Abstract
Protein kinases are key components of most mammalian signal transduction networks and are therapeutically relevant drug targets. Efforts to study protein kinase function would benefit from new technologies that are able to profile kinases in complex proteomes. Here, we describe active site-directed probes for profiling kinases in whole cell extracts and live cells. These probes contain general ligands that stabilize a specific inactive conformation of the ATP-binding sites of protein kinases, as well as trifluoromethylphenyl diazirine and alkyne moieties that allow covalent modification and enrichment of kinases, respectively. A diverse group of serine/threonine and tyrosine kinases were identified as specific targets of these probes in whole cell extracts. In addition, a number of kinase targets were selectively labeled in live cells. Our chemical proteomics approach should be valuable for interrogating protein kinase active sites in physiologically relevant environments.
Introduction
Protein phosphorylation cascades mediate a wide array of intra-cellular signaling events in eukaryotic cells.1, 2 The diversity of processes regulated by this post-translational modification is reflected by the large number of protein kinases (> 500) encoded by the human genome.3 While significant efforts have been made to functionally characterize protein kinases, the roles of many of these enzymes in complex biological environments have yet to be interrogated. For this reason, there is a great deal of interest in the development and application of reagents that allow the global analysis of the protein kinase family. For example, immobilized ATP-competitive inhibitors have proven to be useful chemo-proteomic tools for studying protein kinases because they allow the enrichment of these low abundance enzymes. These reagents have provided valuable insight into specific signaling events and have facilitated exhaustive inhibitor selectivity screens in cell lysates.4–16 Despite the widespread utility of affinity matrices that specifically target kinases, these methods are not ideal because they require cell lysis and the use of homogenous lysate preparations, which may disrupt signaling complexes. Furthermore, non-covalent affinity methods are not able to discriminate between proteins that are directly bound to an inhibitor of interest or are instead associated with an enriched protein complex. Affinity- and activity-based probes that are able to label protein kinases based on conserved active site features overcome many of the limitations of non-covalent affinity reagents.17–22 However, the arsenal of labeling reagents that is available for studying protein kinases is limited compared to other enzyme families (for example, serine hydrolases),23–25 and there is still a clear need for additional chemical tools that allow their functional interrogation. Active site-directed probes that facilitate the in situ labeling of protein kinase active sites are especially needed.
All protein kinases contain a conserved, bi-lobal catalytic core that consists of 250–350 residues.26, 27 The site of phosphate transfer is located between these two lobes, with ATP occupying a narrow hydrophobic cleft. The adenine ring of ATP makes a number of hydrophobic contacts in the ATP-binding cleft and forms at least one hydrogen bond with the backbone of a peptide linker - referred to as the hinge region - that connects the N- and C-terminal lobes. A number of conserved catalytic residues that are necessary for phosphate transfer line the ATP-binding sites of protein kinases. The active sites of protein kinases are highly dynamic and the movement of key residues allows kinases to interconvert between catalytically active and inactive forms.28, 29 While the spatial arrangement of active site residues within the ATP-binding sites of kinases that are catalytically competent are highly conserved, there is more conformational heterogeneity for kinases in an inactive state. Indeed, structural studies of protein kinases have revealed a number of catalytically incompetent ATP-binding site configurations. Despite this variability, there appears to be several distinct classes of inactive conformations that are observed throughout the kinome.30, 31 These inactive forms are distinguished by structural elements that are topologically distinct. One inactive conformation that is accessible to a number of kinases is the DFG-out conformation.32–34 This inactive form is characterized by the movement, and almost 180° rotation, of the highly conserved Asp-Phe-Gly motif (DFG-motif), which is located at the base of the activation loop, relative to the active conformation. This displacement results in the Asp of the DFG-motif being unable to coordinate a magnesium ion that is important for phosphate transfer and prevents the Phe side chain from participating in a regulatory network of hydrophobic residues.35 A number of ligands that specifically complement the topology of kinases in the DFG-out conformation have been identified (Figure 1A).32–34, 36 These inhibitors, commonly referred to as type II inhibitors, contain moieties that make similar contacts as the adenine ring of ATP and hydrophobic substituents that occupy the pocket created by the movement of the Phe side chain in the DFG-motif. In general, type II inhibitors are conformation-selective ligands because they are sterically incompatible with the active forms of protein kinases. This feature has made type II inhibitors useful reagents for probing the ATP-binding site conformations of protein kinases.
