Abstract.
Recent advances in optical coherence tomography (OCT), and the development of image reconstruction algorithms, enabled four-dimensional (4-D) (three-dimensional imaging over time) imaging of the embryonic heart. To further analyze and quantify the dynamics of cardiac beating, segmentation procedures that can extract the shape of the heart and its motion are needed. Most previous studies analyzed cardiac image sequences using manually extracted shapes and measurements. However, this is time consuming and subject to inter-operator variability. Automated or semi-automated analyses of 4-D cardiac OCT images, although very desirable, are also extremely challenging. This work proposes a robust algorithm to semi automatically detect and track cardiac tissue layers from 4-D OCT images of early (tubular) embryonic hearts. Our algorithm uses a two-dimensional (2-D) deformable double-line model (DLM) to detect target cardiac tissues. The detection algorithm uses a maximum-likelihood estimator and was successfully applied to 4-D in vivo OCT images of the heart outflow tract of day three chicken embryos. The extracted shapes captured the dynamics of the chick embryonic heart outflow tract wall, enabling further analysis of cardiac motion.
Keywords: optical coherence tomography, embryonic heart outflow tract, deformable model, robust detection
1. Introduction
This work is motivated by the need to analyze the motion of the embryonic heart and to elucidate relationships between altered cardiac mechanics during early development and congenital heart defects. Blood flow is essential for cardiac development, and alterations in blood flow dynamics can lead to heart defects,1 which affect about 1% of newborn babies. The interaction between cardiac tissue and blood flow dynamics determines the mechanical stimuli (e.g., stresses/strains) to which cardiac cells are subjected, stimuli that modulate cardiac growth and development. Images of the beating heart can then be used not only to visualize cardiac motion but also to quantify mechanical stimuli that modulate intrinsic cardiac genetic programs.
Optical coherence tomography (OCT) has been extensively used to study embryonic and cardiac development.2,3 Recently, the development of spectral domain OCT (SDOCT)4 made it possible to acquire in vivo image sequences of the developing heart5,6 and to obtain four-dimensional (4-D) [three-dimensional (3-D) images over time] OCT images of the beating embryo heart7,8 and blood flow velocities.9,10 Although OCT allows micro-structural and Doppler velocity visualization, quantification and analyses of cardiac motion and blood flow have been hampered by a lack of reliable, automatic procedures to extract the shape and motion of the developing heart from OCT images.
To analyze cardiac motion from in vivo OCT images, most previous studies used manual segmentation and manually extracted measurements.10,11 However, thorough manual analysis of OCT cardiac image sequences, which comprise large volumes of data, can be very time consuming and subject to operator errors. Therefore, to facilitate quantification and comparison of image datasets there is a dire need to develop automated analysis of cardiac motion from OCT images. Here, we present a semi-automatic, robust procedure to extract the dynamic shapes of the beating embryonic heart from 4-D OCT images.
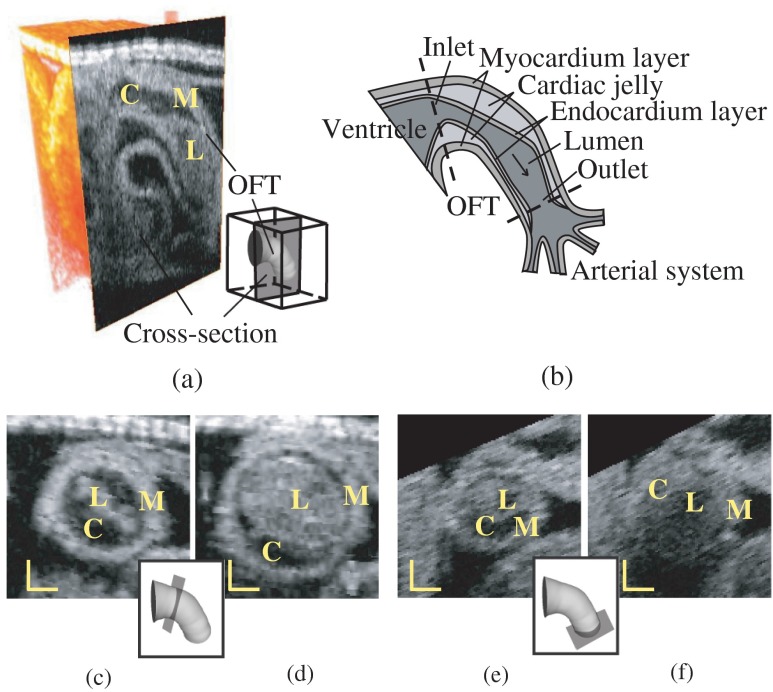
We applied our segmentation procedure to 4-D images of a cardiac segment, the heart outflow tract (OFT), of chick embryos at the Hamburger-Hamilton12 developmental stage 18 (HH18), approximately 3 days of incubation. At HH18, the heart OFT has a tubular structure, with an external myocardial layer, an endocardial layer that is in contact with blood, and a cardiac jelly layer in between the myocardium and endocardium (Fig. 1). From OCT images, the myocardium, cardiac jelly and the blood can be readily distinguished. The endocardium that lines the lumen, however, cannot be distinguished from blood. This is, however, not an important limitation as the endocardium is a thin monolayer of endocardial cells and thus can be assumed to coincide with the lumen-tissue interface. During the cardiac cycle, the OFT myocardium contracts and expands approximately preserving a tubular shape. In contrast, upon contraction, the shape of the endocardium becomes irregular as it “folds” along lines that are approximately aligned with the direction of flow.11 Our developed procedure successfully detects and segments the OFT myocardium and endocardium over the cardiac cycle.
Fig. 1.
Structure of the heart outflow tract (OFT). (a) 3-D image set acquired with OCT, showing a longitudinal OFT section. (b) Sketch of the OFT showing its wall layers, as well as proximal and distal connections. The arrow shows the direction of blood flow. (c) and (d) Cross-sectional images at the inlet of the OFT during maximal contraction and maximal expansion. (e) and (f) Cross-sectional images at the outlet of the OFT during maximal contraction and maximal expansion. M: myocardium; C: cardiac jelly; L: lumen. Scale bars: 100 μm.
Automatic extraction of myocardium and endocardium shapes from 4-D OCT images presents several difficulties, including:
-
(1)
the presence of tissues that are adjacent to the myocardial layer;
-
(2)
image noise and variations in intensity due to weakening of OCT signals with tissue depth [see Fig. 1(e) and 1(f)];
-
(3)
the irregular, folding motion of the endocardium, with a shape that deviates from tubular upon OFT contraction; and
-
(4)
large displacements of OFT tissue layers over time.
To overcome these difficulties, a segmentation procedure must distinguish target tissue from adjacent tissue, even as intensity lowers with tissue depth and must successfully track the large and irregular displacements of target tissues.
