Abstract
Background and Aims
The genus Arachis contains 80 described species. Section Arachis is of particular interest because it includes cultivated peanut, an allotetraploid, and closely related wild species, most of which are diploids. This study aimed to analyse the genetic relationships of multiple accessions of section Arachis species using two complementary methods. Microsatellites allowed the analysis of inter- and intraspecific variability. Intron sequences from single-copy genes allowed phylogenetic analysis including the separation of the allotetraploid genome components.
Methods
Intron sequences and microsatellite markers were used to reconstruct phylogenetic relationships in section Arachis through maximum parsimony and genetic distance analyses.
Key Results
Although high intraspecific variability was evident, there was good support for most species. However, some problems were revealed, notably a probable polyphyletic origin for A. kuhlmannii. The validity of the genome groups was well supported. The F, K and D genomes grouped close to the A genome group. The 2n = 18 species grouped closer to the B genome group. The phylogenetic tree based on the intron data strongly indicated that A. duranensis and A. ipaënsis are the ancestors of A. hypogaea and A. monticola. Intron nucleotide substitutions allowed the ages of divergences of the main genome groups to be estimated at a relatively recent 2·3–2·9 million years ago. This age and the number of species described indicate a much higher speciation rate for section Arachis than for legumes in general.
Conclusions
The analyses revealed relationships between the species and genome groups and showed a generally high level of intraspecific genetic diversity. The improved knowledge of species relationships should facilitate the utilization of wild species for peanut improvement. The estimates of speciation rates in section Arachis are high, but not unprecedented. We suggest these high rates may be linked to the peculiar reproductive biology of Arachis.
Keywords: Arachis, peanut, groundnut, intron sequences, single-copy genes, molecular phylogeny, microsatellites, genetic relationships, speciation rates, genome donors, molecular dating
INTRODUCTION
The genus Arachis is native to South America. It contains 80 described species, assembled into nine sections according to their morphology, geographical distribution and cross-compatibility relationships (Krapovickas and Gregory, 1994; Valls and Simpson, 2005). Section Arachis is the most widely distributed, being found in five countries of the distributional range of the genus (Brazil, Argentina, Bolivia, Paraguay and Uruguay). It includes the cultivated peanut (A. hypogaea) and 30 known wild species. Most species are diploid (2n = 2x = 20), three are aneuploid or dysploid (2n = 2x = 18) and two species, A. hypogaea and A. monticola, are tetraploid (2n = 4x = 40) with a genome formula AABB (Krapovickas and Gregory, 1994; Peñaloza and Valls, 1997; Lavia, 1998; Valls and Simpson, 2005).
In section Arachis, three genome types (A, B and D) have been described for the diploid species with x = 10, according to their chromosome morphology and cross-compatibility (Smartt et al., 1978; Gregory and Gregory, 1979; Singh and Moss, 1982, 1984; Singh, 1986; Stalker, 1991; Fernández and Krapovickas, 1994; Peñaloza and Valls, 2005). Most of these species have an A genome type, characterized by the presence of a so-called A chromosome pair, with a reduced size and a lower level of euchromatin condensation relative to the other chromosomes (Husted, 1936; Seijo et al., 2004). The remaining diploid species with x = 10 do not have the A chromosome pair and have been considered as having a B genome type. The exception is A. glandulifera, with a D genome characterized by the presence of six subtelocentric or submetacentric chromosome pairs, in contrast to the A and B genome species that are mainly composed of metacentric chromosomes (Stalker, 1991; Fernandez and Krapovickas, 1994; Robledo and Seijo, 2008). Recently, two new genome types (F and K) have been described for some of the species formerly considered in the B genome group, based on FISH mapping of rDNA loci and heterochromatin detection (Robledo and Seijo, 2010). Arachis benensis and A. trinitensis are now classified as having an F genome, and A. batizocoi, A. cruziana and A. krapovickasii, a K genome type. These two genomes have centromeric bands on most of the chromosomes, differing from each other in the amount and distribution of heterochromatin.
Peanut ranks fifth among the most important sources of vegetable oils (FAO, 2010). Different types of molecular markers have detected little polymorphism in A. hypogaea (Halward et al., 1991, 1992; Kochert et al., 1996; Subramanian et al., 2000; Gimenes et al., 2002b, 2007; Cuc et al., 2008), and wild Arachis spp. offer novel genes for peanut improvement. Thus knowledge of their genetic relationships is important for their efficient use in breeding programmes for broadening the genetic base of A. hypogaea. It is well known that the transfer of specific genes is easier when the wild species is genetically closely related to the crop, i.e. A. hypogaea. Consequently, a series of studies have been conducted to understand the genetic relationships between Arachis spp., with emphasis on section Arachis, using different markers, including isozymes and proteins (Krishna and Mitra, 1988; Singh et al., 1991; Lu and Pickersgill, 1993; Stalker et al., 1994), RFLP (Kochert et al., 1991, 1996; Paik-Ro et al., 1992; Gimenes et al., 2002a), RAPD (Halward et al., 1991, 1992; Hilu and Stalker, 1995; Subramanian et al., 2000; Dwivedi et al., 2001; Santos et al., 2003; Cunha et al., 2008), AFLP (He and Prakash, 1997, 2001; Gimenes et al., 2002b; Herselman, 2003; Milla et al., 2005; Tallury et al., 2005) and microsatellites (Moretzsohn et al., 2004; Bravo et al., 2006; Gimenes et al., 2007; Koppolu et al., 2010). The genetic relationships among all the 31 described species of section Arachis have been analysed using molecular markers, but few studies have included a large number of accessions from each species. As a consequence, the variation within and among many species remains unclear.
Microsatellites or SSRs (simple sequence repeats) are informative as they are multiallelic, polymorphic and co-dominant markers. The rapid rate of microsatellite evolution also means that reliable information may be gained even for closely related taxa (Goldstein and Pollock, 1997). Additionally, microsatellites have proved to be highly transferable between Arachis spp. (Moretzsohn et al., 2004; Hoshino et al., 2006; Gimenes et al., 2007; Koppolu et al., 2010).
Arachis hypogaea and A. monticola are allotetraploids, whereas most of the species in section Arachis are diploids with the exception of three species (A. decora, A. palustris and A. praecox) which are aneuploids or dysploids. The ideal method for the analysis of genetic relationships between species with different ploidy levels should enable the separation of the two genome components in the tetraploids. By doing so, the species more closely related to each of the two genomes (A and B) of the cultivated peanut can be identified. Recently, markers were developed to amplify orthologous and single-copy genes, used as anchor markers to identify syntenic chromosomal regions across several legume species (Fredslund et al., 2006; Hougaard et al., 2008; Bertioli et al., 2009), and these genes are a valuable tool for the analysis of the phylogenetic relationships between Arachis spp. Theoretical concerns and practical examples have shown that sequences of single- or low-copy nuclear genes are particularly helpful in resolving interspecific relationships and in reconstructing allopolyploidization in plants (Sang, 2002). Sequences of the internal transcribed spacers (ITS) have been used in phylogenetic studies in Arachis (Bechara et al., 2010; Friend et al., 2010; Wang et al., 2011). However, ITS sequence variation may be inadequate for the study of closely related species or intraspecific relationships (Baldwin et al., 1995; Sang, 2002).
The objectives of the present work were to establish the phylogenetic relationships between species of Arachis section Arachis using single-copy gene sequences and to analyse the genetic variation within and between species of section Arachis using microsatellite markers. All but two of the 31 described species of this section were included. We also included accessions of two species of section Arachis that will be described soon and two accessions of A. vallsii with a questionable current classification in section Procumbentes. Single-copy gene sequences were also used to identify the two diploid species involved in the origin of the tetraploids A. hypogaea and A. monticola.
MATERIALS AND METHODS
Plant material and DNA extraction
A total of 161 accessions were included in the analysis with microsatellite markers (Supplementary Data Table S1 available online). These accessions represent 27 of the 31 species described in the section Arachis, two recently collected accessions of species that will be described in this section and two accessions of A. vallsii, which is currently classified in section Procumbentes but with evidence that it should be placed in section Arachis. The same species were included in the phylogenetic analyses using intron sequences, plus the tetraploid species A. hypogaea and A. monticola, with a total of 54 accessions (Supplementary Data Table S1). The seven accessions of A. hypogaea represent both subspecies (hypogaea and fastigiata) and all six varieties, plus an accession collected in Xingu Indigenous Park, which has morphological traits, especially in the pods, exceeding the variation described (Freitas et al., 2007). Accessions of A. subcoriacea and A. pflugeae (section Procumbentes) and A. pintoi (section Caulorrhizae) were included in this analysis as outgroups. The two species of section Procumbentes were included to enable the comparison with A. vallsii, and A. pintoi was included due to its known phylogenetic position as revealed using ITS data (Bechara et al., 2010; Friend et al., 2010). The accessions were obtained from the Brazilian Arachis Germplasm Collection, maintained at Embrapa Genetic Resources and Biotechnology – Cenargen (Brasília-DF, Brazil). All plants were grown under greenhouse conditions at Cenargen prior to DNA extraction. From the known species of section Arachis, only A. trinitensis and A. herzogii were not included due to the unavailability of this material.