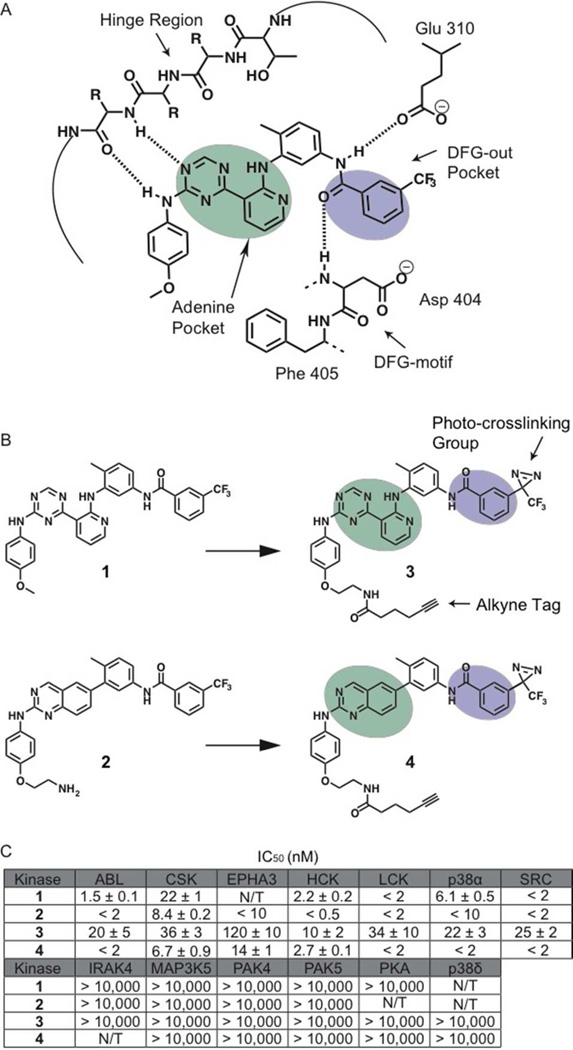
Figure 1.
Binding interactions of type II inhibitors and generation of photo-crosslinking probes. (A) A schematic representation of the interaction of type II inhibitors with the ATP-binding pocket of protein kinases. The residue numbering shown is for human c-SRC. Four hydrogen bonds are made in the binding pocket; a pair of hydrogen bonds to the hinge region, one to the conserved glutamic acid on helix-αC (Glu310) and one to the backbone of the DFG-motif. A heterocyclic moiety occupies the adenine pocket (shown in mint) while the 3-trifluoromethyl phenyl group (shown in purple) accesses the pocket (DFG-out pocket) generated by movement of the DFG-motif. (B) Chemical structures of two general type II inhibitors (1 and 2) and their corresponding probes (3 and 4) that contain a photo-crosslinking group and an alkyne tag. The 3-trifluoromethylphenyl groups (purple) of inhibitors 1 and 2 were replaced with 3-trifluoromethylphenyl diazirine moieties. In addition, the 4-anilino positions of these inhibitors were modified with alkyne tags. (C) IC50 values of 1 –4 against a panel of protein kinases. N/T = not tested. The values shown are the average of assays run in triplicate or quadruplicate +/– SEM.
Here we report the development of a new class of kinase-directed photo-crosslinking reagents. The kinase-targeting elements of these probes are general type II inhibitors that stabilize the DFG-out inactive conformation of protein kinases. To allow labeling of the ATP-binding sites of protein kinases and visualization/enrichment of covalently-modified targets, these probes contain a trifluoromethylphenyl diazirine photo-crosslinker and alkyne tag, respectively. We show that these reagents can be photo-activated to selectively label protein kinases that adopt the DFG-out conformation. Use of these probes in combination with stable-isotope labeling with amino acids in cell culture (SILAC) and mass spectrometry resulted in the identification of a number of selectively enriched protein kinase targets. Furthermore, we demonstrate that a novel kinase target that is specifically labeled by our conformation-selective probes is indeed able to adopt the DFG-out conformation. Finally, we show that these reagents are cell permeable and able to specifically label protein kinases in situ.
