Many human diseases are believed to be caused by undiscovered pathogens1–3. The advent of next-generation sequencing technology presents an unprecedented opportunity to identify pathogens in hitherto idiopathic diseases. Here we present PathSeq, a highly scalable software tool that performs computational subtraction on high-throughput sequencing data to identify non-human nucleic acids that may indicate candidate microbes. PathSeq exhibits high sensitivity and specificity in its ability to discriminate human from non-human sequences using both simulated and experimental transcriptome and whole-genome sequencing data. PathSeq is implemented in a cloud computing environment making it readily accessible by the scientific community.
Previously our group and others have developed a computational approach to pathogen discovery, sequence-based computational subtraction4–8. This method is based on the premise that infected tissues contain both human and microbial nucleic acids and that novel pathogen-derived sequences can be detected after subtracting human sequences. This unbiased approach to pathogen discovery is an advance over targeted PCR or pan-microbial array methods because it requires no sequence information ab initio about the organism being sought (see Lipkin, 2010 for a recent, in-depth review of pathogen discovery methods3). However, performing computational subtraction at any significant scale was initially cost-prohibitive as this method requires a large number of input sequences, given that any pathogen present is likely to have low nucleic acid representation relative to that of the human host.
The recent development of next-generation sequencing methods9–10, however, has made computational subtraction-based pathogen discovery a viable option. For example, massively parallel pyrosequencing combined with computational subtraction has resulted in the discovery of novel viruses in human disease: Merkel cell polyomavirus in Merkel cell carcinoma11 and a novel Old World arenavirus in a cluster of patients with fatal transplant-associated disease12. Indeed, the past few years have seen steep drops in price and increases in throughput for next-generation sequencing technologies, and these trends are expected to accelerate in the near future9–10. However, this advancement in technology brings with it new computational challenges. Analyzing sequence data using the computational subtraction method is computationally expensive relative to most other next-generation sequencing analyses because it requires subtractive alignments to several large reference databases using local alignment algorithms such as BLAST13.
Here we present PathSeq, a comprehensive computational tool for the analysis of the non-host portion of resequencing data that is capable of detecting the presence of both known and novel pathogens as well as any resident microorganisms. PathSeq runs efficiently on sequence datasets of any size in a scalable and completely reproducible fashion because it is developed on a parallel computing architecture and is implemented in a cloud-computing environment. The PathSeq software package is available for public use in the form of a machine image for cloud computing, which can be launched and monitored using no more than a basic laptop computer. We believe that PathSeq opens the way for a new large-scale effort in pathogen discovery by any researcher with access to deep sequencing data from human tissue.