PCR amplification of SSR loci
Total genomic DNA was extracted from young leaflets essentially as described by Grattapaglia and Sederoff (1994). The quality and quantity of the DNA were evaluated in 1 % agarose gel electrophoresis and spectrophotometer (Nanodrop 1000; Thermo Scientific, Wilmington, USA). PCR reactions were performed with the Qiagen Multiplex PCR kit (Qiagen, Hilden, Germany), containing 2·5 µL Master Mix (Taq DNA polymerase, PCR buffer and dNTPs), 0·5 µL Q solution and 0·4–0·7 µL of ultra-pure water (depending on the number of primers amplified in the same PCR), 0·1 µL of each primer (10 µm) and 1 µL genomic DNA (2·5 ng μL−1), in a final volume of 5 µL. Amplifications were carried out in ABI 9700 thermocyclers (Applied Biosystems, Foster City, CA, USA), with the following conditions: 95 °C for 15 min, followed by 35 cycles of 94 °C for 30 s, 52–60 °C for 90 s (annealing temperature depending on primer pair), 72 °C for 90 s, with a final extension for 10 min at 72 °C. The forward primer was labelled with one of three fluorescent dyes, HEX, 6-FAM or NED (Applied Biosystems). The primers were multiplexed according to the fluorescence, annealing temperature and size of the amplified alleles. The PCR products were denatured and size fractioned using capillary electrophoresis on an ABI 3700 automated DNA sequencer (Applied Biosystems). Loading samples contained 1 µL of the PCR product diluted 1 : 10 in ultra-pure water, 8·5 µL of Hi-Di formamide (Applied Biosystems) and 0·5 µL of ROX-labelled size standards. Allele sizing of the electrophoretic data thus obtained was done using Genescan 3·1 and Genotyper 3·1 softwares (Applied Biosystems). These data were exported to Microsoft Excel for further formatting as input files for statistical analysis.
Intron sequencing
We aimed to obtain orthologous sequences from the diploid Arachis spp., from the A and B genomes of the tetraploid A. hypogaea and, as outgroups, from legumes with completely sequenced genomes, Lotus japonicus, Medicago truncatula and Glycine max. Orthologous coding regions tend to be highly conserved and thus are phylogenetically uninformative. Therefore we targeted orthologous intron sequences as described for the development of ‘anchor markers’ by Fredslund et al. (2006). To facilitate the separation of intron sequences from subcomponent genomes of A. hypogaea, we did an initial screen of primer pairs to identify markers that gave PCR products of different sizes for the A and B genomes. From these initial screens, three pairs of intron-amplifying primers were selected:
Leg088-fwd: GCTGCTGTTGGGCAAGATTGTGCTC
Leg088-rev: GTATTGAGRTTGATTCCCATGACGCTCATG
Leg237-fwd: ACTTGTTAACATCWCAAARCAGCGG
Leg237-rev: ACTGGTTCACGTTCAATYGAGAGTGCAGTCCCAAG
Leg242-fwd: GGARCATAACTATCVTGGTTCTARTAAGC
Leg242-rev: CACATGATGAACTGAAAMCCCCCTYGCATGCAC
PCRs were carried out with 25 ng genomic DNA, 5 U Taq DNA polymerase, 1× PCR buffer (200 mm Tris pH 8·4, 500 mm KCl), 1·5 mm MgCl2, 200 µm of each dNTP and 0·4 µm of each primer, in a final reaction volume of 50 µL. Thermocycling was as follows: 32 cycles of 30 s at 96 °C; 45 s at 49 °C, 60 °C or 55 °C (annealing temperatures for Leg088, Leg237 and Leg242, respectively); 1 min at 72 °C, and a final extension for 10 min at 72 °C. PCR products were separated by electrophoresis on polyacrylamide gels stained with silver nitrate (Creste et al., 2001).
For diploid species that produced single products in PCR reactions, sequencing was performed directly. For A. hypogaea and A. monticola, from which two PCR products were amplified, bands were excised from the dried silver-stained gel and rehydrated in 100 µL water at 92 °C for 5 min, and then at room temperature for several hours. The individual A and B genome bands were then amplified using the PCR conditions cited above and 2 µL of the gel slice water as template. Sequencing was done using the BigDye Terminator sequencing kit (Applied Biosystems) on ABI 377 or 3730 sequencers (Applied Biosystems). Sequences were processed using the Staden Package, with base calling using Phred (Staden, 1996; Ewing and Green, 1998). Each sample was sequenced in both directions and at least twice. All single nucleotide polymorphisms were confirmed by manual inspection.
Sequence alignment and phylogenetic analysis
Arachis introns were used in BLAST similarity searches against the genomes of Lotus japonicus, Medicago truncatula and Glycine max. Sequences were aligned using Spin from the Staden package (Staden, 1996), Jalview (Waterhouse et al., 2009) and Muscle (Edgar, 2004), with manual editing using Seaview (Gouy et al., 2010), and concatenated in BioEdit v7·0·9 (Tom Hall; Ibis Biosciences). The loci were found to be rich in indels, which were coded as binary characters using the simple coding scheme of Simmons and Ochoterena (2000) as implemented in the program Seqstate (Muller, 2006) and appended to the data matrix. The alternative modified complex indel coding (MCIC) method of the same authors was also implemented in Seqstate for comparison. The Leg 088, Leg 237 and Leg 242 markers comprised 442, 290 and 384 aligned characters, respectively. The simple indel coding (SIC) provided a total of 38 characters.
A phylogenetic analysis of the three combined DNA loci and gap characters was obtained under the maximum parsimony (MP) criterion using PAUP* (version 4·0b10; Swofford, 2003). Heuristic searches comprised 1000 repeats of five cycles of random taxon addition, holding one tree per cycle. Gaps were treated as missing data and character-state optimization was by accelerated transformation (ACCTRAN). Branch swapping was by tree bisection reconnection (TBR) and the optimal trees from the first phase of the search were subjected to a further round of TBR branch swapping to widen tree space. Branch support for trees was assessed using 1000 bootstrap pseudoreplicates (Felsenstein, 1985), utilizing the same heuristic search strategy as above. To reduce the effects of homoplasy in the data matrices, progressive character reweighting was applied, optimizing for the maximum fit to the rescaled consistency index (RCI), and including TBR branch swapping after each reweighting. The RCI was found to stabilize after up to two cycles of reweighting. Branch support for trees based on the reweighted matrices was again evaluated using 1000 bootstrap pseudoreplicates. Bootstrap support for bipartitions was calculated and transferred to consensus trees using Bootscore v3·11 (Sukumaran, 2007).
Evolutionary divergence time estimates were conducted in MEGA5 (Tamura et al., 2007), based on alignments of Leg088 intron and Leg242 intron sequences from all Arachis spp. included in the phylogenetic analysis and orthologous sequences from the sequenced genomes of Lotus japonicus and Glycine max. The latter yielded two variant sequences for each marker, reflecting a polyploidization event dated at 13 Mya (Schmutz et al., 2010). Diversification rates were estimated assuming a Yule (pure birth) process according to the methods described in Scherson et al. (2008) and using the following formula: r = [ln(n) − ln(2)]/t, where r = rate of species diversification, n = number of species in the clade and t = age of crown node. Calibrations for the Lotus–Glycine and Lotus–Arachis divergences were based on previous estimates (Lavin et al., 2005). The number of base substitutions per site between sequences were calculated using the Tamura–Nei model (Tamura and Nei, 1993), excluding positions with gaps. Sequences were aligned with both MUSCLE (Edgar, 2004) and MAFFT (Katoh et al., 2009), since inclusion of the more divergent Lotus and Glycine sequences introduced significant ambiguity.
Analysis of microsatellite data
A total of 30 microsatellite markers (Palmieri et al., 2002; He et al., 2003; Ferguson et al., 2004; Moretzsohn et al., 2004, 2005; Proite et al., 2007; Cuc et al., 2008) were included in this analysis. Allelic data obtained from the microsatellite markers were subjected to AlleloBin (Prasanth et al., 2006) to classify observed allele sizes into representative discrete alleles based on the repeat units using the least-square minimization algorithm of Idury and Cardon (1997). Pairwise genetic similarities were estimated from the binned allelic data using the band-sharing coefficient of Lynch (1990). The resulting diagonal matrix was then submitted to cluster analysis using UPGMA (unweighted pair-group method analysis). To verify the consistency of the resulting dendrogram, the cophenetic correlation – r (Mantel, 1967) was calculated. All these analyses were performed using the software NTSYS 2·21 (Rohlf, 2009).
For each of the 30 microsatellite loci, the total number of amplified alleles (A), observed (Ho) and expected heterozygozities (He) and the polymorphism information content (PIC; Botstein et al., 1980) were estimated using the software PowerMarker 3·25 (Liu and Muse, 2005).
RESULTS
Phylogenetic analyses using intron sequences
High-quality sequences were obtained for Leg088, Leg237 and Leg242 for 54 different Arachis accessions and the two outgroups (A. pintoi and A. pflugeae). As expected, single sequences were obtained for these three Leg markers for the diploid species, and pair of sequences (A and B) were obtained for the tetraploids A. hypogaea and A. monticola.
BLAST similarity searches of the genome sequences of L. japonicus, M. truncatula and G. max identified clearly homologous regions to the Arachis Leg introns as follows: Leg088, single sequences from L. japonicus and M. truncatula, and a pair of sequences from G. max; Leg237, sequences that may be homologous to the Arachis intron were identified in the three species; Leg242, a single homologous sequence from L. japonicus, and a pair of sequences from G. max were identified. Satisfactory multi-sequence alignments with all legume genera could be obtained for Leg088 and Leg242 but not for Leg237. Single sequences are expected from the diploid Arachis spp. used, L. japonicus and M. truncatula because the last polyploidy event in these species was some 59 million years ago, and since then they have become highly diploidized (Cannon et al., 2010). Pairs of sequences are expected from the palaeotetraploid G. max, the genomes of which diverged about 13 million years ago (Schmutz et al., 2010). Sequence alignments are provided as Supplementary Data.