Results and Discussion
Design and synthesis of photo-affinity probes derived from type II ATP-competitive inhibitors
Affinity reagents that are able to selectively label the active sites of protein kinases must contain three components: 1) a high affinity ligand that selectively recognizes the active sites of protein kinases, 2) a reactive group that is able to covalently modify probe-bound kinases, and 3) a tag that allows visualization and/or purification. We first focused on identifying general ATP-competitive inhibitors that are able to target the active sites of protein kinases over other ATP-binding proteins. Ligands that are able to stabilize specific active site conformations of protein kinases are of particular interest because they have the potential to provide information on the functional state of these dynamic enzymes. Therefore, we pursued two general scaffolds based on type II kinase inhibitors, which stabilize an inactive form of the ATP-binding sites of protein kinases called the DFG-out conformation (Inhibitors 1 and 2 in Figure 1B).37–39 Both of these ligands contain structural features that make specific contacts with kinases that are in the DFG-out conformation, including a hetero-aromatic moiety that mimics adenine, and additional functionalities that make a characteristic set of hydrogen bonds with a conserved glutamate in helix-αC and the backbone amide of aspartate in the catalytically-important DFG-motif (Figure 1B). Furthermore, 1 and 2 both possess 3-trifluoromethylphenyl groups that occupy the hydrophobic pocket created by the movement of the Phe side chain from the DFG-motif. Because protein kinases do not possess any conserved residues that are highly nucleophilic, a photo-crosslinker was incorporated into our active site-directed probes. A trifluoromethylphenyl diazirine was selected due to the relatively small size of this moiety and its ability to be photo-activated at a wavelength (365 nm) that causes minimal photochemical damage to proteins and cells.40 To allow the introduction of this crosslinking group without disrupting the interaction of the core scaffold with protein kinase ATP-binding sites, the trifluoromethyl groups of inhibitors 1 and 2 were replaced with trifluoromethyl diazirines. Substitution at this position places the crosslinking moiety deep within the hydrophobic pocket created by the movement of the DFG-motif. Finally, the solvent-exposed 4-anilino moieties of both inhibitors were derivatized with an alkyne tag that allows visualization and enrichment of probe-labeled proteins.
To determine whether probes 3 and 4 retain their ability to interact with the ATP-binding sites of protein kinases that adopt the DFG-out conformation, their inhibitory activities against a small panel of protein kinases was tested (Figure 1C). ABL, CSK, EPHA3, HCK, LCK, p38α, and SRC have been structurally characterized in the DFG-out conformation and are sensitive to type II inhibitors.33, 39, 41–46 In contrast, IRAK4, MAP3K5, PAK4, PAK5, PKA, and p38δ have not been observed in this inactive conformation and are not inhibited by type II inhibitors.45, 46 Like 1 and 2, probes 3 and 4 inhibit kinases that adopt the DFG-out conformation and have very little activity against IRAK4, MAP3K5, PAK4, PAK5, PKA, and p38δ (Figure 1C). While probes 3 and 4 have similar potencies as their parent ligands 1 and 2 for some kinases, they generally have > 5-fold higher IC50s for most. The observed loss in potency is most likely due to the trifluoromethyl diazirine groups of 3 and 4 being less well accommodated in the hydrophobic pocket created by the movement of the DFG-motif, relative to the trifluoromethyl groups of 1 and 2. Regardless, the affinity of both probes for DFG-out adopting kinases is sufficient for their use as kinase-directed probes.