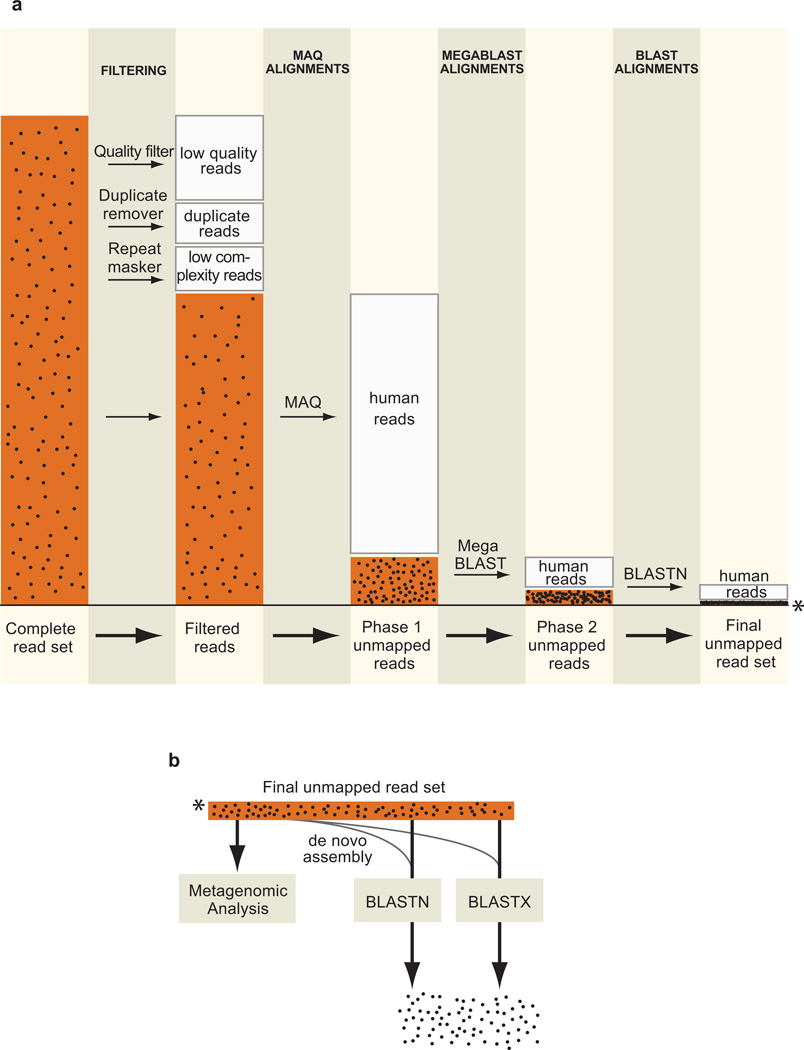
The PathSeq process begins with a subtractive phase in which input reads are subtracted by alignment to human reference sequences (Fig. 1a), and continues with an analytic phase in which the remaining reads are aligned to microbial reference sequences and assembled de novo. The input reads are first filtered to remove low quality, duplicate, and repetitive sequences. The initial subtractive alignments are performed using the rapid short read aligner MAQ14 against five reference human sequence databases, including both genomic DNA and transcriptome references (see Methods). At the end of each subtractive alignment step, mapped reads are discarded and unmapped reads are subjected to further subtractive analyses. In the final steps, the residual reads are aligned to two additional human reference databases first using the Mega BLAST algorithm15 and then BLASTN13. This identifies alignable reads with additional mismatches and/or short gaps that are not aligned by MAQ. The set of reads which remain unmapped after the subtractive phase are candidate non-human, pathogen-derived reads. A similar schema may be used for other host organisms by substituting the appropriate reference genome databases.
Figure 1. The PathSeq workflow.
(a) Conceptual workflow of the subtractive phase of PathSeq. The size of the read set (orange bars) is proportional to the number of reads at the indicated step in a typical run of the method. The black dots in the bars represent pathogen-derived sequences which become progressively concentrated. The steps in this conceptual workflow have been reordered for concision (see Methods for actual ordering). (b) Conceptual workflow of the analytic phase of PathSeq. The asterisk indicates the unmapped readset that is carried over from the subtractive phase.
The analytic phase of PathSeq is composed of several steps that are performed in parallel (Fig. 1b). To identify previously sequenced microbes, all unmapped reads are aligned to reference viral, bacterial, and fungal sequence databases by BLASTN and BLASTX. To assess the bacterial composition of a sample containing a rich microbiome, PathSeq performs a metagenomic analysis by aligning all unmapped reads to the complete collection of currently sequenced whole bacterial genomes and quantifying bacterial representation by a measure of both the total number of aligned reads and the bacterial genome coverage (see Supplementary Methods, Supplementary Tables 1 and 2). To increase the likelihood of discovering a novel organism, all unmapped reads are de novo assembled using the short read assembler Velvet16. The formation of large contigs composed of several unmapped reads that do not possess significant alignment similarity to any sequence in the reference databases may be suggestive of a previously undetected organism.