For calibration of the molecular clock, nucleotide substitutions per site were calculated for the intron regions for the genome divergences with previously known time estimates: 50 Mya Lotus–Glycine; 55 Mya Lotus–Arachis; 55 Mya Arachis–Glycine (Lavin et al., 2005) and 13 Mya Glycine–Glycine (Schmutz et al., 2010). Using this calibration together with the observed nucleotide substitutions per site it was possible to make age estimations for the major divergences in the Arachis intron phylogenetic tree (Fig. 1 and Table 1). Using the divergence age of the Arachis A and B genomes as the crown age for section Arachis with the number of known species in the section (31), the diversification rate in section Arachis could be calculated as 0·95 speciation events per million years.
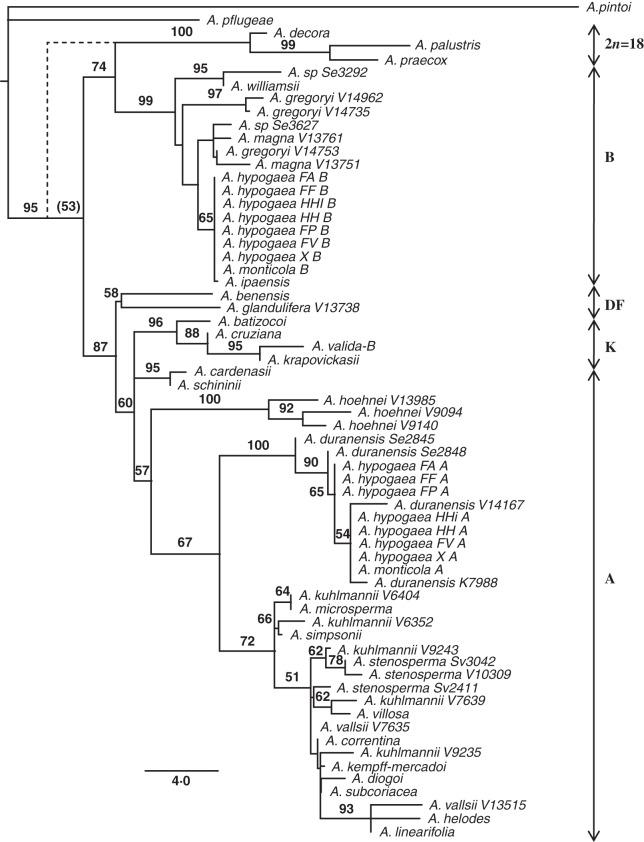
Fig. 1.
Phylogenetic tree (one of 2000) obtained under the maximum parsimony (MP) criterion, and two cycles of progressive character reweighting, based on intron sequence data, for 54 wild and cultivated accessions of Arachis. The data matrix included indel characters coded using the SIC indel coding. The broken line represents the alternative topology of the 2n = 18 clade under MCIC indel coding (Simmons and Ochoterena, 2000).
Table 1.
DNA base substitution rates (SR) and divergence times in millions of years (Mya) for four known divergences and estimates of evolutionary divergence times for three genomes in Arachis, based on Leg088 intron and Leg242 intron sequences
| Substitutions per site† |
|||||||
|---|---|---|---|---|---|---|---|
| Evolutionary divergence | Leg088 MUSCLE | Leg088 MAFFT | Leg242 MUSCLE | Leg242 MAFFT | Mean SR | Divergence (SR/Mya) | Divergence (Mya) |
| Lotus–Arachis pintoi | 0·33988 | 0·33887 | 0·23377 | 0·40663 | 0·3297875 | 0·005950261 | 55 |
| Glycine–A. pintoi | 0·31203 | 0·36140 | 0·39999 | 0·43607 | 0·3773725 | 0·007158330 | 55 |
| Lotus–Glycine | 0·25326 | 0·19528 | 0·30121 | 0·25803 | 0·2519450 | 0·005032325 | 50 |
| Glycine–Glycine | 0·07068 | 0·07956 | 0·20141 | 0·02537 | 0·0942550 | 0·007703750 | 13 |
| A. pintoi–A. duranensis | 0·01127 | 0·02814 | 0·04566 | 0·04399 | 0·0322650 | 0·006461166* | 4·99 |
| A. pintoi–A. ipaënsis | 0·01716 | 0·01618 | 0·04578 | 0·04411 | 0·0308075 | 0·006461166* | 4·77 |
| A. ipaënsis–A. duranensis (genomes A–B) | 0·01716 | 0·02814 | 0·01485 | 0·01432 | 0·0186175 | 0·006461166* | 2·88 |
| A. duranensis–A. benensis (genomes A–F) | 0·01150 | 0·02814 | 0·01485 | 0·01069 | 0·0162950 | 0·006461166* | 2·52 |
| A. duranensis–A. batizocoi (genomes A–K) | 0·01716 | 0·02814 | 0·00738 | 0·00712 | 0·0149500 | 0·006461166* | 2·31 |
Numbers in bold are inferred values.
* Mean of the four known divergences.
† Alignments for analysis were made using both MUSCLE (Edgar, 2004) and MAFFT (Katoh et al., 2009).
Of 1157 total characters (including 38 SIC characters), 950 were constant, 94 variable characters were parsimony uninformative and 113 were potentially parsimony informative. The heuristic MP analysis found 1455 equally parsimonious trees [length 300; confidence interval (CI) = 0·7200; retention index (RI) = 0·8930; RCI = 0·6430]. Following two rounds of progressive character reweighting (Felsenstein, 1985), 51 potentially parsimony informative characters had weights other than one. The heuristic search found >2000 equally parsimonious trees (score 195·8; CI = 0·8972; RI = 0·9547; RCI = 0·8566). This phylogenetic analysis showed a clustering of species according to their genome types (Fig. 1), corroborating the cytogenetic data. The 2n = 18 species formed a clade, with a bootstrap value of 100 %, closely related (bootstrap value of 74 %) to a clade containing the B genome species (supported by a bootstrap of 99 %). The other superclade (II) contained all the A, D, F and K genome species and A. vallsii and A. subcoriacea, with a bootstrap support of 87 %. The A genome component of A. hypogaea and A. monticola grouped with the four A. duranensis accessions (bootstrap value of 100 %), and the B component grouped in a well-defined B genome group of species (bootstrap value 99 %), most closely with the only known accession of A. ipaënsis (bootstrap value 65 %). The topologies of the strict consensus and most-parsimonious trees, using either simple or modified complex indel coding methods (Simmons and Ochoterena, 2000), were essentially congruent, except for the position of the A. decora–A. palustris–A. praecox clade, which under MCIC was sister to superclades I and II combined (broken line in Fig. 1). Bootstrap support for most internal nodes was, however, significantly greater under SIC.
Genetic relationships based on microsatellite data
Genetic similarities were estimated by the band-sharing coefficient (Lynch, 1990) in pairwise comparisons of 161 accessions of wild diploid species belonging to section Arachis (Supplementary Data Table S1), using 30 microsatellite loci. It has been shown that, in Arachis, the variation among alleles differed not only by increments of the repeat motif, but also by insertion/deletions (indels) occurring in regions near the simple sequence repeat (Barkley et al., 2007). Therefore, an infinite allele model or a genetic distance measure that assumes all alleles are equally related, such as the band-sharing coefficient (Lynch, 1990), might be appropriate to analyse microsatellite data in peanut. Genotyping data are provided in Supplementary Data Table S2. Genetic similarity values ranged from 0·0 to 0·93. Therefore all the accessions were differentiated using the 30 loci. A dendrogram based on UPGMA was constructed for the 161 accessions (Fig. 2). The tetraploid species, A. hypogaea and A. monticola, were not included in the analysis, since the alleles of the A and B genomes cannot be analysed separately. Their inclusion would result in their grouping with the more closely related species with the A or the B genome species, instead of both components separately.