Algorithms have been developed to automatically extract the shape of tubular structures from vascular images.13–19 In particular, deformable models,20,21 such as active-contour (snake), have been widely used to detect the boundary of target tissues and extract their shapes.22–25 These models, however, typically fail to correctly detect target tissues when adjacent tissues with similar intensity are present. To correctly segment target tissue layers, piece-wise deformable models (a type of template-matching model) adjust their shape to match the shape of the tissue layer by using intensity values from inside and outside of the model.26–28 Here, we designed a robust two-dimensional (2-D) piece-wise deformable model, the double-line model (DLM), to locally detect the myocardial layer and the endocardial layer from 2-D cross-sectional images of the developing heart. A robust likelihood estimator is used to achieve accurate detection of the target tissue layer even with weakening intensities of OCT signals (lower signal-to-noise ratios) with tissue depth, and in the presence of adjacent tissues. Tracking of tissue layers was performed in space and time to achieve 4-D segmentations. The extracted cardiac shapes accurately captured the geometrical configuration of the OFT tissue layers and their motion over the cardiac cycle.
2. 4-D OCT Imaging of the Chick Embryonic Heart Outflow Tract
In this section we briefly describe the acquisition of in vivo imaging data using OCT, and image data reconstruction into 4-D images. Prior to imaging, white leghorn eggs were incubated to HH18 (). The eggs were then removed from the incubator and placed in an organic glass cube in which temperature was controlled to approximately 37.5°C to keep the cardiac cycle at a physiological range (around 370 ms). The egg shell and part of the membrane overlaying the embryo were removed to provide optical access to the chick heart. A spectral domain OCT system was then used to image the OFT of the embryonic chick heart.8,9 The system acquired images (256 A-scans) at a rate of 140 frames per second (fps), with axial and lateral resolutions of and . Following our previously described 4-D imaging strategy,8 we acquired 2-D image sequences that included about four cardiac cycles (200 frames at speed of 140 fps) at different image planes along the OFT, separated by 7.5 μm. Our imaging procedure resulted in 120 frame sequences along the OFT (see Fig. 2). We also imaged longitudinal frame sequences (for reconstruction purposes). Raw image acquisition took about 20 minutes.
Fig. 2.
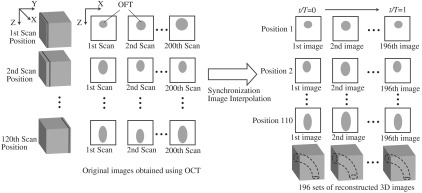
Schematic representation of OCT imaging strategy and reconstruction procedure. Cross-sectional OCT images were obtained by scanning the heart at 120 positions (separated by 7.5 μm) along the OFT. At each position, 200 sequential 2-D scans were obtained at 140 fps (about four cardiac cycles). A transverse position was also imaged (not shown). Acquired image sequences, however, start at different phases (timings) over the cardiac cycle. To synchronize the images, cross-sectional sequences are first pooled to one cardiac cycle, phases between image sequences are calculated, and then image sequences are interpolated so that sequences from all positions start at the same time and have the same number of image frames (196 frames over the cardiac cycle). 3-D images that show the position of the OFT over the cardiac cycle are then constructed to obtain 196 sets of 3-D images, with each 3-D image consisting of 110 positions along the OFT. in the right image is time, and is the period of the cardiac cycle (370 ms).
To obtain 4-D images of the OFT, the acquired OCT image frames were combined using our developed reconstruction procedure.8 Since image acquisition was not gated, we used similarity of lines from neighboring images to synchronize frame sequences acquired at different image planes along the OFT, and then we calibrated the synchronization using time-point lags estimated from longitudinal image frames. To generate the 4-D image dataset, we pooled the acquired image frames to one cardiac cycle interpolating the images to obtain several 3-D reconstructions over the cardiac cycle (see Fig. 2). Each reconstructed 4-D image dataset consisted of 3-D image datasets at 196 different time points (over one cardiac cycle), with each 3-D image dataset consisting of . We applied our shape extraction algorithm to these reconstructed 4-D OFT image datasets.
3. Developed 4-D Shape Extraction Procedure
We designed an algorithm to extract the dynamic shapes of the heart myocardium and endocardium from reconstructed 4-D OCT images. We extracted three surfaces over time: the external and internal surfaces of the myocardium layer and the endocardium (or tissue-lumen interface) surface. Here, we first present a brief overview of the extraction procedure. Then, we explain in more detail the DLM model and the segmentation strategy employed.
3.1. Overview of the 4-D Extraction Algorithm
To extract the shape of the cardiac tissue layers, we used 2-D cross-sectional planes swept along the centerline of the heart OFT (see Fig. 3 and Sec. 3.4 for a detailed description of how the centerline and cross-sectional planes are obtained), and a robust 2-D segmentation algorithm. The advantages of using 2-D cross-sectional planes are:
-
(1)
for tubular structures tissue layer surfaces form a closed contour on the plane and
-
(2)
image processing is simpler on 2-D frames (than 3-D or 4-D volume data).
The 2-D segmentation algorithm was successively applied to 2-D cross-sectional planes over space and time to render 4-D segmentations. We used this strategy, together with smoothing and regularization procedures, to extract the shapes of the myocardium and endocardium over the cardiac cycle.
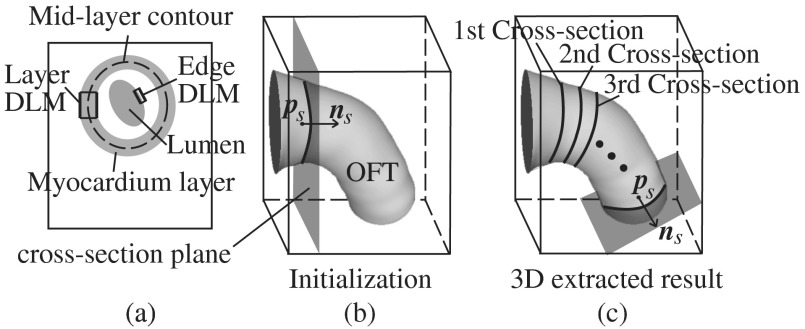
Fig. 3.
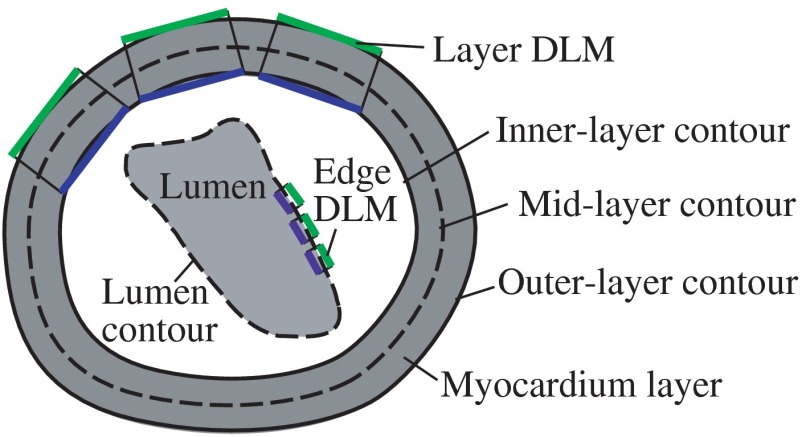
Sketch showing the OFT shape extraction procedure from OCT images. (a) Cross-sectional plane depicting the myocardium and endocardium layers, the myocardium mid-layer contour, a layer DLM placed on the myocardium and an edge DLM placed on the endocardium. (b) Initial cross-sectional plane. (c) Extraction of the cardiac shape is performed by sweeping cross-sectional planes along the OFT.