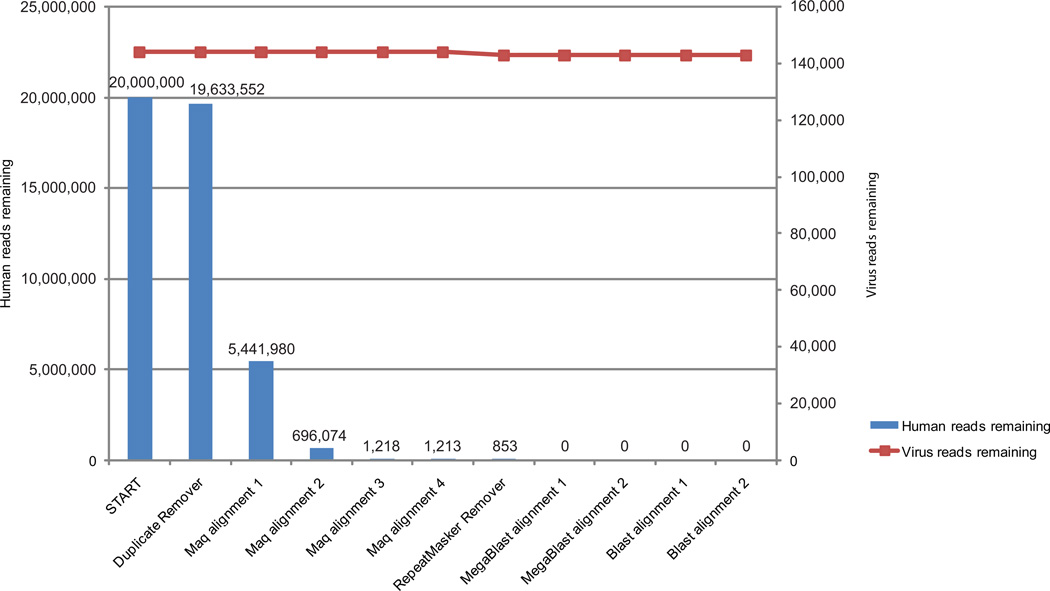
To demonstrate the utility of PathSeq, we used simulated data to assess the ability of the method to (i) efficiently subtract human-derived sequences and (ii) minimize the subtraction of microbe-derived sequences (Fig. 2). We created a simulated sequence dataset by combining sequences generated from a reference human transcriptome database and several virus genomes (Supplementary Fig. 1). Twenty million 100-mers were randomly generated from the reference transcriptome. The simulated virus reads were generated from twelve viral genomes; each viral genome was substitutionally mutated randomly at twelve distinct rates (0, 1, 5, 10, 20, 30, 40, 50, 60, 70, 80, and 90 percent), to simulate unknown viruses at different evolutionary distances from known viruses, producing 1,000 reads per mutated genome for a total of (12 × 12 × 1,000) 144,000 virus reads.
Figure 2. PathSeq performance on artificially generated sequence data.
Reads were generated by sampling random 100-mer sequences from a human transcriptome database to produce 20 million reads, and from a set of twelve virus genomes each substitutionally mutated at twelve distinct rates, generating 144,000 reads (see Supplementary Fig. 1). The blue bars represent the number of human reads remaining after the indicated step in the PathSeq workflow, and the red squares connected by a line represent the remaining viral reads.
After the subtractive steps of the PathSeq pipeline, all 20 million human transcriptome-derived reads were correctly identified as human, and only 1,122 (0.78%) virus genome-derived reads were subtracted (Fig. 2). Of these 1,122 reads, 1,120 were identified as repetitive sequences and the remaining 2 reads were subtracted because of alignment similarity to the human genome (Supplementary Table 3).
To model the performance of PathSeq on low-quality sequence data, we introduced “sequencing errors” into this same readset based on the distribution of actual Illumina sequencing errors and found that this did not significantly affect the performance of PathSeq (Supplementary Fig. 2, Supplementary Table 4, Supplementary Methods). With the exception of human rhinovirus A, which contained many repetitive sequences, greater than 97% of all non-mutated, virus-derived reads were correctly identified by alignment to the viral nucleotide database, and over 50% were still correctly identified at a substitutional mutation rate of over 20% (Supplementary Fig. 3a). Sequence alignment following de novo assembly allowed the identification of sequences with an even higher mutation rate (Supplementary Fig. 3b). We note that the presence of large, unidentifiable contigs in experimental sequence data could suggest the presence of a novel microbe lacking sequence homology to known microbes, and propose that such a result should justify follow-up by PCR, 3’- or 5’-RACE, and Sanger sequencing.