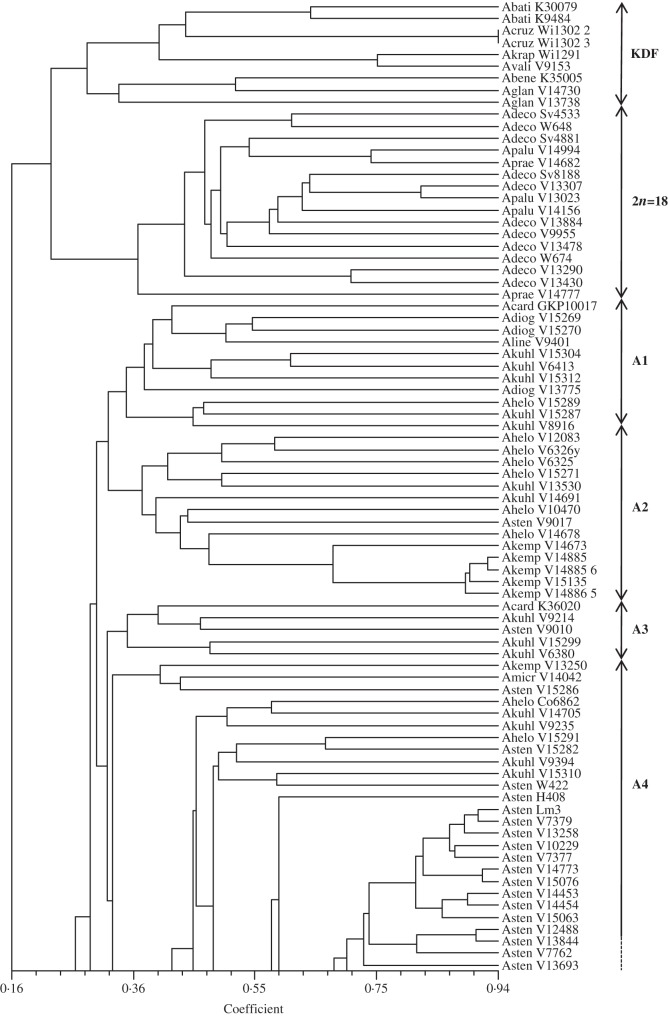
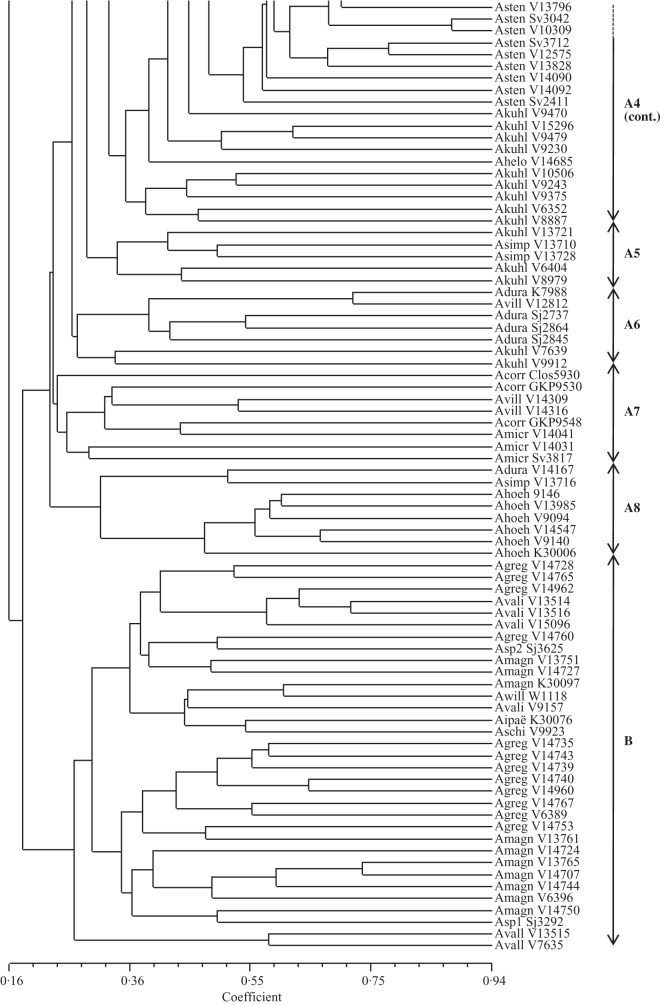
Fig. 2.
Dendrogram based on genetic similarities estimated by the band coefficient (Lynch, 1990) of 161 accessions of species of section Arachis generated by UPGMA. The coefficient of cophenetic correlation (r) was 0·81 (significant at 1% probability by the Mantel test).
Cluster analysis also showed the grouping of accessions according to their genome types, with some few exceptions. The upper group (DFK group) had two subgroups: (1) comprised the five accessions of the K genome species (A. batizocoi, A. cruziana and A. krapovickasii) and one accession of A. valida (V9153); (2) contained the only accession of the F genome (A. benensis) and the two accessions of A. glandulifera that has a D genome type. The 2n = 18 species (A. decora, A. palustris and A. praecox) formed a clearly differentiated group (2n = 18 group). The 13 of the 14 known A genome species clustered in a large group (A group) with eight subgroups, comprising in most cases the accessions of the same species (Fig. 2). The B genome species (B group) were separated into four subgroups. One subgroup was composed of A. magna and the accession of A. sp_Se3292, closely associated with a subgroup mainly composed of A. gregoryi accessions. The other subgroup contained four of the five accessions of A. valida included, the unique accessions of A. ipaënsis, A. williamsii and A. sp_Se3625 and two accessions each of A. magna and A. gregoryi. The two accessions of A. vallsii, which was described in section Procumbentes, were sister to the B group.
Microsatellites showed that most species with more than one analysed accession have a high genetic variability. Despite this, accessions of the same species tended to group together. For example, well-defined groups of A. stenosperma, A. kempff-mercadoi and A. gregoryi are evident in the dendrogram (Fig. 2). The main exception was A. kuhlmannii, with 28 accessions (including four A. aff. kuhlmannii and one A. cf. kuhlmannii) scattered throughout the A genome group.
The 30 microsatellite markers were developed for A. hypogaea or A. stenosperma and analysed in 161 accessions belonging to 31 species. The same PCR conditions were used to avoid the amplification of non-specific fragments. The transferability ranged from 0·42 to 0·98 with an average of 0·79. These microsatellites amplified an average of 16·8 alleles per locus, ranging from three (RM14B11) to 34 (PM3) (Table 2). The average observed heterozygosity was low (0·101) as compared with the expected heterozygosity (0·776). This low proportion of heterozygotes could indicate most species analysed are autogamous. PIC values ranged from 0·33 (RM14B11) to 0·95 (PM3) with an average of 0·76 (Table 2).
Table 2.
Number of alleles (A), expected heterozygosity (He), observed heterozygosity (Ho), polymorphism information content (PIC) and transferability estimates based on the analysis of 161 accessions of 31 Arachis species for 30 microsatellite markers
| Marker loci | A | He | Ho | PIC | Transferability | Reference |
|---|---|---|---|---|---|---|
| Ah-275 | 9 | 0·5963 | 0·3092 | 0·5386 | 0·9441 | Moretzsohn et al., 2004 |
| Ah3 | 27 | 0·9457 | 0·1517 | 0·9430 | 0·9006 | Bravo et al., 2006 |
| Ap40 | 23 | 0·8872 | 0·1453 | 0·8788 | 0·7267 | Palmieri et al., 2002 |
| gi-385 | 22 | 0·8683 | 0·1250 | 0·8586 | 0·7453 | Moretzsohn et al., 2004 |
| gi-623 | 21 | 0·8976 | 0·1545 | 0·8901 | 0·6832 | Moretzsohn et al., 2004 |
| IPAHM123 | 26 | 0·8424 | 0·1469 | 0·8313 | 0·8882 | Cuc et al., 2008 |
| IPAHM406 | 24 | 0·8362 | 0·0803 | 0·8239 | 0·8509 | Cuc et al., 2008 |
| PM137 | 15 | 0·4499 | 0·1871 | 0·4350 | 0·9627 | He et al., 2003 |
| PM204 | 16 | 0·8597 | 0·0000 | 0·8485 | 0·8509 | He et al., 2003 |
| PM3 | 34 | 0·9549 | 0·1765 | 0·9531 | 0·8447 | He et al., 2003 |
| PM32 | 20 | 0·9112 | 0·1159 | 0·9045 | 0·8571 | He et al., 2003 |
| PM36 | 25 | 0·9088 | 0·1268 | 0·9022 | 0·8820 | He et al., 2003 |
| RI2A06 | 10 | 0·6686 | 0·0748 | 0·6447 | 0·9130 | Moretzsohn et al., 2004 |
| RM14B11 | 3 | 0·3940 | 0·0098 | 0·3303 | 0·6335 | Proite et al., 2007 |
| RM6F03 | 11 | 0·7492 | 0·0196 | 0·7132 | 0·9503 | Proite et al., 2007 |
| RN10F09 | 14 | 0·8592 | 0·2254 | 0·8443 | 0·8820 | Moretzsohn et al., 2004 |
| RN12E01 | 12 | 0·8081 | 0·0970 | 0·7838 | 0·8323 | Moretzsohn et al., 2004 |
| RN25B01 | 8 | 0·5807 | 0·1275 | 0·5539 | 0·9255 | Proite et al., 2007 |
| RN27A10 | 7 | 0·6789 | 0·0741 | 0·6230 | 0·6708 | Proite et al., 2007 |
| RN34H10 | 25 | 0·8944 | 0·1679 | 0·8854 | 0·8137 | Proite et al., 2007 |
| RN35H04 | 12 | 0·8188 | 0·0345 | 0·7985 | 0·5404 | Proite et al., 2007 |
| RN36A01 | 6 | 0·4368 | 0·0526 | 0·4146 | 0·8261 | Proite et al., 2007 |
| Seq18G09 | 19 | 0·7053 | 0·0000 | 0·6912 | 0·9814 | Ferguson et al., 2004 |
| Seq3D09 | 11 | 0·8189 | 0·0851 | 0·8020 | 0·5839 | Ferguson et al., 2004 |
| Seq4B09 | 10 | 0·6355 | 0·0625 | 0·6130 | 0·6957 | Moretzsohn et al., 2004 |
| Seq4F10 | 28 | 0·9213 | 0·1239 | 0·9163 | 0·7019 | Moretzsohn et al., 2004 |
| TC3B05 | 23 | 0·9234 | 0·0725 | 0·9186 | 0·4286 | Moretzsohn et al., 2004 |
| TC3G05 | 14 | 0·8166 | 0·0067 | 0·7982 | 0·9317 | Moretzsohn et al., 2004 |
| TC4D09 | 15 | 0·7581 | 0·0095 | 0·7258 | 0·6522 | Moretzsohn et al., 2004 |
| TC4H02 | 13 | 0·8473 | 0·0789 | 0·8309 | 0·7081 | Moretzsohn et al., 2004 |
| Mean | 16·77 | 0·7758 | 0·1014 | 0·7565 | 0·7936 |
DISCUSSION
Genetic diversity and relationships of species of Arachis section Arachis
In the present study two complementary methods were used to analyse the genetic relationships of species in Arachis section Arachis. Microsatellite markers were chosen to provide efficient analyses of intraspecific variability and relationships among closely related species, and single-copy gene sequences to elucidate the phylogenetic relationships between species.