Photo-labeling of protein kinases in mammalian cell lysate
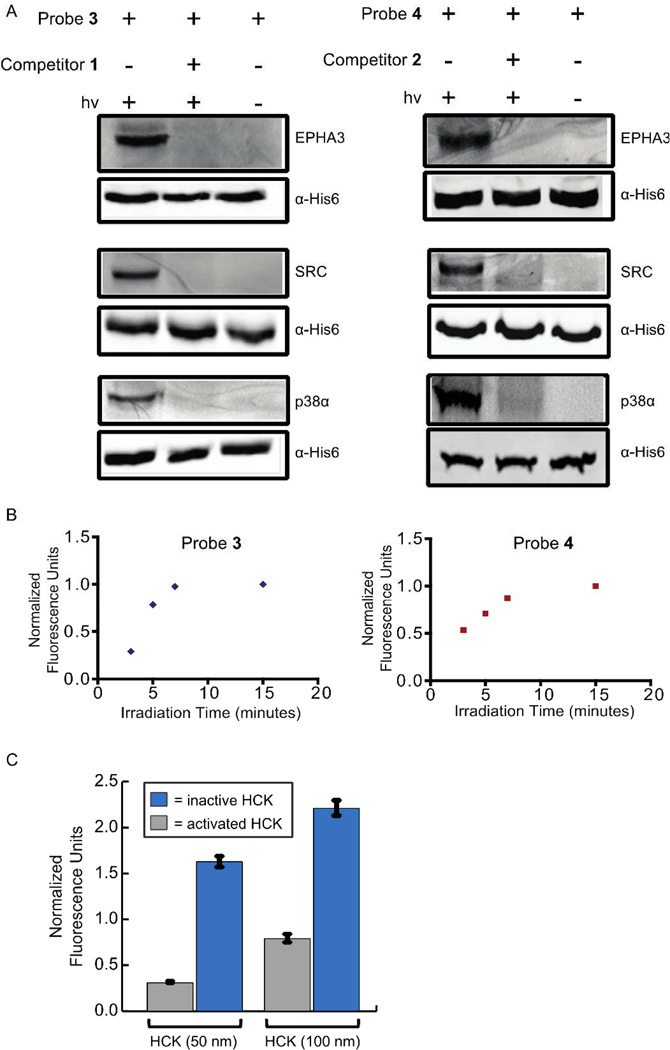
Probes 3 and 4 were first tested for their ability to label protein kinases in mammalian cell lysate (Figure 2A). The DFG-out-adopting protein kinases SRC, p38α, and EPHA3, were incubated with either probe 3 (1 µM) or 4 (1 µM). Samples were then irradiated with 365 nm light, conjugated to rhodamine-azide using copper-mediated click chemistry conditions, separated by SDS-PAGE, and analyzed by in-gel fluorescence scanning. Robust labeling of SRC, p38α and EPHA3 was observed with both probes (Figure 2A, lane 1). In contrast, no fluorescent signal was observed if photo-crosslinking was performed in the presence of an excess of the active site competitors 3 or 4 (10 µM) (Figure 2A, lane 2), indicating specific labeling of kinase active sites. In addition, the type II inhibitors Ponatinib and Rebastinib, which are based on different chemical scaffolds, are able to block active site labeling of SRC (SI, Figure S1). Furthermore, no labeling was observed in the absence of UV light (Figure 2A, lane 3). To determine whether probes 3 and 4 are able to selectively label kinases that adopt the DFG-out conformation, photo-crosslinking experiments were performed in the presence of two kinases, MAP3K5 and PAK5, that have not been characterized to adopt this inactive form and are not sensitive to type II inhibitors (Figure S2). The presence of only a single fluorescent band, corresponding to SRC, EPHA3, or p38α, under these conditions further demonstrates the specificity of probes 3 and 4 (Figure S2). Before performing experiments in more complex protein mixtures, the effect of irradiation time on kinase crosslinking efficiency was explored. For both probes, labeling was found to be linear up to 7 minutes, with maximum signal observed within 10 minutes (Figure 2B).
Figure 2.
Labeling of DFG-out adopting kinases with probes 3 and 4. (A) His6-tagged protein kinases (90 nM) in mammalian cell lysate (0.2 mg/mL) were incubated with probes 3 (1 µM) or 4 (1 µM) in the absence (lanes 1 and 3) or presence (lane 2) of an active site competitor (10 µM). The samples shown in lanes 1 and 2 were then irradiated with UV light for 15 minutes, while those shown in lane 3 were not. All samples were then tagged with rhodamine-azide, resolved by SDS-PAGE, and labeled proteins were detected with in-gel fluorescence scanning (top blots). Immunoblots were performed with an anti-His6 tag antibody (Cell Signaling) to ensure equal amounts of kinase were present (bottom blots). (B) The effect of UV irradiation time on photocross-linking efficiency. The tyrosine kinase SRC was incubated with probes 3 (1 µM) or 4 (1 µM) and then irradiated with 365 nm light for 3, 5, 7, or 15 minutes. Normalized fluorescence units versus irradiation time are shown. (C) Labeling of an activated and an inactive HCK construct with probe 3. Activated or inactive HCK (50 or 100 nM) in mammalian cell lysate (0.2 mg/mL) were incubated with probe 3 (1 µM). Samples were then irradiated with UV light for 15 minutes, conjugated to rhodamine-azide, resolved by SDS-PAGE, and labeled proteins were detected with in-gel fluorescence scanning. Fluorescence intensities were quantified and normalized to a fluorescent standard. The values shown are the average of assays run in triplicate ± SEM.