This notion of identifying the presence of microbes by contig formation prompted us to ask how many reads are required to form sufficiently large contigs. The probability of forming contigs from reads originating from a single genome is a function of two variables: (i) the size in base-pairs of the genome in question; and (ii) the number of reads derived from the genome. We simulated the ability of Velvet, the short read assembler that is used in PathSeq, to form contigs that are at least 1.75 times the size of the input reads from a genome by randomly generating reads from genomes of varying length (Supplementary Fig. 4). We found that there is a >75% chance of forming contigs from genomes as large as 20kb when only 20 reads are derived from the genome (using 100bp reads). This suggests that relying on contig assembly to indicate the presence of a novel genome may be a practical approach.
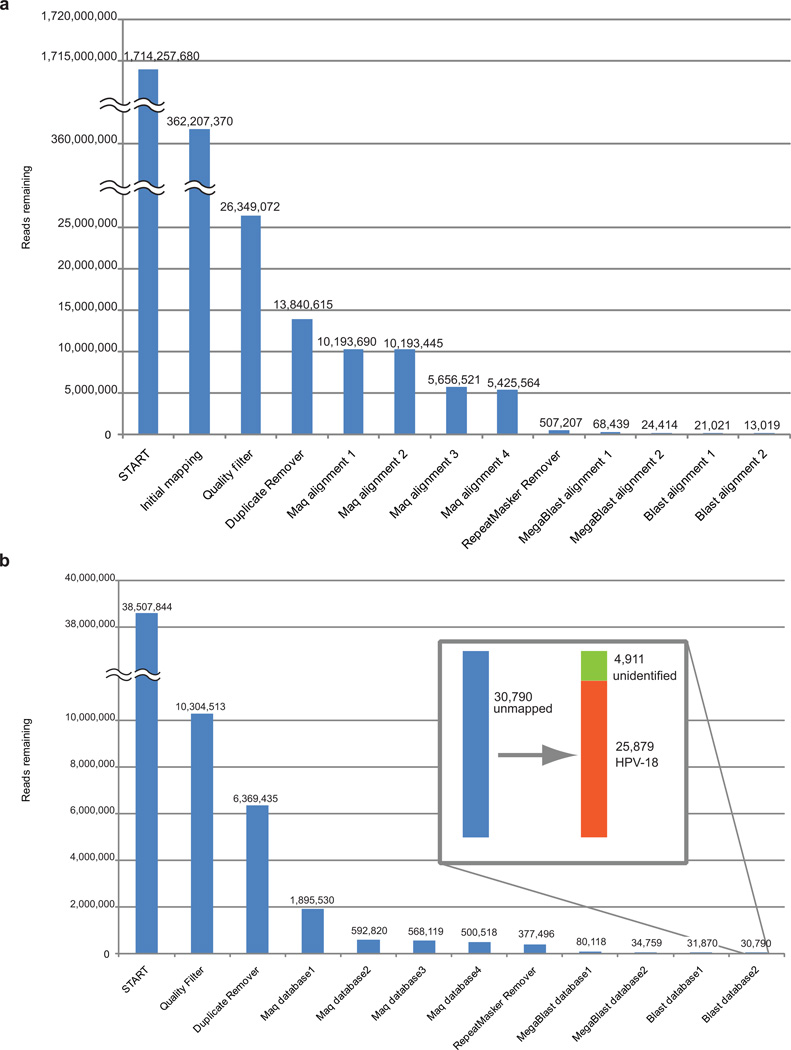
We then tested the performance of PathSeq on a set of sequences representing many-fold coverage of the whole genome of a serous ovarian carcinoma tumor that was sequenced as part of The Cancer Genome Atlas (Supplementary Methods). Starting with slightly more than 1.7 billion reads of 101 bp each, we first removed mapped reads following initial alignment to the reference human genome, and then performed stringent quality filtering to yield 26.3 million reads. We ran these sequences on PathSeq and were left with a final set of 13,019 reads after subtraction, less than 0.001% of the original reads (Fig. 3a). The analytic phase of PathSeq did not yield any evidence that the remaining reads were derived from a pathogen; rather they likely represented yet-uncharacterized regions of the human genome or sequencing artifacts. This substantial subtraction efficiency, 99.9992%, demonstrates that the performance of PathSeq on simulated data can indeed be extended to real, human whole-genome sequencing.