The 2-D segmentation algorithm was based on a local, piece-wise deformable double-line model [DLM; see Fig. 3(a)]. Two slightly different DLM formulations were developed: the layer DLM, and the edge DLM. To detect the myocardium layer (internal and external surfaces) we used the layer DLM, and to capture the endocardium layer (essentially a thin interface between the tissue and the lumen), we used the edge DLM [see Fig. 3(a)]. In our algorithm implementation, to detect cardiac tissues, several layer DLMs are placed around the myocardium and several edge DLMs are placed around the endocardium. The DLMs on each tissue layer are then linked together using active-contour techniques, and detection of the myocardial and endocardial layers is guided by a robust maximum-likelihood estimator. Linking the DLMs with active-contour facilitates detection and tracking of tissue motion.
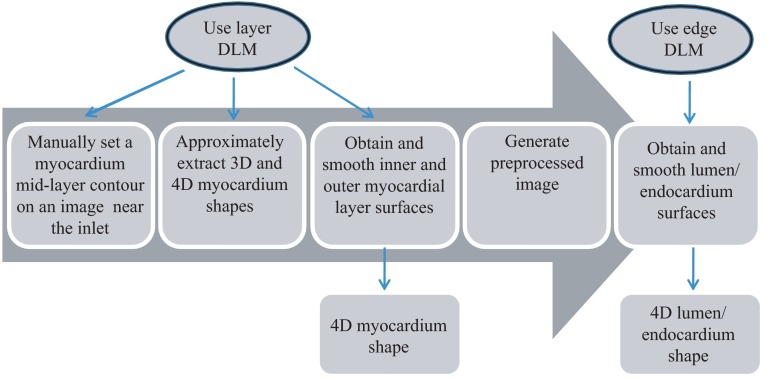
Our extraction procedure segments the myocardium layer and then the endocardium layer (see Fig. 4). Extraction of 3-D surface shapes from the first 3-D image volume dataset (time ) starts with a manual setting of the myocardium mid-layer contour [a contour that divides the thickness of the myocardium layer in half, see Fig. 3(a)] on an initial plane. Layer DLMs are arranged around the mid-layer contour, and linked with an active-contour model. Then, guided by a maximum-likelihood estimator, the layer DLM thickness and position are adjusted to match the myocardium layer [see Fig. 3(b)]. The procedure is repeated for each cross-sectional plane until the entire 3-D volume data is spanned [see Fig. 3(c)]. To segment the myocardium at the next time-step , the configuration of the layer DLMs at is copied to the 3-D volume data corresponding to , and the myocardium layer is detected in the new time step. The procedure is repeated for all time steps, until the entire cardiac cycle is spanned. Smoothing in space and time is then performed to yield the myocardial shape.
Fig. 4.
Steps to extract the 4-D OFT myocardial and endocardial layer surfaces.
Once the myocardium has been segmented, we used the segmentation to mask the regions of the cross-sectional images that lie outside the inner myocardial surface (see Fig. 4). This is done to facilitate detection of the endocardium. To reduce image noise, we then generate a binary image that increases lumen contrast. Edge DLMs are then placed around the lumen-tissue interface, and linked with an active-contour. The position of the edge DLMs is then adjusted to match the endocardium layer on the preprocessed image. This is repeated in space and time until the endocardium from the whole 4-D dataset is segmented. Smoothing of the lumen/endocardium surface is then performed in space and time.
3.2. 2-D Detection of the Myocardial Layer
The layer DLM is a local 2-D template deformable model that we designed to track the myocardium layer, and is key to our myocardial shape extraction procedure. In this section, we will introduce the layer DLM, its main parameters, and the maximum-likelihood estimator used to match the DLM to the myocardial layer. We will also describe how DLMs are linked together with an active-contour model to smooth the contour and guide myocardium detection.
3.2.1. Layer DLM basic parameters
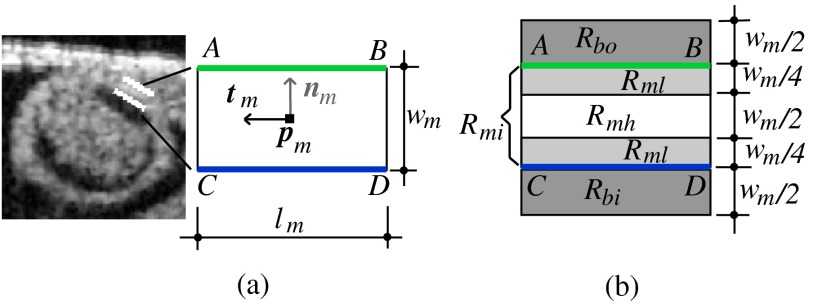
The DLM consists of two parallel lines: AB and CD [Fig. 5(a)], which are the outer and inner DLM edges, respectively. Basic parameters of the DLM describe its size and location: width ; length ; position (DLM center point position vector); and direction (a unit vector parallel to the DLM edges). These parameters also define a binormal vector , a unit vector perpendicular to and pointing from the DLM center to the DLM outer edge. represents the state of the DLM. To detect the OFT myocardium, layer DLMs are scaled by changing and ; rotated by adjusting ; and shifted by changing .
Fig. 5.
The layer double-line model (layer DLM). The layer DLM consists of two parallel lines AB (outer edge) and CD (inner edge), which can be adjusted to detect a tissue layer. (a) DLM parameters that define the DLM state : width, ; length, ; position, ; and direction, . (b) Regions of the layer DLM: the DLM is divided into an internal region, , which is in turn subdivided into two subregions, and ; and two external regions, and . These regions are used to calculate foreground and background intensity levels from OCT images.
The layer DLM was designed to
-
(1)
detect and track target, high-intensity tissue layers and
-
(2)
accurately capture the thickness of the tissue layers (avoiding adjacent tissues).
Local DLM intensity values are used to determine adaptive intensity levels, which, within a maximum-likelihood estimator, guide adjustment of DLM parameters to match the myocardial layer locally.
3.2.2. Foreground and background intensity estimation
To distinguish the target tissue from adjacent tissues, foreground and background intensity levels are estimated. The foreground intensity () is defined as the intensity of the target tissue; the background intensity () is defined as the intensity around the target tissue. In OCT images, the intensity at the center of the myocardial tissue layer is higher than the intensity near the tissue boundaries; also the intensity around the outer edge of the DLM may be different from the intensity around its inner edge. Thus, we define two levels of foreground intensity: high (), and low (); and two levels of background intensity: outer (), and inner (). To estimate and we use pixel intensities from regions inside and outside of the layer DLM, respectively [see Fig. 5(b)]. We divide the internal DLM region, , into two subregions, and , corresponding to the middle portion of the DLM, and the portions adjacent to the edges, respectively [see Fig. 5(b)]. Thus is a disjoint region. Similarly, we define two external regions, and , which are adjacent to the DLM outer and inner edges, respectively. The equations used to calculate foreground and background intensities are listed in Table 1. Foreground and background intensities are adaptive and depend on the local intensities of the region where the DLM is located. This allows successful detection of cardiac layers even when image intensity varies.
Table 1.