Phylogenetic analysis showed that the 2n = 18 species formed a monophyletic group, closely associated with a clade containing the B genome species (Fig. 1). The 2n = 18 species usually form a well-differentiated group in different genetic relationship analyses using molecular markers including our microsatellite-based data and have been associated with the A genome group (Tallury et al., 2005; Bravo et al., 2006; Koppolu et al., 2010) or, more often, with the B, D, F and K genome species (Moretzsohn et al., 2004; Gimenes et al., 2007; Bechara et al., 2010; Friend et al., 2010). Using AFLP marker data, Milla et al. (2005) suggested they are as closely related to the B (including the species that are now classified as F and K genome types) and D genome species as to the A genome species. These accumulated results raise questions about the evolutionary history of these species. Our sequencing analyses of single-copy genes proved to be a valuable tool for phylogenetic reconstruction, and added weight to the evidence that the 2n = 18 species were derived from the B genome sensu stricto species.
The B genome sensu stricto species formed a well-defined clade. Microsatellite data indicate three subgroups, with some intraspecific variability. No cytological or cross-compatibility assays have been published for the species A. sp_Se3292 and A. sp_Se3627 which will be described formally soon (G. Seijo, Instituto de Botánica del Nordeste, Argentina, pers. comm.) and nothing is known about their relationships with species of section Arachis. Our microsatellite and gene sequence results strongly suggest both species are closely related to the B genome species.
The analysis of intron sequences indicated that species with K, F and D genomes grouped together with A genome species in superclade II (bootstrap value of 87 %; Fig. 1). The classification of the D genome for A. glandulifera is well established and is based on its asymmetric karyotype (Stalker, 1991). The definition of F and K genomes is a more recent reassignment of species formerly considered to belong to the B genome group. The K genome was described for A. batizocoi, A. cruziana and A. krapovickasii, and the F genome for A. benensis and A. trinitensis (Robledo and Seijo, 2010). Arachis trinitensis was not included in our study, because no accession was available at the Cenargen germplasm collection. The placement of the three K genome species in a clade strongly supports the validity of the genome assignment made by Robledo and Seijo (2010), although the inclusion of the B genome species A. valida in this clade is unexpected and warrants further investigation. The placement of A. benensis distant from the B genome clade also gives support to the validity of the F genome assignment. The placement of the K, F and D genomes closer to the A than to the B genome is worthy of further discussion. The species with K, D and F genomes, and all the B genome species have a substantially shorter Leg088 sequences due to a approx. 210-bp deletion compared with the A genome species. Conversely, an 86-bp deletion in Leg242 is restricted to the A and K genome species. Evidence from other studies is also conflicting, some indicate an association of the K genome species with the B genome species (Moretzsohn et al., 2004; Bechara et al., 2010; Friend et al., 2010) and others with the A genome species (Tallury et al., 2005; Robledo et al., 2009; Robledo and Seijo, 2010). Arachis batizocoi, A. cruziana and A. krapovickasii generate highly sterile hybrids when crossed with A and B genome species (Krapovickas and Gregory, 1994; Valls and Simpson, 2005; Burow et al., 2009). Arachis benensis (F genome) and A. glandulifera (D genome) clustered together in both of our analyses (Figs 1 and 2) and were closer to the K and A genome groups, but have the large deletion typical of the B genome species (in Leg088). Therefore, we consider that the exact phylogenetic relationships of the F, K and D genomes with the A and the B genomes needs further study.
Both intron sequences and microsatellite data indicated high genetic variability of the A genome species, with similar groupings and clades being apparent. The microsatellite analysis showed eight subgroups, comprising in most cases the accessions of the same species (Fig. 2). Subgroup A1 contained accessions of A. cardenasii, A. cf. helodes, A. diogoi, A. linearifolia and five of the 28 accessions of A. kuhlmannii included. Arachis linearifolia shows strong morphological similarities to A. diogoi (Valls and Simpson, 2005). The six accessions of A. helodes grouped together in the subgroup A2. The other five, classified as Arachis aff. helodes or Arachis cf. helodes, were dispersed throughout the A genome group raising questions about their affinities with A. helodes. In subgroup A2 the five A. kempff-mercadoi accessions also clustered. Therefore, both analyses showed the close genetic relationship of the species A. linearifolia, A. helodes, A. diogoi and A. kempff-mercadoi and some accessions currently classified as A. kuhlmannii.
The 28 accessions of A. kuhlmannii (including four A. aff. kuhlmannii and one A. cf. kuhlmannii) were scattered throughout the A genome group, in at least four subgroups. These minor groups were not related to the collection sites or any known characteristics. The intron sequence analysis also indicated that A. kuhlmannii is polyphyletic, and the five accessions included were dispersed among five clades or subclades (Fig. 1). The polyphyly of A. kuhlmannii has also been observed using AFLP (Milla et al., 2005) and microsatellite markers (Koppolu et al., 2010). This species grows throughout the Pantanal Matogrossense, in the states of Mato Grosso and Mato Grosso do Sul (Brazil), the major centre of morphological, cytogenetic and genetic variation for Arachis (Gregory et al., 1980; Fernández and Krapovickas, 1994). Nothing is known about the reproductive system of A. kuhlmannii and cross-pollination with sympatric species could explain its high genetic variability. Krapovickas and Gregory (1994) mentioned some morphological differences between accessions of A. kuhlmannii, and more taxonomic studies seem to be necessary for the material currently classified as A. kuhlmannii.
In the small subgroup A3, five accessions of three different species grouped together, but with low coefficient values. Subgroup A4 was composed of A. stenosperma, with 27 out of the 29 accessions included, and A. kuhlmannii, with 13 accessions. Arachis stenosperma is the only species of section Arachis that grows on the Atlantic coast, being found from Rio de Janeiro to Paraná (Krapovickas and Gregory, 1994). It is also found in the state of Mato Grosso in central Brazil. These areas are separated by >1000 km, but this material is morphologically similar. Additionally, as shown for the first time with a substantial number of accessions, it is also closely related genetically.
The two accessions identified as A. aff. simpsonii grouped together, with three accessions of A. kuhlmannii (subgroup A5). These five accessions were collected close to Porto Esperidião, in Mato Grosso state (Brazil). The only accession of A. simpsonii included in the analysis (V13716), also collected in Porto Esperidião, was located in subgroup A8. These results suggested the accessions identified as A. aff. simpsonii (V13710 and V13728) and A. simpsonii (V13716) could be different species. Arachis simpsonii is morphologically similar to A. villosa and also has some leaflet similarities with A. diogoi (Krapovickas and Gregory, 1994). However, both our analyses showed that these three species are genetically distinct, in agreement with previous studies based on microsatellites (Moretzsohn et al., 2004; Gimenes et al., 2007) and ribosomal ITS sequence data (Bechara et al., 2010).
Accessions of A. duranensis grouped together in the microsatellite analysis, with the exception of V14167 (subgroup A6), whereas subgroup A7 contained two accessions of A. villosa, all three accessions of A. correntina and three of the four accessions of A. microsperma. The close relationship of these three perennial species was also observed in a recently published study based on ITS sequence data (Bechara et al., 2010).
The external subgroup (A8) of the A group contained the six accessions of A. hoehnei, and three accessions of this species also clustered together in the intron sequence analysis, being clearly distinct from other A genome species. This species was thought not to have the small ‘A’ chromosome pair (Fernández and Krapovickas, 1994), but a recent analysis showed it has a karyotype that matches the A genome species (Robledo and Seijo, 2010). This species was associated with the 2n = 18 species based on AFLP markers (Tallury et al., 2005) or it clustered outside the A–B genome groups based on AFLP and RFLP markers (Milla et al., 2005; Burow et al., 2009). However, A. hoehnei grouped with the A genome species based on microsatellites (Bravo et al., 2006; Gimenes et al., 2007), RAPD markers (Cunha et al., 2008) and sequencing of ITS and trnT-trnF regions (Bechara et al., 2010; Friend et al., 2010), in accordance with our data. Crossings between A. hoehnei and several species of section Arachis are currently underway and will shed more light on the classification and relationships of this species.