Figure 3. PathSeq performance on experimental sequence data.
(a) We applied whole-genome sequencing data from a human ovarian tumor and (b) one lane of total-RNA transcriptome sequencing from HeLa cell lines to PathSeq. The inset in panel b shows that the 30,790 reads remaining after the subtractive phase of PathSeq are predominantly composed of HPV-18 sequences.
We next generated sequence data from HeLa cervical cancer cell lines with the expectation of finding human papillomavirus (HPV) type18. We sequenced a cDNA library generated from total RNA isolated from HeLa cells on a single lane of Illumina sequencing, generating 10.3 million quality- and purity-filtered 76-bp reads. We applied this sequence data to PathSeq (Fig. 3b). Human-derived reads were efficiently subtracted during the subtractive phase such that 0.30% of input reads remained unmapped. Out of these 30,790 reads, 25,879 were identified as HPV-18, leaving 4,911 non-human, non-HPV-18 reads. We then collected all of the HPV-18 reads and searched for those whose pair-mate aligned to the human reference genome. This allowed us to identify the integration site of the viral genome in a region of chromosome 8q24 between positions 128,300,300 and 128,310,400 just upstream of the MYC oncogene, as previously reported17.
One longstanding goal of computational subtraction is the identification and characterization of every read in a dataset. Although the sensitivity and specificity data for PathSeq are impressive, it still leaves 0.00076% of the ovarian whole-genome sequence reads and 0.013% of the HeLa RNA-Seq reads unaccounted for. This shortfall might be explained by error-ridden sequences passing the quality filter, reads that map to the splice junction of rare splice variants in RNA-Seq data, or reads that map to regions of the human genome that have not yet been characterized. A number of groups have recently reported novel human genome sequences by de novo assembly of next-generation whole-genome sequence data18–19 or by using a fosmid end-sequence pair mapping approach20. Ideally, these new sequences could form a subtractive database for PathSeq and help reduce the total number of unaccounted reads. However, by performing a simple Mega BLAST alignment of these sequences to microbial databases we find that many sequences from all three above-mentioned studies have perfect matches to known bacteria, fungi, and viruses (Supplementary Data Set 1–3), raising the question of whether they may contain novel microbes as well. Therefore, an essential next step in the improvement of computational subtraction-based methods is the creation of a reliable database of human genome scaffolds that extend the current human reference genome.
Taken together, our results demonstrate the ability of PathSeq to identify both known and novel microorganisms in high throughput human resequencing data. Just as current metagenomic surveys of the world’s oceans and soils are yielding remarkable new organisms, so too do we expect to reveal new viruses, bacteria, and fungi in human tissue with important medical implications. We are making PathSeq available for public use at http://www.broadinstitute.org/software/pathseq/, and it is our hope that investigators will use this tool to join our efforts in pathogen discovery.
METHODS
RNA-seq library construction from HeLa cells and Sequencing
RNA was extracted from cultured HeLa cells according to the RNeasy Kit (Qiagen) protocol. cDNA sequencing library construction was performed as described previously21, with noted modifications (Supplementary Methods). The cDNA library was sequenced on the Illumina Genome Analyzer II (GAII) platform. The mean fragment length was approximately 350 base pairs. One lane of paired-end, 76 base pair sequencing was performed, producing 38.5 million purity filtered reads, which yielded 10,304,513 high quality reads following quality filtering.