Equations used to calculate foreground and background intensity levels.
| Intensity level | Formula |
|---|---|
| , | |
| , and | |
| , and | |
| , | |
| , |
: intensity of the pixel at position .
: median value.
3.2.3. Maximum-likelihood estimator
It has been shown that maximizing a likelihood estimator that uses intensity information from inside and outside of a deformable model can be successfully used to differentiate target tissues from adjacent tissues.17,26,29 Inspired by these studies, we define the normalized likelihood function in discrete form by:
| (1) |
where is the intensity of a pixel located at position in the cross-sectional image, and is the state of the layer DLM. To reduce mis-detection of target tissue layers, the likelihood function was normalized with and the DLM area, , defined as the number of pixels in the DLM region considered.
To detect the target tissue, we find the DLM state that maximizes . Because the likelihood function uses local intensity information from inside and outside of the DLM, the detection algorithm is robust and can locally find the target tissue even when the local image is noisy and intensity is low.
3.2.4. Linking layer DLMs with the active-contour model
The layer DLMs are used to detect the myocardium locally. To mitigate local uncertainties, and facilitate smoothing, the DLMs placed around the myocardium layer (see Fig. 6) are linked with an active-contour (snake) model. The snake model is a classical method to detect the contour of an object in an image by means of energy minimization.22 The energy function, , of the active-contour model that we used here is,
| (2) |
where is the parametric curve of the myocardium mid-layer contour, with ; and are the first-order and second-order derivatives of ; denotes an external energy term, which in our implementation was equal to the negative of the intensity of the local pixel (maximum in the center of the myocardium layer, where the mid-layer contour of the DLM is placed); , and are constant weights.
Fig. 6.
Localization of DLMs around the myocardial layer and lumen interface.
3.2.5. Implementation and constraints
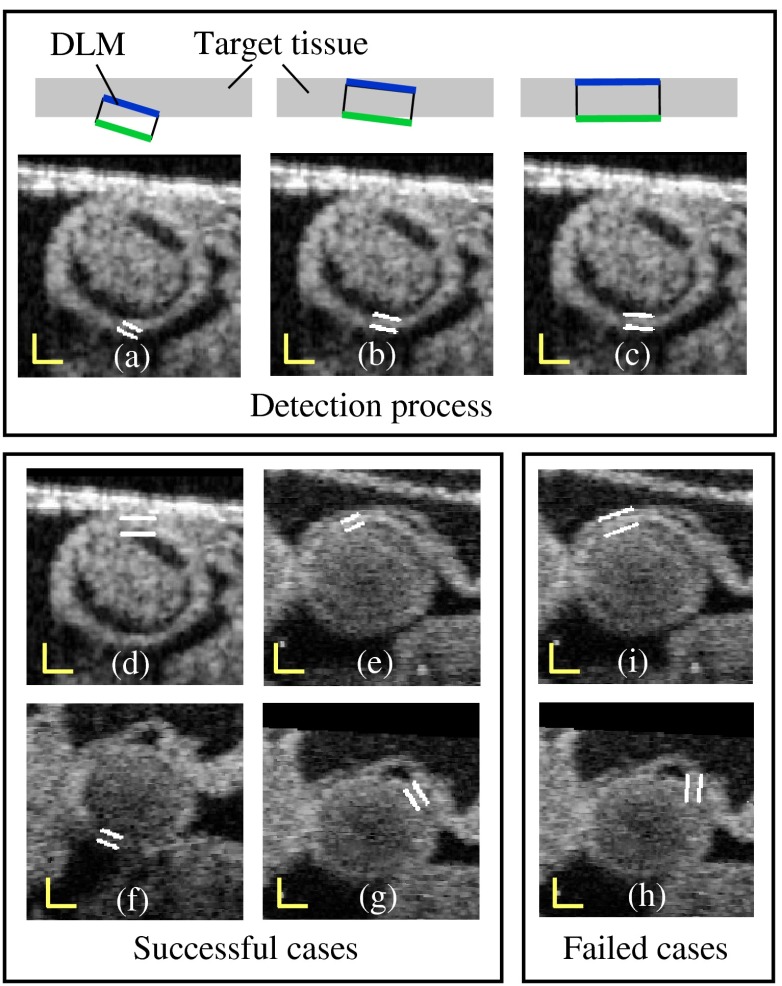
The layer DLM can be used to locally find the target tissue, by changing its size, position and direction to approach the tissue layer and, finally, match the tissue layer [see Fig. 7(a)–7(c)]. However, sometimes detection fails, especially in situations when there is: 1. close proximity of target and adjacent tissue layers [e.g., Fig. 7(i)]; 2. incorrect initial orientation of the layer DLM [e.g., Fig. 7(h)]; and 3. overlap of layer DLMs placed around the myocardium layer. To avoid these issues and increase the robustness of the extraction procedure, we constrained the layer DLM state parameters. Constraint values were found empirically from images, but we could use them in several image datasets without change.
Fig. 7.
Detection of the myocardial layer (target tissue) from OCT images of the embryonic heart OFT using the layer DLM. (a–c) Detection process. (d–g) Successful segmentation results: the layer DLM can successfully detect the myocardial layer even when there are adjacent tissues (in d and e) and intensity is weak (in f and g). (h) and (i) Examples of failed detection of the myocardium: adjacent and target tissues detected together (in i) and the orientation of the DLM is incorrect (in h). Scale bars: 100 μm.
To prevent simultaneous detection of both the target and adjacent tissue layers, we set bounds on the value of the DLM width . The maximum was constrained to be larger than the actual thickness of the target tissue layer, but smaller than about twice the target layer thickness. From analysis of OCT cross-sectional images, the thickness of the OFT myocardial layer was about 14 pixels (70 μm) near the OFT inlet, and 10 pixels (50 μm) near the OFT outlet. Thus, we set the maximum to be 16 pixels near the OFT inlet, and 14 pixels near the OFT outlet. We also set the minimum to be 4 pixels (20 μm) everywhere. To avoid incorrect orientation of layer DLMs, the DLM direction, , was aligned to the tangent of the active-contour curve that links the DLMs around the myocardium (see Fig. 6). Finally, to prevent overlapping of layer DLMs, DLM shifts were allowed only in the direction of the DLM binormal (that is perpendicular to the active-contour curve). Placing 40 layer DLMs around the myocardial layer in a cross-sectional image (see Fig. 6) was found to be enough to achieve an accurate segmentation of the myocardium.
Parameters of the active-contour model [see Eq. (2)] that linked the layer DLMs were also found empirically. Because the second term in Eq. (2), , is related to the contour curve smoothness, was chosen to be larger than and . Setting about 1.5 times larger than and (, ) worked well in implementation in several image datasets.
3.3. 2-D Detection of the Endocardial Layer
The procedure to extract the endocardium layer from the cross-sectional images is similar to the procedure used to extract the myocardium layer. In OCT images, the endocardium layer cannot be distinguished, but the lumen-wall interface appears as the “edge” between the low-intensity cardiac jelly and the high-intensity endocardium and lumen. However, the lumen edge is frequently blurred in OCT images and to facilitate detection, we preprocessed the images to increase contrast and reduce noise. In the preprocessed images, the lumen-wall edge shows as a region of finite width in which intensity changes continuously (from dark to bright), therefore suited for detection using a DLM. Changes to the DLM formulation to detect the lumen-wall interface (the “edge” DLM) included redefining the foreground and background intensities and the DLM constraints and changing the definition of the associated maximum-likelihood estimator.