The two accessions of A. vallsii were located in the clade with the A genome species in the intron sequence-based tree (Fig. 1) and possess four indels characteristic of the A genome species (Supplementary Data). In contrast, A. vallsii formed a small group at the base of B group in the microsatellite analysis (Fig. 2). To our knowledge, only one study has included A. vallsii in a genetic relationship analysis using molecular data (Koppolu et al., 2010). The only accession analysed in that study did not group in any cluster or subcluster. Arachis vallsii was classified in section Procumbentes by Krapovickas and Gregory (1994), despite being morphologically different from the other species of that section. Recently, considering morphological and chromosomal features, it was suggested that A. vallsii should be moved to section Arachis (Lavia et al., 2009). Unpublished data from our research group showed that A. vallsii produced hybrids when crossed with different species of section Arachis, including A. hypogaea. Therefore, our results based on microsatellites, gene sequence comparisons and cross-compatibility assays corroborate this proposal.
One accession of A. subcoriacea, section Procumbentes, was included in our gene sequence analysis as an outgroup, as well as accessions of A. pintoi and A. pflugeae. The latter two species were located outside the clade containing all the other species included in the present study. However, A. subcoriacea was located in the clade of the A genome species and appears to be closely related to A. diogoi. Arachis subcoriacea has morphological similarities to A. diogoi (Krapovickas and Gregory, 1994), but it has a karyotype characteristic of section Procumbentes and does not display the A chromosome pair (Lavia, 2000, 2001). No studies of cross compatibilities including A. subcoriacea have been published to date, and our results raise questions about the taxonomic status of this species.
Genome donors of Arachis hypogaea
For each of the intron sequences, two PCR products of the tetraploid species A. hypogaea and A. monticola were individually sequenced. This enabled the comparison of the wild diploid species with the two genome components of the tetraploids separately. All previously published studies have made direct comparison of the diploids and tetraploids, which allows the identification of just one (A or B) of the most probable genome donors of A. hypogaea and A. monticola (Kochert et al., 1991; Hilu and Stalker, 1995; Moretzsohn et al., 2004; Bravo et al., 2006; Gimenes et al., 2007; Bechara et al., 2010; Koppolu et al., 2010; Ren et al., 2010). The only known exception was a phylogenetic study based on ITS and trnT-trnF sequences, recently published by Friend et al. (2010). However, the polymorphism detected in that study was low, resulting in the presence of polytomies in both A and B genome lineages. The tetraploids appeared as part of the A and B genome clades with nine and four species, respectively, and the identification of the donor species was not possible.
In the present study, all but two of the 29 described diploid and aneuploid/dysploid species of section Arachis were included. The relationships among the subspecies of A. hypogaea could not be inferred, since no polymorphism was observed in the B genome component and only one indel in a poly-A sequence was observed in the A genome component differentiating three of the seven samples of A. hypogaea. These samples included the six varieties and an accession from the Xingu Indigenous Park. Arachis monticola was also identical to the accessions of A. hypogaea. The high genetic similarity of these two species has also been supported by the analysis of the genetic relationships using different molecular markers (Kochert et al., 1991; Paik-Ro et al., 1992; Lu and Pickersgill, 1993; Hilu and Stalker, 1995; Singh et al., 2002; Milla et al., 2005; Barkley et al., 2007; Gimenes et al., 2007; Cunha et al., 2008; Ren et al., 2010). Some of those authors suggested that they should not be considered as separate species. Physical mapping of rDNA loci by FISH (Seijo et al., 2004) and GISH analyses (Raina and Mukai, 1999; Seijo et al., 2007) also resulted in identical patterns for these two species. Moreover, hybrids between A. monticola and A. hypogaea are fertile (Krapovickas and Gregory, 1994; Pattee et al., 1998). Arachis monticola is considered a distinct species from A. hypogaea based mainly on its fruit structure, which has an isthmus separating each seed. This trait is not observed in any cultivated peanut, and is considered a primitive feature in the genus. These lines of evidence support the hypothesis that A. monticola is the immediate wild ancestor of A. hypogaea, as suggested by some authors (Gregory and Gregory, 1976; Krapovickas and Gregory, 1994; Moretzsohn et al., 2004; Seijo et al., 2004, 2007; Koppolu et al., 2010).
The A genome component of A. hypogaea and A. monticola was placed in a clade that also contained the four accessions of A. duranensis (Fig. 1). No other A genome wild species was located in this well-supported subclade. The B genome component of A. hypogaea and A. monticola formed a subclade, closely related to the only known accession of A. ipaënsis (Fig. 1). These results strongly support the hypothesis that A. duranensis and A. ipaënsis were the A and B genome donors of A. hypogaea (Kochert et al., 1991; Seijo et al., 2004, 2007; Fávero et al., 2006). The results also effectively exclude the possibility that either A. correntina or A. villosa are the A genome ancestors of cultivated peanut, exclusions that could previously only be made based on geographical location (Seijo et al., 2004, 2007). In addition, they corroborate the assumption of Seijo et al. (2004) that the tetraploid species originated from a single event of allopolyploidization or, if from multiple events, always involving the same diploid parental species. To the best of our knowledge, the evidence presented here provides the strongest support yet presented indicating A. duranensis and A. ipaënsis as the diploid ancestral species of cultivated peanut.
The species most closely related to A. duranensis and the A genome of A. hypogaea/A. monticola could not be identified, since all the other A genome species were located in a distinct sister clade. In contrast, accessions of A. magna, A. gregoryi and A. sp_Se3627 were placed in a subclade close to A. ipaënsis. These species, most closely related to A. hypogaea and its wild progenitors, are attractive as sources for increasing genetic variability in peanut breeding programmes. To date, various difficulties, both biological and technical, have led to these resources being underutilized. Our improved understanding of the species relationships in the genus and improved tools for genetic and genomic studies should enable a more efficient use of the available genetic resources.
Speciation and evolution in section Arachis
Using the observed intron nucleotide substitutions, it was possible to estimate the major divergence dates in the Arachis phylogeny (Table 1). These estimates place the divergence of sections Arachis and Caulorrhizae (the latter being the section including A. pintoi) at just under 5 million years ago, and the first (A–B) divergence in section Arachis at just under 3 million years ago. These time frames are consistent with the high degree of synteny between genetic maps of the A and B genomes (Burow et al., 2001; Moretzsohn et al., 2009) and with the relatively high divergence of the repetitive DNAs in A and B genome species (Seijo et al., 2007, Nielen et al., 2010, 2011). This is because synteny of low-copy regions of genomes degrades slowly over time and in legumes is still detectable after >50 million years of species divergence (Bertioli et al., 2009). However, the repetitive fractions of plant genomes show high birth rates and rapid degradation and elimination. As a consequence, most easily datable plant retrotransposons are less than three million years old (e.g. Wicker and Keller, 2007). Furthermore, the Arachis divergence dates allow the diversification rate of section Arachis to be estimated at about 0·95 speciation events per million years. Although not unprecedented (Scherson et al., 2008), this rate can be considered as high and much higher than the average for legumes in general, estimated at 0·15 (Magallon and Sanderson, 2001). It seems possible that this high rate of speciation may be linked to the peculiar reproductive biology of Arachis spp., all of which bear their fruits underground. Deposited below the soil surface, Arachis seeds are afforded protection from many pests and predators, favourable conditions for germination and privileged access to soil moisture. However, a buried seed cannot be efficiently dispersed and in natural conditions dispersal is mostly limited to the area covered by the maternal plant. More rarely, seeds may be deposited further afield by water-driven soil erosion or animals (including man). In these cases, a single or few seeds then found a new population, resulting in natural populations that are ‘patches’ with typically only tens to hundreds of individuals. The combination of multiple recurrent severe genetic bottle-necks, small population sizes and a typically high rate of self-fertilization provides the perfect conditions for genetic drift and the evolution of genetic isolation. These same mechanisms may have also caused the remarkable degree of sexual incompatibility observed between different collections classified as the same species of wild Arachis (Krapovickas and Gregory, 1994).
Conclusions
The phylogenetic relationships of species of section Arachis were analysed based on DNA sequence information for three single-copy gene introns and provided new information about the basic structure of this section. Results were consistent with the current classification, since clades contained species with the same genome types. The species with 2n = 18 were shown to be genetically closely related to the B sensu stricto genome species, whereas D, F and K genome species were closer to the A genome species. The accessions of A. vallsii were placed in the same clade as the A genome species, providing support for this species being included in section Arachis. The divergences based on nucleotide substitution rates between the A, B and K genomes were estimated and suggested these three genomes diverged between 2·3 and 2·9 million years ago. Our phylogenetic analyses provide strong evidence for the hypothesis that A. duranensis and A. ipaënsis were the A and B genome donors to A. hypogaea. Microsatellite markers results showed that most species have high genetic variability. Arachis kuhlmannii accessions were hypervariable and appear to be polyphyletic suggesting that this species needs deeper taxonomic study. Estimates for the age of section Arachis indicate that speciation rates are high, a phenomenon that we suggest may be linked to the peculiar geocarpic reproductive biology of Arachis.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by the host institutions Embrapa Recursos Genéticos e Biotecnologia and the Universidade de Brasília and the Generation Challenge Programme. We also acknowledge CNPq for a Research Fellowship for David Bertioli and José Valls and CAPES for a Student Fellowship for Ediene Gouvea. Special thanks to Guillermo Seijo (IBONE) for providing the “Se” Arachis accessions.