The PathSeq workflow and cloud implementation
The PathSeq pipeline is designed using the Apache Hadoop implementation of the MapReduce programming framework (http://hadoop.apache.org/mapreduce) and can be run on the Amazon Elastic Compute Cloud (EC2) (http://aws.amazon.com/ec2/)22. The workflow is comprised of three modules: pre-subtraction, subtraction, and post-subtraction. The pre-subtraction module is simply a quality filtering step and is run on the user’s local machine, whereas the subtraction and post-subtraction modules are executed on a Hadoop-based cluster (19 worker nodes and 1 master node) built using the Amazon Elastic Compute Cloud (Amazon EC2).
Amazon’s Simple Storage Service (S3) file system (http://aws.amazon.com/s3/) is used to store the reference sequences and readset, and the config files and scripts are distributed across all nodes on the cluster using the Hadoop Distributed File System (HDFS). The reference sequences are continuously updated on the PathSeq system and users are given the option of substituting any built-in database with a database of their choice; however we provide data download dates for reference sequences used in experiments reported in this paper below.
All processes are run on the Hadoop cluster in multiple map phases. The subtraction module comprises of two mappers. First, subtractive alignments are performed with MAQ (Release 0.5.0, default settings) against a set of six human sequence databases: the 1000 Genomes Project female reference (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/, downloaded 2009-04-11), the Ensembl Homo sapiens cDNA database (ftp://ftp.ensembl.org/pub/current/fasta/homo_sapiens/cdna/, downloaded 2009-04-22), the human genome and transcriptome BLAST database (ftp://ftp.ncbi.nih.gov/blast/db/, downloaded 2009-05-25), and the set of three assembled human genomes available on NCBI (hs_alt_Celera, hs_alt_HuRef, hs_ref_GRCh37, ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/Assembled_chromosomes/, downloaded 2009-06-19). The next map phase is composed of three steps: RepeatMasker (http://www.repeatmasker.org/), MegaBlast, and BLASTN. First, the reads are applied to RepeatMasker (version open-3.2.8, libraries dated 2009-06-04), and any reads with three or more masked nucleotides are discarded. Subtractive alignments are next performed using MegaBlast (Blast Tools version 2.2.23, cut-off expect value 10−7, word size 16) to two human sequence databases: the NCBI Homo sapiens RNA database (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/, downloaded 2009-11-20), and the Ensembl human genome reference (ftp://ftp.ensembl.org/pub/current/fasta/homo_sapiens/dna/, downloaded 2009-10-22). The final set of subtractive alignments are then performed with BLASTN (Blast Tools version 2.2.23, cut-off expect value 10−7, word size 7, nucleotide match reward 1, nucleotide mismatch reward −3, gap open cost 5, gap extension cost 2) to the same two databases. A reduce phase gathers all remaining reads into one consolidated file which serves as input to the post-subtraction module.
The post-subtraction module is also comprised of two mappers. The first mapper is a set of BLASTN (parameters as above) and BLASTX alignments (Blast Tools version 2.2.23, cut-off expect value 10-4, word size 3, matrix: BLOSUM62, gap open cost 11, gap extension cost 1) to viral (downloaded from NCBI Nucleotide (http://www.ncbi.nlm.nih.gov/nucleotide) using search term “‘viruses’[porgn:__txid10239]” on 2010-02-26), fungal (downloaded using the term “’fungi’[porgn:__txid4751]” on 2009-11-23), bacterial and archaeal (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/, downloaded 2010-03-30), and non-redundant protein (ftp://ftp.ncbi.nih.gov/blast/db/nr, downloaded 2010-04-05) reference sequences. This mapper also performs a de novo assembly (Velvet 0.7.31, k-mer size 21) on the full set of reads remaining from the previous map phase. The complete post-subtraction BLAST output files as well as the full set of unmapped reads and contigs are then uploaded and stored on the S3 storage system.
