). In addition, when working with a multicopy plasmid, attention has to be paid to the copy number of the regulator(s) involved in the promoter control. Here, multicopies of the main regulator RcnR of the used promoter (PrcnA) were provided from the same plasmid (Figure 3).
Limitations, possible modifications
To ensure a perfect reproductibility, the adherence test has always to be performed in the same brand of polystyrene plates (see Table). Fluorescence microscopy is easier with fluorescent-tagged strains, but this requirement can be overcome by using fluorescent dyes such as Syto, or plasmid carrying lac-GFP fusions 23. Bacterial attachment to abiotic surfaces such as polystyrene and glass depends on the ionic strength or osmolarity of the medium. Best results are usually obtained in minimal medium or diluted LB 22, 24.
Future applications or directions after mastering this technique
The method described here to quantify bacterial adherence is rapid and cheap. However, to characterize a large number of strains or culture conditions, and/or to obtain a detailed structural characterization of biofilms, high throughput method based on confocal laser scanning microscopy combined with the use of 96-well microtiter plates has to be considered 25.
The MBEC screening system, formerly Calgary Biofilm Device 26 can also be used. This system is based on the use of a microtiter plate with a removable lid that hold 96 pegs substratum allowing microscopic observations of biofilm structure 27, or quantification of cells in biofilm after disruption by sonication on a water table sonicator (The MBEC High-throughput (HTP) Assay, Innovotech, Edmonton, Canada). Another system, the Biofilm Ring Test, monitors how inert paramagnetic beads included in the culture medium are immobilized during the formation of the biofilm 28, and can be used to quantify biofilm formation. MBEC and Biofilm Ring Test require specific materials that are more expensive than basic consumables used in this study.
Significance of the technique with respect to existing methods
To create a single independent curli operon, we tried two methods in parallel: a synthetic approach described here, and a classic method involving mutagenesis to remove internal EcoRI and PstI restriction sites, and PCR steps followed by ligations. Both approaches were initiated at the same time, but sequencing analysis of the operon obtained by the classical method revealed undesirable mutations. Therefore, synthesis of the device has been more efficient and more rapid than classical cloning and mutagenesis procedures.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank other members of the Lyon INSA-ENS iGEM team (Viviane Chansavang, Mathilde Dumond, Alexandre Duprey, Mélanie Geffroy, Clémence Gonthier, Margaux Jaulin, Aurélie Haag, Goki Ly, Thomas Poinsot, Béryl Royer-Bertrand, Julie Soula, Michael Vonzy, Pierre Yves Zundel, Soufiane Bouhmadi, Olivier Brette, Gaël Chambonnier, Laura Izard, Aurianne Kroiss, Philippe Lejeune, Agnès Rodrigue, Arnaud Rondelet, Sylvie Reverchon and Valérie Desjardin), our sponsors for their financial support (bioMérieux, Assystem, EDF, Fondation INSA, ENS-Lyon and the Department of Biosciences INSA-Lyon), F. Wisniewski-Dyé for critical reading of this manuscript and Dr C.C. Sze for strain gift. B. Drogue receives a Ph.D. fellowship from Région Rhône-Alpes.
References
- O'Toole GA. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011. p. e2437. [DOI] [PMC free article] [PubMed]
- Melamed S, Elad T, Belkin S. Microbial sensor cell arrays. Curr. Opin. Biotechnol. 2011. [DOI] [PubMed]
- Melamed S. A printed nanolitre-scale bacterial sensor array. Lab Chip. 2011;11:139–146. doi: 10.1039/c0lc00243g. [DOI] [PubMed] [Google Scholar]
- Raghu G, Balaji V, Venkateswaran G, Rodrigue A, Maruthi Mohan P. Bioremediation of trace cobalt from simulated spent decontamination solutions of nuclear power reactors using E. coli expressing NiCoT genes. Appl. Microbiol. Biotechnol. 2008;81:571–578. doi: 10.1007/s00253-008-1741-6. [DOI] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, et al. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal O. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo G, Chen X, Hay AG, Lion LW. Curli produced by Escherichia coli PHL628 provide protection from Hg(II) Appl. Environ. Microbiol. 2010;76:6939–6941. doi: 10.1128/AEM.01254-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon-Rodriguez W, Yatsimirsky AK, Glabe CG. Binding of Zn(II), Cu(II), and Fe(II) ions to Alzheimer's A beta peptide studied by fluorescence. Bioorg. Med. Chem. Lett. 1999;9:2243–2248. doi: 10.1016/s0960-894x(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Taylor JD, et al. Atomic resolution insights into curli fiber biogenesis. Structure. 2011;19:1307–1316. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E, Dorel C, Zehnder AJ, Landini P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- Pesavento C, et al. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueholm MS, et al. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry. 2011;50:8281–8290. doi: 10.1021/bi200967c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, et al. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostaert AS, Higgins MJ, Fukuma T, Rindi F, Jarvis SP. Nanoscale mechanical characterisation of amyloid fibrils discovered in a natural adhesive. J. Biol. Phys. 2006;32:393–401. doi: 10.1007/s10867-006-9023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Blaha D. The Escherichia coli metallo-regulator RcnR represses rcnA and rcnR transcription through binding on a shared operator site: Insights into regulatory specificity towards nickel and cobalt. Biochimie. 2011;93:434–439. doi: 10.1016/j.biochi.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Miao H, Ratnasingam S, Pu CS, Desai MM, Sze CC. Dual fluorescence system for flow cytometric analysis of Escherichia coli transcriptional response in multi-species context. J. Microbiol. Methods. 2009;76:109–119. doi: 10.1016/j.mimet.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin C. Nickel promotes biofilm formation by Escherichia coli K-12 strains that produce curli. Appl. Environ. Microbiol. 2009;75:1723–1733. doi: 10.1128/AEM.02171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg GV, Wijfjes AH, Lamers GE, Stuurman N, Lugtenberg BJ. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: new perspectives for studying microbial communities. Mol. Plant Microbe Interact. 2000;13:1170–1176. doi: 10.1094/MPMI.2000.13.11.1170. [DOI] [PubMed] [Google Scholar]
- Landini P, Jubelin G, Dorel C. In: Biological Adhesives. Callow, J, Smith AM, editors. Springer-Verlag; 2006. [Google Scholar]
- Bridier A, Dubois-Brissonnet F, Boubetra A, Thomas V, Briandet R. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J. Microbiol. Methods. 2010;82:64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Ceri H, et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JJ, et al. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary Biofilm Device. Biol. Proced. Online. 2006;8:194–215. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavant P, Gaillard-Martinie B, Talon R, Hebraud M, Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J. Microbiol. Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Miller JE. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]