LITERATURE CITED
- Baldwin B, Sanderson M, Porter J, Wojciechowski M, Campbell C, Donoghue M. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Barkley NA, Dean RE, Pittman RN, Wang ML, Holbrook CC, Pederson GA. Genetic diversity of cultivated and wild-type peanuts evaluated with M13-tailed SSR markers and sequencing. Genetic Research. 2007;89:93–106. doi: 10.1017/S0016672307008695. [DOI] [PubMed] [Google Scholar]
- Bechara M, Moretzsohn MC, Palmieri D, et al. Phylogenetic relationships in genus Arachis based on ITS and 5·8S rDNA sequences. BMC Plant Biology. 2010;10(255) doi: 10.1186/1471-2229-10-255. http://dx.doi.org/10.1186/1471-2229-10-255 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D, Moretzsohn M, Madsen L, et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10(45) doi: 10.1186/1471-2164-10-45. http://dx.doi.org/10.1186/1471-2164-10-45 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Bravo JP, Hoshino AA, Angelici C, Lopes CR, Gimenes MA. Transferability and use of microsatellite markers for the genetic analysis of the germplasm of some Arachis section species of the genus Arachis. Genetics and Molecular Biology. 2006;29:516–524. [Google Scholar]
- Burow MD, Simpson CE, Starr JL, Paterson AH. Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics. 2001;159:823–837. doi: 10.1093/genetics/159.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow MD, Simpson CE, Faries W, Starr JL, Paterson AH. Molecular biogeography study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome. 2009;52:107–119. doi: 10.1139/g08-094. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Ilut D, Farmer AD, Maki SL, May GD, Singer SR, Doyle JJ. Polyploidy did not predate the evolution of nodulation in all legumes. PLoS ONE. 2010;5:e11630. doi: 10.1371/journal.pone.0011630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creste S, Yulmann Neto A, Figueira A. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Molecular Biology Reporter. 2001;19:299–306. [Google Scholar]
- Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biology. 2008;8(55) doi: 10.1186/1471-2229-8-55. http://dx.doi.org/10.1186/1471-2229-8-55 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha FB, Nobile PM, Hoshino AA, Moretzsohn MC, Lopes CR, Gimenes MA. Genetic relationships among Arachis hypogaea L. (AABB) and diploid species with AA and BB genomes. Genetic Resources and Crop Evolution. 2008;55:15–20. [Google Scholar]
- Dwivedi SL, Gurtu S, Chandra S, Yuejin W, Nigam SN. Assessment of genetic diversity among selected groundnut germplasm. I. RAPD analysis. Plant Breeding. 2001;120:345–349. [Google Scholar]
- Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred II: error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations) 2010 Available at http://faostat.fao.org/ (accessed 8 March 2012) [Google Scholar]
- Fávero AP, Simpson CE, Valls FMJ, Velo NA. Study of evolution of cultivated peanut through crossability studies among Arachis ipaënsis, A duranensis and A hypogaea. Crop Science. 2006;46:1546–1552. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferguson ME, Burow MD, Schulze SR, et al. Microsatellite identification and characterization in peanut (A. hypogaea L.) Theoretical and Applied Genetics. 2004;108:1064–1070. doi: 10.1007/s00122-003-1535-2. [DOI] [PubMed] [Google Scholar]
- Fernández A, Krapovickas A. Cromosomas y evolucion en Arachis (Leguminosae) Bonplandia. 1994;8:187–200. [Google Scholar]
- Fredslund J, Madsen L, Hougaard B, et al. A general pipeline for the development of anchor markers for comparative genomics in plants. BMC Genomics. 2006;7(207) doi: 10.1186/1471-2164-7-207. http://dx.doi.org/10.1186/1471-2164-7-207 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas FO, Moretzsohn MC, Valls JFM. Genetic variability of Brazilian Indian landraces of Arachis hypogaea L. Genetics and Molecular Research. 2007;6:675–684. [PubMed] [Google Scholar]
- Friend SA, Quandt D, Tallury SP, Stalker HT, Hilu KW. Species, genomes, and section relationships in the genus Arachis (Fabaceae): a molecular phylogeny. Plant Systematics and Evolution. 2010;290:185–199. [Google Scholar]
- Gimenes MA, Lopes CR, Galgaro ML, Valls JF, Kochert G. RFLP analysis of genetic variation in species of section Arachis, genus Arachis (Leguminosae) Euphytica. 2002a;123:421–429. [Google Scholar]
- Gimenes MA, Lopes CR, Valls JFM. Genetic relationships among Arachis species based on AFLP. Genetics and Molecular Biology. 2002b;25:349–353. [Google Scholar]
- Gimenes MA, Hoshino AA, Barbosa AVG, Palmieri DA, Lopes CR. Characterization and transferability of microsatellite markers of the cultivated peanut (Arachis hypogaea) BMC Plant Biology. 2007;7(9) doi: 10.1186/1471-2229-7-9. http://dx.doi.org/10.1186/1471-2229-7-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Pollock DD. Launching microsatellites: a review of mutation processes and methods of phylogenetic inference. Journal of Heredity. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular and Biological Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grattapaglia D, Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137:1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MP, Gregory WC. Exotic germ plasm of Arachis L. interspecific hybrids. Journal of Heredity. 1979;70:185–193. [Google Scholar]
- Gregory W, Gregory M. Groundnut. In: Simmonds N, editor. Evolution of crop plants. London: Longman; 1976. [Google Scholar]
- Gregory W, Krapovickas A, Gregory M. Structure, variation, evolution and classification in Arachis. In: Summerfield R, Bunting A, editors. Advances in legume science. Kew, London: Royal Botanic Gardens; 1980. [Google Scholar]
- Halward TM, Stalker HT, Larue EA, Kochert G. Genetic variation detectable with molecular markers among unadapted germ-plasm resources of cultivated peanut and related wild species. Genome. 1991;34:1013–1020. [Google Scholar]
- Halward T, Stalker T, LaRue E, Kochert G. Use of single-primer DNA amplifications in genetic studies of peanut (Arachis hypogaea L.) Plant Molecular Biology. 1992;18:315–325. doi: 10.1007/BF00034958. [DOI] [PubMed] [Google Scholar]
- He G, Prakash CS. Identification of polymorphic DNA markers in cultivated peanut (Arachis hypogaea L.) Euphytica. 1997;97:143–149. [Google Scholar]
- He G, Prakash CS. Evaluation of genetic relationships among botanical varieties of cultivated peanut (Arachis hypogaea L.) using AFLP markers. Genetic Resources and Crop Evolution. 2001;48:347–353. [Google Scholar]
- He G, Meng R, Newman M, Gao G, Pittman R, Prakash C. Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.) BMC Plant Biology. 2003;3(3) doi: 10.1186/1471-2229-3-3. http://dx.doi.org/10.1186/1471-2229-3-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herselman L. Genetic variation among southern African cultivated peanut (Arachis hypogaea L.) genotypes as revealed by AFLP analysis. Euphytica. 2003;133:319–327. [Google Scholar]
- Hilu KW, Stalker HT. Genetic relationships between peanut and wild species of Arachis sect. Arachis (Fabaceae): evidence from RAPDs. Plant Systematics and Evolution. 1995;198:167–178. [Google Scholar]
- Hoshino AA, Bravo JP, Angelici CMLCD, Barbosa AVG, Lopes CR, Gimenes MA. Heterologous microsatellite primer pairs informative for the whole genus Arachis. Genetics and Molecular Biology. 2006;29:665–675. [Google Scholar]
- Hougaard BK, Madsen LH, Sandal N, et al. Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics. 2008;179:2299–2312. doi: 10.1534/genetics.108.090084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted L. Cytological studies on the peanut Arachis. II. Chromosome number, morphology and behavior, and their aplication to the problem of the origin of the cultivated forms. Cytologia. 1936;7:396–423. [Google Scholar]
- Idury RM, Cardon LR. A simple method for automated allele binning in microsatellite markers. Genome Research. 1997;7:1104–1109. doi: 10.1101/gr.7.11.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Kochert G, Halward T, Branch WD, Simpson CE. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theoretical and Applied Genetics. 1991;81:565–570. doi: 10.1007/BF00226719. [DOI] [PubMed] [Google Scholar]
- Kochert G, Stalker HT, Gimenes M, Galgaro L, Lopes CR, Moore K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae) American Journal of Botany. 1996;83:1282–1291. [Google Scholar]
- Koppolu R, Upadhyaya HD, Dwivedi SL, Hoisington DA, Varshney RK. Genetic relationships among seven sections of genus Arachis studied by using SSR markers. BMC Plant Biology. 2010;10(15) doi: 10.1186/1471-2229-10-15. http://dx.doi.org/10.1186/1471-2229-10-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapovickas A, Gregory W. Taxonomia del genero Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- Krishna TG, Mitra R. The probable genome donors to Arachis hypogaea L. based on arachin seed storage protein. Euphytica. 1988;37:47–52. [Google Scholar]
- Lavia GI. Karyotypes of Arachis palustris and A. praecox (Section Arachis), two species with basic chromosme number x = 9. Cytologia. 1998;63:177–181. [Google Scholar]
- Lavia GI. Chromosome studies in wild Arachis (Leguminosae) Caryologia. 2000;53:277–281. [Google Scholar]
- Lavia GI. Chromosomal characterization of germplasm of wild species of Arachis L. belonging to sections Trierectoides, Erectoides and Procumbentes. Caryologia. 2001;54:115–119. [Google Scholar]