An interesting application of photo-affinity probes based on type II inhibitors is their use to interrogate how specific post-translation modifications and protein-protein interactions affect the active site conformation of protein kinases. It has previously been demonstrated that some protein kinases, like the tyrosine kinase ABL, have lower affinities for ligands that stabilize the DFG-out conformation when their activation loops are phosphorylated.47 While this is not true for all kinases (for example, the phosphorylation status of the activation loop of p38α does not affect its sensitivity to most type II inhibitors)44, our photo-affinity probes can serve as valuable reagents for testing this relationship. To explore this possibility, we determined the labeling efficiency of probe 3 for two different activation states of the tyrosine kinase HCK (Figure 2C). An activated construct of HCK, which contains a phosphorylated activation loop and two mutations that disrupt an autoinhibitory intra-molecular SH3 domain interaction, and an inactive HCK construct, which contains an unphosphorylated activation loop and two mutations that strengthen an autoinhibitory intra-molecular SH3 domain interaction, were tested at two different concentrations (50 and 100 nM). At both kinase concentrations tested, probe 3 more readily labeled the inactive HCK construct than the activated HCK construct. This result is consistent with the ATP-binding site of the inactive HCK construct showing an increased sampling of the DFG-out conformation. The ability to discriminate between kinase activation states should allow our probes to profile the inter-relationship between protein kinase activation states and ATP-binding site conformations.
Proteome-wide labeling of protein kinases in A431 cell lysate
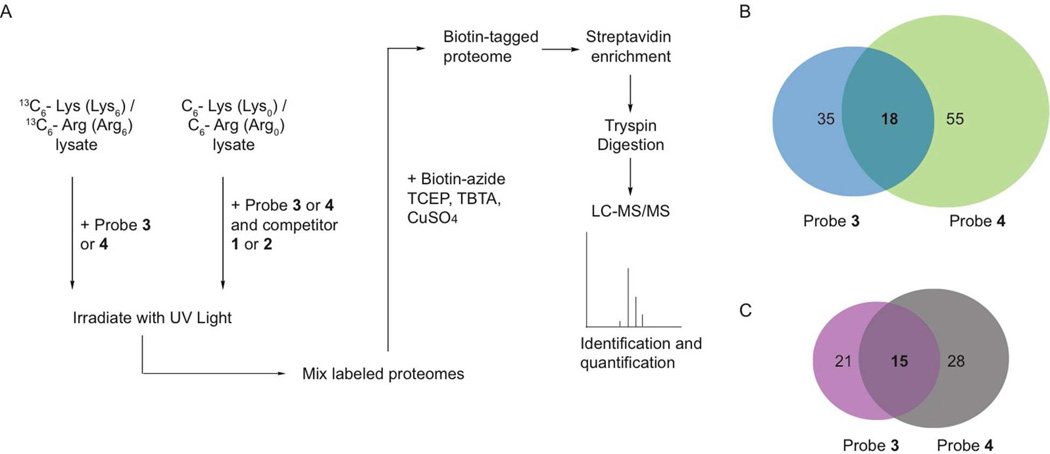
To determine the full labeling profiles of both probes, photo-crosslinking experiments were performed in lysate derived from A431 cells (Figure 3A). A431 cells are a human epithelial carcinoma cell line that overexpress epidermal growth factor receptor and are a model for studying growth factor-stimulated signaling. Because kinases are low abundance enzymes that can often be obscured during proteomic analysis by more abundant cellular proteins, we used a quantitative proteomics approach to identify the specific targets of probes 3 and 4. SILAC methodology was used to generate lysate proteins that contain either “light” (natural isotopic abundance) arginine and lysine or “heavy” (U-13C) arginine and lysine.48 Photo-crosslinking was performed in the heavy lysate with either probe 3 (1 µM) or 4 (1 µM). The same labeling experiments were performed with the light lysate in the presence of an excess of an active site competitor (10 µM). After UV irradiation, both the heavy and light lysate were combined, conjugated to biotin-azide, and enriched with streptavidin-conjugated beads. The streptavidin-conjugated beads were then washed extensively and subjected to on-bead digestion with trypsin. Eluted peptides were separated by reverse-phase chromatography and online analyzed in a hybrid LTQ-Orbitrap mass spectrometer. Mass spectrometric analysis revealed peptide identities and relative peptide abundances between the heavy and light samples. Proteins with a heavy-to-light ratio (H/L) greater than 2-fold were deemed as specific targets of the probes. Due to the stringent wash conditions (1% SDS (3x) and 6M urea (3x)) used in the pull-down experiments, we felt it was unlikely that any protein targets would be enriched non-covalently. However, a model study was performed to confirm that this is most likely the case (Figure S3). A cell lysate supplemented with 90 nM HCK was incubated with probes 3 (1 µM) or 4 (1 µM) but not irradiated with UV light. Samples were then conjugated to biotin-azide, subjected to the pull-down and wash protocol described above, and bound proteins were eluted from the streptavidin beads with SDS. Under these conditions, no HCK could be detected in the eluted samples by Western blot analysis (Figure S3). In contrast, HCK could readily be detected when samples were irradiated prior to performing the same pull-down experiment. This indicates that any proteins that are enriched in the SILAC experiments described above have most likely been covalently modified by probes 3 and 4.