3.3.1. Image preprocessing
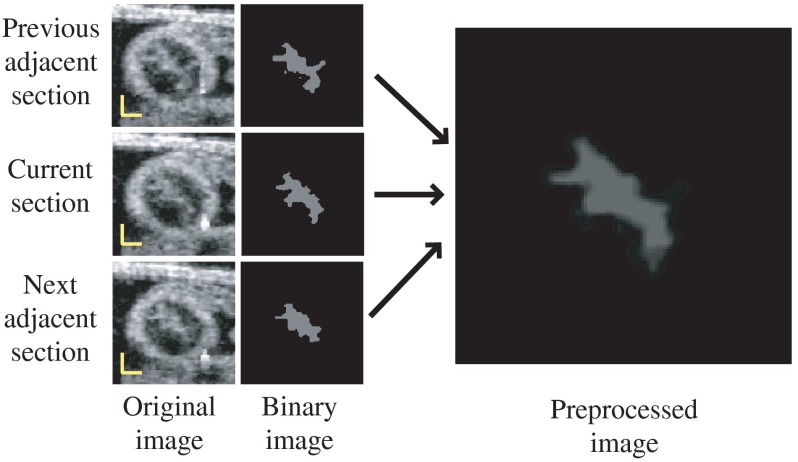
To generate preprocessed OCT images, we used an adaptive intensity threshold on the previously identified 2-D cross-sectional images and created a binary image with intensities either set to a fix high value, , or 0, with lumen areas showing with high-intensity. Because only the tissues inside the region delineated by the inner contour of the myocardium layer (the inner-layer contour, see Fig. 6) are useful for extracting the lumen-wall interface, the intensity of the pixels that are outside of the inner-layer contour are set to 0 (masked out) in the binary image. To increase image contrast, the intensity of the pixels within the inner-layer contour is calculated using the median intensity of a small image region (window) centered at each pixel. If the intensity of the pixel is smaller than the median intensity of the region, pixel intensity is set to 0; otherwise, it is set to . To reduce noise, we then calculate the mean intensity of three binary images: the image of interest and the two images that are adjacent to it in a 3-D image dataset (see Fig. 8). This operation reduces the sharpness of the lumen “edge” allowing use of a formulation that employs DLMs. We then use this preprocessed image to extract the lumen-wall interface.
Fig. 8.
Image preprocessing for extracting the 2-D lumen contour. Scale bars: 100 μm.
3.3.2. Edge DLM and maximum-likelihood estimator
To detect the lumen-wall interface, we slightly modified the formulation of the DLM, resulting in what we called an edge DLM formulation. The edge DLM is similar to the layer DLM, except that:
-
(1)
its width is set to a small, fixed constant;
-
(2)
background intensities are constant (, ,);
-
(3)
only one foreground intensity level is used, and its value is constant () rather than adaptive.
The maximum-likelihood estimator function used for detection of the lumen-wall interface, , is defined as follows:
| (3) |
The edge DLMs on the lumen contour are connected by an active-contour model. Similar to how layer DLMs are used to detect the myocardium, in detecting the lumen-wall interface at each cross-sectional preprocessed image, the likelihood function is locally maximized.
3.3.3. Implementation and constraints
The values of parameters needed to obtain an accurate segmentation of the lumen were determined empirically and tested on several image datasets. For the binary image, we used . Further, to make the lumen visible and yet accurate for extraction, the size of the window used to calculate the binary intensity of pixels was set to 31 pixels (that is about 165 μm) in the image vertical and horizontal directions, centered on the pixel of interest. These values resulted in preprocessed images that accurately showed the lumen domain (see Fig. 8).
To detect the lumen-wall interface, we set to be 4 pixels (20 μm), and background and foreground intensities to be constant: , , . Due to the irregular shape of the lumen contour, edge DLMs easily overlap when using an active-contour model to smooth the contour. Instead, we smooth the contour by averaging the position of adjacent DLMs on the lumen contour and, like for the layer DLM, use the active-contour to constrain edge DLM orientation and shifts. We found that placing 80 edge DLMs around the lumen contour in a cross-sectional plane was sufficient to accurately extract the lumen-wall interface.
3.4. Extraction of 4-D OFT Shapes
Extracting 4-D OFT shapes is performed as summarized in Sec. 3.1. We first determine the orientation of cross-sectional planes along the OFT, and extract the contours of the myocardial layer from cross-sectional images, using layer DLMs linked by active-contour models (see Fig. 4). Then, the lumen contours are extracted using edge DLMs applied to preprocessed cross-sectional images. It is worth noting here that we have not implemented a global optimization function on our procedure. Instead, our approach is to find an overall solution by successively solving local optimization problems:
-
(1)
Maximum-likelihood estimators to find local parameters of each DLM (DLM states are independent of each other at this step), and then
-
(2)
An active-contour algorithm to smooth and further refine the contour made by linking individual DLMs.
Step 2 changes the position of DLMs, and thus the procedure is iterated to find accurate solutions. Further, smoothing in space (outside of the plane) and time is also performed, and the successive procedure described above applied again after smoothing.
In this section, we describe in detail how we determine the orientation of cross-sectional planes, how the iterative segmentation procedure is applied, and how we smooth the extracted contours in space and time.
3.4.1. Determination of cross-sectional planes
A cross-sectional plane is defined here as a plane approximately perpendicular to the 3-D centerline of the OFT myocardium, and is represented by a position vector, , and a normal vector, (see Fig. 3). Changing shifts the plane, and changing rotates the plane. In our implementation, is the centroid of the myocardium mid-layer contour. Identification of cross-sectional planes in 4-D starts with a manual setting of the myocardium mid-layer contour on an initial plane on the first 3-D image volume dataset (). Layer DLMs are arranged around the mid-layer contour, linked with an active-contour model, and used to detect the myocardium layer on the plane. The initial plane chosen, however, is not necessarily a cross-sectional plane. To find a cross-sectional plane, the initial plane is rotated. DLMs are used to match the myocardium layer after each rotation, until a plane that has the minimum mid-layer contour perimeter (the cross-sectional plane) is found. The DLM configuration is then copied to a new plane that is parallel to the cross-sectional plane found but slightly shifted in the direction of the plane normal. DLMs then are used to match the myocardium layer, and the plane is rotated until a new cross-sectional plane is found. Plane shifting and rotation are repeated until the entire OFT from the first 3-D volume data is spanned [see Fig. 3(c)]. The centroids of the cross-section mid-layer contours are then extracted to form the OFT centerline, which is smoothed in space. After centerline smoothing, cross-sectional planes are adjusted, so that they remain perpendicular to the centerline and detection of the myocardium layer is performed on adjusted planes. This procedure results in an initial segmentation of the myocardium layer at time .