The Amazon Machine Image (AMI) required to build the PathSeq Hadoop cluster is accessible from Amazon Web Services (http://www.broadinstitute.org/software/pathseq/).Pathseq is implemented in Python, Java, C++ and C shell, and has been tested on a Linux 2.6.18-194.8.1.e15 X86_64 system.
Generation and analysis of simulated sequencing data
Simulated human transcriptome and virus sequence data
Twenty million 100-mers were randomly generated from a reference human transcriptome (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/, downloaded 2009-11-20). Simulated virus reads were derived from twelve virus sequences: NCBI Nucleotide accession AY740741, CY000455, EU643590, FJ356716, FJ464337, GQ290462, GQ415051, NC_000883, NC_001405, NC_001806, NC_005179, and NC_007815. For each of these sequences, substitutional mutations were introduced at a frequency of 0%, 1%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, and 90%. In this process, nucleotides along the sequence are chosen at random with replacement (i.e. the same nucleotide can be chosen twice at random) and converted to a different nucleotide (for example, A is converted to C, G, or T). This produces 144 sequences (12 input sequences, each mutated at 12 frequencies). For each of these 144 sequences, 1,000 “reads” of length 100bp were produced at random. The resulting 144,000 simulated reads were pooled with the 20 million simulated human reads and analyzed on PathSeq.
Contig formation simulations
In this experiment, “genomes” of size 200bp to 20,000bp in increments of 200bp were generated from the Human herpesvirus 5 genome (accession GQ466044). For each of these 100 “genomes”, two to twenty 100-mer sequences (“reads”) were generated at random. This process was performed eleven times. For each “genome” size, “read” number pair, Velvet assembly with k-mer size 21 was performed. The frequency with which contigs of at least 175bp were generated was recorded.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute (RC2CA148317), the Starr Cancer Consotium, and the Claudia Adams Barr Program in Innovative Cancer Research. A.D.K. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship. A.I.O. was supported by the Rebecca Ridley Kry Fellowship of the Damon Runyon Cancer Research Foundation. We thank R. Verhaak, M. Berger, C. Mermel, J. Levin, V. Fusaro and J. Shendure for helpful discussions and S. Hart for illustrations.
Footnotes
AUTHOR CONTRIBUTIONS
A.D.K., A.I.O., J.J., G.G. and M.M. conceived the study. A.D.K. implemented the method and C.S.P. made it available in a cloud computing environment. A.D.K. performed the RNA-Seq. A.D.K., A.I.O., C.S.P., J.J., G.G. and M.M. designed and A.D.K. and C.S.P. implemented simulation and validation experiments. A.D.K., C.S.P. and M.M. wrote the manuscript.
References
- 1.Relman DA. Science. 1999;284:1308–1310. doi: 10.1126/science.284.5418.1308. [DOI] [PubMed] [Google Scholar]
- 2.Lipkin WI. PLoS Pathog. 2008;4:e1000002. doi: 10.1371/journal.ppat.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipkin WI. Microbiol Mol Biol Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Nat Genet. 2002;30:141–142. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, et al. Genomics. 2003;81:329–335. doi: 10.1016/s0888-7543(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 6.Tengs T, et al. Nucleic Acids Res. 2004;32:e121. doi: 10.1093/nar/gnh120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, et al. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacConaill L, Meyerson M. Nat Genet. 2008;40:380–382. doi: 10.1038/ng0408-380. [DOI] [PubMed] [Google Scholar]
- 9.Mardis ER. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 10.Shendure J, Ji H. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 11.Feng H, Shuda M, Chang Y, Moore PS. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacios G, et al. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Ruan J, Durbin R. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Schwartz S, Wagner L, Miller W. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 16.Zerbino DR, Birney E. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dürst M, C M Croce CM, L Gissmann L, E Schwarz E, K Huebner K. Proc Natl Acad Sci U S A. 1987;84:1070–1074. doi: 10.1073/pnas.84.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler DA, et al. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 19.Li R, et al. Nat Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 20.Kidd JM, et al. Nat Methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, et al. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall DP, et al. BMC Bioinformatics. 2010;11:259. doi: 10.1186/1471-2105-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.