- Lavia G, Ortiz A, Fernández A. Karyotypic studies in wild germplasm of Arachis (Leguminosae) Genetic Resources and Crop Evolution. 2009;56:755–764. [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Systematic Biology. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse S. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lu J, Pickersgill B. Isozyme variation and species relationships in peanut and its wild relatives (Arachis L. – Leguminosae) Theoretical and Applied Genetics. 1993;85:550–560. doi: 10.1007/BF00220913. [DOI] [PubMed] [Google Scholar]
- Lynch M. The similarity index and DNA fingerprinting. Molecular Biology and Evolution. 1990;7:478–484. doi: 10.1093/oxfordjournals.molbev.a040620. [DOI] [PubMed] [Google Scholar]
- Magallon S, Sanderson M. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Mantel NA. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Milla SR, Isleib TG, Stalker HT. Taxonomic relationships among Arachis sect. Arachis species as revealed by AFLP markers. Genome. 2005;48:1–11. doi: 10.1139/g04-089. [DOI] [PubMed] [Google Scholar]
- Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM, Ferreira ME. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biology. 2004;4(11) doi: 10.1186/1471-2229-4-11. http://dx.doi.org/10.1186/1471-2229-4-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretzsohn M, Leoi L, Proite K, et al. A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae) Theoretical and Applied Genetics. 2005;111:1060–1071. doi: 10.1007/s00122-005-0028-x. [DOI] [PubMed] [Google Scholar]
- Moretzsohn M, Barbosa A, Alves-Freitas D, et al. A linkage map for the B-genome of Arachis (Fabaceae) and its synteny to the A-genome. BMC Plant Biology. 2009;9(40) doi: 10.1186/1471-2229-9-40. http://dx.doi.org/10.1186/1471-2229-9-40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K. Incorporating information from length-mutational events into phylogenetic analysis. Molecular Phylogenetic and Evolution. 2006;38:667–676. doi: 10.1016/j.ympev.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Nielen S, Campos-Fonseca F, Leal-Bertioli S, et al. FIDEL – a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut. Chromosome Research. 2010;18:227–246. doi: 10.1007/s10577-009-9109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielen S, Vidigal B, Leal-Bertioli S, et al. Matita, a new retroelement from peanut: characterization and evolutionary context in the light of the Arachis A-B genome divergence. Molecular Genetics and Genomics. 2011;287:21–38. doi: 10.1007/s00438-011-0656-6. [DOI] [PubMed] [Google Scholar]
- Paik-Ro OG, Smith RL, Knauft DA. Restriction fragment length polymorphism evaluation of six peanut species within the Arachis section. Theoretical and Applied Genetics. 1992;84:201–208. doi: 10.1007/BF00224001. [DOI] [PubMed] [Google Scholar]
- Palmieri DA, Hoshino A, Bravo J, Lopes C, Gimenes M. Isolation and characterization of microsatellite loci from the forage species Arachis pintoi (genus Arachis) Molecular Ecology Notes. 2002;2:551–553. [Google Scholar]
- Pattee HE, Stalker HT, Giesbrecht FG. Reproductive efficiency in reciprocal crosses of Arachis monticola with A. hypogaea subspecies. Peanut Science. 1998;25:7–12. [Google Scholar]
- Peñaloza A, Valls J. Contagem do número cromossômico em acessos de Arachis decora (Leguminosae) In: Veiga R, Bovi M, Betti J, editors. Simpósio Latino-Americano de Recursos Genéticos Vegetais. Campinas. Voltan RBQ: IAC/Embrapa-Cenargen; 1997. [Google Scholar]
- Peñaloza APS, Valls JFM. Chromosome number and satellite chromosome morphology of eleven species of Arachis (Leguminosae) Bonplandia. 2005;14:65–72. [Google Scholar]
- Prasanth VP, Chandra S, Jayashree B, Hoisington D. 2006. AlleloBin – A program for allele binning of microsatellite markers based on the algorithm of Idury and Cardon (1997) International Crops Research Institute for the Semi-Arid Tropics. [Google Scholar]
- Proite K, Leal-Bertioli S, Bertioli D, et al. ESTs from a wild Arachis species for gene discovery and marker development. BMC Plant Biology. 2007;7(7) doi: 10.1186/1471-2229-7-7. http://dx.doi.org/10.1186/1471-2229-7-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina SN, Mukai Y. Genomic in situ hybridization in Arachis (Fabaceae) identifies the diploid wild progenitors of cultivated (A. hypogaea) and related wild (A. monticola) peanut species. Plant Systematics and Evolution. 1999;214:251–262. [Google Scholar]
- Ren X, Huang J, Liao B, Zhang X, Jiang H. Genomic affinities of Arachis genus and interspecific hybrids were revealed by SRAP markers. Genetic Resources and Crop Evolution. 2010;57:903–913. [Google Scholar]
- Robledo G, Seijo G. Characterization of the Arachis (Leguminosae) D genome using fluorescence in situ hybridization (FISH) chromosome markers and total genome DNA hybridization. Genetics and Molecular Biology. 2008;31:717–724. [Google Scholar]
- Robledo G, Seijo G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theoretical and Applied Genetics. 2010;121:1033–1046. doi: 10.1007/s00122-010-1369-7. [DOI] [PubMed] [Google Scholar]
- Robledo G, Lavia G, Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theoretical and Applied Genetics. 2009;118:1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- Rohlf F. Exeter Software: Setauket. New York: 2009. NTSYSpc: numerical taxonomy system. ver. 2·21c. [Google Scholar]
- Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Critical Reviews in Biochemistry and Molecular Biology. 2002;37:121–147. doi: 10.1080/10409230290771474. [DOI] [PubMed] [Google Scholar]
- Santos VSE, Gimenes MA, Valls JFM, Lopes CR. Genetic variation within and among species of five sections of the genus Arachis L. (Leguminosae) using RAPDs. 2003;50:841–848. [Google Scholar]
- Scherson RA, Vidal R, Sanderson MJ. Phylogeny, biogeography, and rates of diversification of New World Astragalus (Leguminosae) with an emphasis on South American radiations. American Journal of Botany. 2008;95:1030–1039. doi: 10.3732/ajb.0800017. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Seijo G, Lavia GI, Fernandez A, et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. American Journal of Botany. 2007;94:1963–1971. doi: 10.3732/ajb.94.12.1963. [DOI] [PubMed] [Google Scholar]
- Seijo JG, Lavia GI, Fernandez A, Krapovickas A, Ducasse D, Moscone EA. Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaënsis are the wild diploid progenitors of A. hypogaea (Leguminosae) American Journal of Botany. 2004;91:1294–1303. doi: 10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- Simmons M, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Singh AK. Utilization of wild relatives in the genetic improvement of Arachis hypogaea L. Part 8. Synthetic amphidiploids and their importance in interspecific breeding. Theoretical and Applied Genetics. 1986;72:433–439. doi: 10.1007/BF00289523. [DOI] [PubMed] [Google Scholar]
- Singh AK, Moss JP. Utilization of wild relatives in genetic improvement of Arachis hypogaea L. Part 2. Chromosome complements of species of section Arachis. Theoretical and Applied Genetics. 1982;61:305–314. doi: 10.1007/BF00272846. [DOI] [PubMed] [Google Scholar]
- Singh AK, Moss JP. Utilization of wild relatives in the genetic improvement of Arachis hypogaea L. Part 5. Genome analysis in section Arachis and its implications in gene transfer. Theoretical and Applied Genetics. 1984;68:355–364. doi: 10.1007/BF00267889. [DOI] [PubMed] [Google Scholar]
- Singh AK, Sivaramakrishnan S, Mengesha MH, Ramaiah CD. Phylogenetic relations in section Arachis based on seed protein profile. Theoretical and Applied Genetics. 1991;82:593–597. doi: 10.1007/BF00226795. [DOI] [PubMed] [Google Scholar]
- Singh KP, Singh A, Raina SN, Singh AK, Ogihara Y. Ribosomal DNA repeat unit polymorphism and heritability in peanut (Arachis hypogaea L.) accessions and related wild species. Euphytica. 2002;123:211–220. [Google Scholar]
- Smartt J, Gregory W, Gregory M. The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica. 1978;27:665–675. [Google Scholar]
- Staden R. The Staden sequence analysis package. Molecular Biotechnology. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Stalker HT. A new species in section Arachis of peanuts with a D genome. American Journal of Botany. 1991;78:630–637. [Google Scholar]
- Stalker HT, Phillips TD, Murphy JP, Jones TM. Variation of isozyme patterns among Arachis species. Theoretical and Applied Genetics. 1994;87:746–755. doi: 10.1007/BF00222901. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Gurtu S, Nageswara Rao RC, Nigam SN. Identification of DNA polymorphism in cultivated groundnut using random amplified polymorphic DNA (RAPD) assay. Genome. 2000;43:656–660. [PubMed] [Google Scholar]
- Sukumaran J. Bootscore: a bootstrap tree scoring utility. Version 3·0. 2007 http://sourceforge.net/projects/bootscore . [Google Scholar]
- Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods), 4·04 Beta. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Tallury SP, Hilu KW, Milla SR, et al. Genomic affinities in Arachis section Arachis (Fabaceae): molecular and cytogenetic evidence. Theoretical and Appled Genetics. 2005;111:1229–1237. doi: 10.1007/s00122-005-0017-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Valls JFM, Simpson CE. New species of Arachis L. (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia. 2005;14:35–63. [Google Scholar]
- Wang CT, Wang XZ, Tang YY, et al. Phylogeny of Arachis based on internal transcribed spacer sequences. Genetic Resources and Crop Evolution. 2011;58:311–319. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Research. 2007;17:1072–1081. doi: 10.1101/gr.6214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.