To segment the myocardium at the next time-step, , the configuration of the layer DLMs at is copied to the 3-D volume data corresponding to , and the segmentation procedure is used to change the DLM thickness and position to match the myocardial layer in the new volume dataset. The OFT centerline is then extracted, smoothed in space, and cross-sectional planes are adjusted. This is followed by detection of the myocardial layer on the adjusted cross-sectional planes. The procedure is then repeated for all time steps until the entire cardiac cycle is spanned. This strategy provides a good initial estimation of the position and orientation of cross-sectional planes in 4-D.
After the initial estimation of cross-sectional planes described above, the location and orientation of the planes is adjusted once more. To avoid spurious oscillations near the boundaries (the inlet and outlet of the outflow tract), the first and last cross-sectional planes along the OFT are fixed in space ( and do not change over time). For these planes, and are the mean of the previously estimated normal and position vectors over time. Plane positions in between the first and last planes are adjusted by first smoothing the centerlines in space and time. Smoothing in space and time uses data from adjacent planes: suppose that denotes a quantity to smooth on the ’th cross-sectional plane at time ; the new, smoothed value of is then caculated as the mean of , , , and . This procedure is performed for each component of and , and for each plane (except the first and last), and each time-point. Cross-sectional planes are then arranged so that distance within planes is as uniform as possible, while avoiding cross-sectional planes from crossing each other (such that a nonoverlapping mesh can be constructed directly from extracted results). In regions where the OFT centerline is rather straight, plane positions are approximately equidistant along the centerline, and in regions where the radius of curvature of the OFT centerline is small, distance between planes is larger (proportional to the contour mean radius and the angular change of ). This last step determines the final position and orientation of cross-sectional planes in 4-D.
3.4.2. Correction of inaccurate segmentations
Inaccurate segmentation could happen in our procedure because, like with other deformable models, layer detection using DLMs depends on the DLM initial state. Even after combining DLMs with active-contour models, and setting constraints for the DLM parameters (thickness, orientation), we still found a few sections that were inaccurately segmented (especially close to the OFT outlet, where signal-to-noise ratios diminish due to tissue depth). To solve this problem, an interactive interface was designed to allow the user to manually adjust cross-sectional contours and thus reset the state of the DLMs when needed. With this procedure, accurate segmentation of the 4-D cardiac OFT wall layer was achieved. In our implementation, inaccurate segmentations occurred at a very low frequency, with at most 0.2% frames showing failed segmentations. Nevertheless, even after detection failed, the DLM accurately found the target tissue again at the next adjacent section or time step.
3.4.3. Extraction and smoothing of the myocardium layer
Myocardium layer detection presents local fluctuations in space and time that need to be further smoothed. We proceed by first smoothing the myocardium mid-layer surface, which is described by the position vectors, , of the layer DLMs. Smoothing in space and time was performed (as in Sec. 3.4.1) by calculating the mean position of four spatiotemporally adjacent layer DLMs: the corresponding positions of two adjacent cross-sectional planes of the 3-D image set and the corresponding positions of two adjacent time points (in plane smoothing was performed using the active-contour). After smoothing the myocardium mid-layer, layer DLMs are used once more to accurately find the myocardium thickness at the new mid-layer positions. Outer and inner surface layers of the myocardium are then obtained by linking the outer and inner edges of the layer DLMs (see Fig. 6). Smoothing of the outer-layer and inner-layer surfaces is also carried out by calculating the mean positions of the four spatiotemporally adjacent points on the surfaces.
3.4.4. Extraction and smoothing of the endocardium/lumen surface
Once the myocardium layer is extracted, we proceed to extract endocardium/lumen surfaces. To this end, we first generate preprocessed images of each cross-sectional plane as described in Sec. 3.3.1. For the very first cross-sectional plane, edge DLMs are placed manually around the lumen edge on the preprocessed image, and the DLM parameters are optimized by maximizing , Eq. (3). Edge DLM parameters are then copied to the adjacent cross-sectional plane and then optimized. This is repeated until the whole 3-D dataset is segmented. For subsequent 3-D datasets, edge DLM parameters are copied from the previous time step, and then optimized. Like in the case of myocardial surfaces, after obtaining the lumen surfaces, surface smoothing is performed by calculating mean positions of four spatiotemporally adjacent edge DLM positions. This step completes the extraction procedure of 4-D surfaces from 4-D OCT images of the chicken heart outflow tract.
4. Results
We semi-automatically extracted the shapes of HH18 OFT cardiac tissue layers from 4-D in vivo OCT images of the chicken heart. More precisely, we extracted 4-D surfaces of the myocardium and tissue-lumen interface over 196 time points spanning one cardiac cycle (approximately 370 ms), with each 3-D OFT shape consisting of 50 cross-sections spanning around 700 μm along the OFT centerline. While we analyze here the extraction from one set of 4-D images in detail, we have so far successfully extracted data from seven HH18 OFTs.
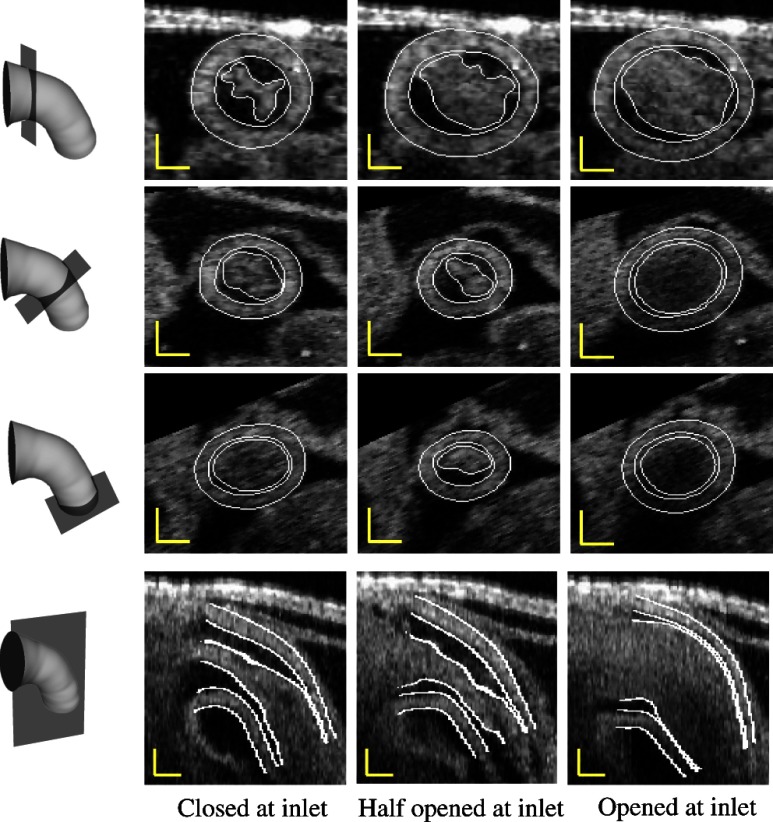
To evaluate the accuracy of the extracted results, we compared segmentations of 2-D cross-sectional images performed with our extraction algorithm and manually. For this study, manual segmentation was performed at three positions along the OFT: near the inlet, approximately in the middle, and near the outlet; at three different time points during the cardiac cycle: when the OFT walls are closed, half-closed, or opened (see Fig. 9, first to third rows). To compare results we used the areas enclosed by contours obtained by manual segmentation, ; contours obtained by automatic segmentation, ; and , which is the area of the intersection between the automatic and manually extracted regions. Two metrics, recall and precision, were used to evaluate the accuracy of the automatic segmentation. The recall, defined as , measures the ability of the automatic extracted result to match the target tissue; the precision, defined as , measures the ability of the automatic extracted result to contain only the target tissue. The average recall of the myocardium from the nine selected 2-D cross-sectional images was 84.7%, with a standard deviation of 3.3%, and its precision was 92.8%, with a standard deviation of 4.4%. The average recall of the lumen from the nine selected cross-sectional images was 91.9%, with a standard deviation of 3.3%, and its precision was 93.2%, with a standard deviation of 5.1%. To further confirm the overall accuracy of the segmentation, we visually inspected automatic segmentation results on OFT longitudinal sections at three different time points (see last row in Fig. 9). We found that in most places deviations between visual estimations of the location of a boundary and segmentation lines were smaller than 1 pixel (around 5 μm). The comparison of segmentation results on the 2-D cross-sections and longitudinal sections demonstrated that our segmentation strategy can accurately extract the OFT surfaces of the myocardium and lumen from 4-D OCT images of the developing heart even when images are noisy and intensity levels (as well as signal-to-noise ratio) diminish with depth.
Fig. 9.
Automatic extracted myocardial layer and lumen surfaces. (First to third rows) Segmentations of cross-sectional planes perpendicular to the 3-D centerline of the OFT at three time points over the cardiac cycle. (Bottom row) Segmentations of OFT longitudinal sections. Scale bars: 100 μm.
Figure 10 shows extracted OFT surfaces from 4-D OCT images (myocardial surfaces and the lumen-tissue interface surface) as well as volume data (see also Videos 1, 2, and 3). The extracted OFT shapes showed the peristaltic-like motion of the myocardial and endocardial layers of the OFT over the cardiac cycle, characterized by a contraction wave that travels through the OFT. During contraction, the OFT lumen appears to close, preventing blood flow, and folding of the endocardium layer becomes apparent. Folding and unfolding of the endocardium layer are observed from the extracted endocardium surfaces (as well as in raw images). The extracted shapes are therefore useful not only to visualize cardiac motion over the cardiac cycle but also in the analysis of cardiac motion.
Fig. 10.
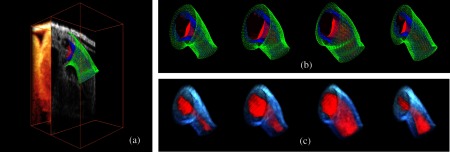
We successfully extracted OFT shapes from six other 4-D image datasets of embryonic chick OFTs at HH18 (not shown). These chick embryo heart OFTs had similar size, but 4-D OCT images differed in intensity and level of noise. In segmenting the OFTs from these datasets, and due to the robustness of our extraction procedure, there was no need to adjust DLM parameters in our algorithm. However, a parameter of the active-contour model, [see Eq. (2)] was adjusted on a case-by-case basis to avoid deviations of the active-contour from the target tissue layer. The range of values used was from about 1.3 to 1.9 times the value of the other two parameters and (), which were not modified. Larger values of were used when adjacent tissues outside of the myocardium had higher intensity levels than the myocardium tissue because this choice of penalized deviations from the myocardium layer by reducing curvature changes and increasing smoothing. For other cases, the smaller values of were chosen. All seven extracted 4-D chick heart shapes showed similar peristaltic-like motion of the myocardial and endocardial layers of the OFT, and endocardial folding upon contraction.
5. Discussion
In this paper, we present a semi-automatic procedure to extract the shape and motion of developing heart tissues from 4-D OCT images. The extracted tissue layers included the myocardium (internal and external surfaces) and the endocardium (which also represents the lumen-tissue interface). While the method has been applied to 4-D OCT images of the heart outflow tract of HH18 chick embryos, the procedure could also be applied to other stages of development, other portions of the heart, and also to other animal models and images acquired with different imaging modalities. This is, however, outside the scope of this work. The main characteristic of the presented procedure is that it can accurately track the dynamics of tissue layer motion over time, even when motion and deformation are large, and in the presence of image noise, gaps, and variations in intensity.
The main drawbacks of the extraction procedure presented are that the procedure is sensitive to initial conditions, and it has a relatively high computational cost. Like with other deformable models, procedures to match the layer and edge DLMs to tissue layers are sensitive to the initial state (e.g., position, orientation). To increase the robustness of the procedure, we linked DLMs with active-contour models and constrained some of its parameters (thickness, orientation). To increase the accuracy of the endocardium detection, which presents “folding” patterns, in our procedure we preprocessed cross-sectional images. Nevertheless, we found a few sections that were inaccurately segmented (). We solved this problem by designing an interactive interface that allows the user to first check the accuracy of the segmentations, and, when needed, manually correct segmentations that are not accurate. Manual correction allows “resetting” the state of DLMs, improving the initial conditions and facilitating accurate detection.
Our segmentation strategy differs from other methods to extract tissue layers. Most methods13,14 detect tissue layers using intensity variations, but detection becomes difficult when adjacent tissue layers are present. Unlike other template-matching deformable models used to detect tissue layers,20 which only consider intensities inside of the model, our procedure uses intensities from inside and outside of the DLM; further intensity levels are adaptive to adjust for the diminishing intensities with tissue depth.
Robustness of the procedure was achieved at the cost of a modest increase in computations. To achieve robustness, the layer DLM uses adaptive foreground and background intensity levels. Further, layer DLMs are linked with an active-contour model. Nevertheless, in our system (Dell T7400 workstation with eight core CPUs at 2.0 GHz and 10.0 GB RAM), extracting the OFT cardiac shape and motion from 4-D OCT images took about 3 h. Comparing this performance to that of manual segmentations, which can take more than a week for the whole 4-D image dataset, the proposed procedure enables faster and accurate analysis of cardiac motion. Thus, while computationally expensive, 4-D segmentation of the OFT of HH18 chick embryos is feasible.
The main advantage of our proposed semi-automatic procedure, is that it achieves robust segmentation of cardiac tissues from highly dynamic 4-D OCT images. This was confirmed by applying the algorithm to several 4-D OCT image reconstructions of the chick heart OFT, images that differ in the level of noise, image intensity and embryo orientation, and were also affected by biological variations, and possibly inaccuracies in embryo staging. The presented extraction procedure successfully segmentated all datasets. We are confident that we could also successfully apply the algorithm to other developmental stages and sections of the heart, enabling the study of changes in cardiac motion over developmental stages and after interventions.
The extracted 4-D results provide detailed information on the motion of the OFT wall, which could be used to quantify motion, tissue strains, or as input for subject-specific computational models of the developing heart OFT, among other applications. This work then provides a foundation to later work that aims at understanding how biomechanical stimuli affect cardiac development.
Acknowledgments
This work has been supported in part by Grants NIH R01 HL094570 and NSF DBI-1052688. The content is solely the responsibility of the authors and does not necessarily represent the official views of grant-giving bodies. The authors would like to thank Dr. Zhenhe Ma (Northeastern University at Qinhuangdao) for setting up the OCT system employed; and Dr. Cindy Grimm (Washington University in St. Louis) for carefully reviewing this paper and providing insightful suggestions.
References
- 1.Hove J. R., et al. , “Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis,” Nature 421, 172–177 (2003). 10.1038/nature01282 [DOI] [PubMed] [Google Scholar]
- 2.Boppart S. A., et al. , “Investigation of developing embryonic morphology using optical coherence tomography,” Develop. Biol. 177, 54–63 (1996). 10.1006/dbio.1996.0144 [DOI] [PubMed] [Google Scholar]
- 3.Yelbuz T. M., et al. , “Optical coherence tomography: a new high-resolution imaging technology to study cardiac development in chick embryos,” Circulation 106(22), 2771–2774 (2002). 10.1161/01.CIR.0000042672.51054.7B [DOI] [PubMed] [Google Scholar]
- 4.Leitgeb R., Hitzenberger C. K., Fercher A. F., “Performance of fourier domain vs. time domain optical coherence tomography,” Opt. Express 11(8), 889–894 (2003). 10.1364/OE.11.000889 [DOI] [PubMed] [Google Scholar]
- 5.Davis A. M., et al. , “In vivo spectral domain optical coherence tomography volumetric imaging and spectral doppler velocimetry of early stage embryonic chicken heart development,” J. Opt. Soc. Am. A 25, 3134–3143 (2008). 10.1364/JOSAA.25.003134 [DOI] [PubMed] [Google Scholar]
- 6.Rugonyi S., et al. , “Changes in wall motion and blood flow in the outflow tract of chick embryonic hearts observed with optical coherence tomography after outflow tract banding and vitelline-vein ligation,” Phys. Med. Biol. 53(18), 5077–5091 (2008). 10.1088/0031-9155/53/18/015 [DOI] [PubMed] [Google Scholar]
- 7.Jenkins M. W., et al. , “In vivo gated 4-D imaging of the embryonic heart using optical coherence tomography,” J. Biomed. Opt. 12, 030505 (2007). 10.1117/1.2747208 [DOI] [PubMed] [Google Scholar]
- 8.Liu A., et al. , “Efficient postacquisition synchronization of 4-D nongated cardiac images obtained from optical coherence tomography: application to 4-D reconstruction of the chick embryonic heart,” J. Biomed. Opt. 14(4), 044020 (2009). 10.1117/1.3184462 [DOI] [PubMed] [Google Scholar]
- 9.Ma Z., et al. , “Measurement of absolute blood flow velocity in outflow tract of HH18 chicken embryo based on 4-D reconstruction using spectral domain optical coherence tomography,” Biomed. Opt. Exp. 1, 798–811 (2010). 10.1364/BOE.1.000798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis A., Izatt J., Rothenberg F., “Quantitative measurement of blood flow dynamics in embryonic vasculature using spectral doppler velocimetry,” The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 292, 311–319 (2009). 10.1002/ar.v292:3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garita B., et al. , “Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping,” Am. J. Physiol. Heart. Circ. Physiol. 300, H879–H891 (2011). 10.1152/ajpheart.00433.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamburger V., Hamilton H. L., “A series of normal stages in the development of the chick embryo,” J. Morphol. 88, 49–92 (January 1951). 10.1002/(ISSN)1097-4687 [DOI] [PubMed] [Google Scholar]
- 13.Kirbas C., Quek F., “A review of vessel extraction techniques and algorithms,” ACM Comput. Surveys 36(2), 81–121 (2004). 10.1145/1031120 [DOI] [Google Scholar]
- 14.Lesage D., et al. , “A review of 3-D vessel lumen segmentation techniques: models, features and extraction schemes,” Med. Image Anal. 13, 819–845 (2009). 10.1016/j.media.2009.07.011 [DOI] [PubMed] [Google Scholar]
- 15.Geiger D., et al. , “Dynamic programming for detecting, tracking and matching deformable contours,” IEEE Trans. Pattern Anal. Mach. Intell. 17, 294–302 (1995). 10.1109/34.368194 [DOI] [Google Scholar]
- 16.Mikic I., Krucinski S., Thomas J. D., “Segmentation and tracking in echocardiographic sequences: active contours guided by optical flow estimates,” IEEE Trans. Med. Imag. 17, 274–284 (1998). 10.1109/42.700739 [DOI] [PubMed] [Google Scholar]
- 17.Florin C., Paragios N., Williams J., “Particle filters, a quasi-monte carlo solution for segmentation of coronaries,” Med. Imag. Comput. Comput.-Assist. Interven., 246–253 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Marsousi M., et al. , “Endocardial boundary extraction in left ventricular echocardiographic images using fast and adaptive B-spline snake algorithm,” Int. J. Comput. Assist. Radiol. Surg. 5(5), 501–513 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Metaxas D. N., et al. , “Hybrid deformable models for medical segmentation and registration,” in 9th Internat. IEEE Conf. on Control, Automation, Robotics and Vision, ICARCV 2006, pp. 1–6 (2006). [Google Scholar]
- 20.McInerney T., Terzopoulos D., “Deformable models in medical image analysis: a survey,” Med. Imag. Anal. 1, 91–108 (1996). 10.1016/S1361-8415(96)80007-7 [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Hoyos M., et al. , “A deformable vessel model with single point initialization for segmentation, quantification and visualization of blood vessels in 3-D MRA,” Med. Imag. Comput. Comput.-Assist. Intervent., 735–745 (2000). [Google Scholar]
- 22.Kass M., Witkin A., Terzopoulos D., “Snakes: active contour models,” Int. J. Comput. Vis. 1, 321–331 (1988). 10.1007/BF00133570 [DOI] [Google Scholar]
- 23.Xu C., Prince J. L., “Snakes, shapes, and gradient vector flow,” IEEE Trans. Image Process. 7, 359–369 (1998). 10.1109/83.661186 [DOI] [PubMed] [Google Scholar]
- 24.Luo H., et al. , “Robust snake model,” Proc. IEEE Conf. Comput. Vis. Pattern Recog., 452–457 (2000). [Google Scholar]
- 25.Laptev I., et al. , “Automatic extraction of roads from aerial images based on scale space and snakes,” Mach. Vis. Appl. 12, 23–31 (2000). 10.1007/s001380050121 [DOI] [Google Scholar]
- 26.Tyrrell J. A., et al. , “Robust 3-D modeling of vasculature imagery using superellipsoids,” IEEE Trans. Med. Imag. 26, 223–237 (2007). 10.1109/TMI.2006.889722 [DOI] [PubMed] [Google Scholar]
- 27.Hu X., Zhang Z., Tao C. V., “A robust method for semi-automatic extraction of road centerlines using a piecewise parabolic model and least square template matching,” Photogramm. Eng. Remote Sens. 70(12), 1393–1398 (2004). [Google Scholar]
- 28.Friman O., Hindennach M., Peitgen H.-O., “Template-based multiple hypotheses tracking of small vessels,” 5th IEEE Internat. Symp. on Biomed. Imaging: : From Nano to Macro, pp. 1047–1050 (2008). [Google Scholar]
- 29.Poor H. V., An Introduction to Signal Detection and Estimation, 2nd ed., Springer-Verlag, USA: (1994). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.