Figure 3.
Labeling of endogenous protein kinases in A431 cell lysate with probes 3 and 4. (A) A schematic representation of the photo-labeling experiments that were performed in A431 cell lysate with probes 3 and 4. A431 cells were metabolically labeled with arginine (13C6-Arg) and lysine (13C6-Lys) and the lysate generated was photo-crosslinked with probes 3 or 4. In parallel, the same photo-labeling experiments were performed with probes 3 or 4 in the presence of an ATP-binding site competitor (inhibitors 1 or 2) with cell lysate that contains “light” Lys and Arg. After photo-crosslinking, both labeling experiments were combined and the subsequent mixture was subjected to copper-mediated click chemistry with biotin-azide. Streptavidin-conjugated beads were used to enrich biotin-labeled proteins. These beads were then washed extensively and subjected to digestion with trypsin. Tryptic peptides were identified by tandem mass spectrometry and quantified to obtain a heavy-to-light peptide ratio. The results of these experiments are shown in Table 1. (B) Venn diagram showing total and overlapping proteins enriched by probes 3 and 4. (C) Venn diagram showing total and overlapping protein kinases enriched by probes 3 and 4.
A summary of the data that was obtained in the A431 cell lysate labeling experiments is shown in Figure 3 and Table 1. For probe 3, 35 proteins were specifically labeled, with 21 of those being protein kinases. For probe 4, 55 proteins were specifically labeled, with 28 of those being protein kinases. There is a 45% overlap in the protein kinases that were enriched by the two probes. A diverse group of protein kinases were specifically labeled, with tyrosine kinases representing the largest percentage (42%) of enriched kinases. Of the 15 protein kinases that were selectively enriched by both probes 3 and 4, 10 are tyrosine kinases. Specifically enriched tyrosine kinases include several SRC-family members, such as SRC, FRK and SRM, which are known to adopt the DFG-out conformation. With probe 3, a 10-fold or higher heavy-to-light peptide ratio was observed for both SRC and SRM, while only the heavy peptides were detected for FRK. Similar results were observed with probe 4. In addition, probes 3 and 4 also selectively labeled a number of Ephrin-activated receptor tyrosine kinases including EPHA2, EPHB2, EPHB3, and EPHB4. This family of receptor tyrosine kinases has been structurally characterized to adopt the DFG-out conformation and is believed to play prominent roles in many forms of cancer.46 Of the 18 remaining enriched protein kinase targets that are not tyrosine kinases, 44% are from the STE Group. Unlike the tyrosine kinases, a majority of the serine/threonine kinase targets were labeled by only one probe. For example, only probe 3 selectively enriched the mitogen-activated protein kinase p38α (H/L ratio = 5.1), which has been well characterized to adopt the DFG-out conformation. These results demonstrate that probes 3 and 4 are able to selectively label expected as well as novel protein kinase targets. While selective labeling of a kinase target is not definitive proof that a kinase can adopt the DFG-out conformation, these studies provide a number of interesting candidates for future biochemical and structural characterization. In addition to protein kinases, a number of other diverse protein targets were selectively enriched by probes 3 and 4 (Table S2, SI). Unlike the protein kinases, there are not multiple members of any one protein family represented. Perhaps the most interesting of the enriched non-kinase targets are retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta (PDE6D), reticulon-3 (RTN-3), and a member of the cytoplasmic epoxide hydrolase 2 family (EPHX2), which are all selectively labeled by probes 3 and 4. We are currently exploring whether type II inhibitors perturb the functions of these targets and if these proteins contain binding sites that are similar to the DFG-out conformation of protein kinases.
Table 1.
Kinases enriched in A431 lysate with probes 3 and 4.
| Protein | Probe | H/L Ratio (Probe 3) |
# of Peptides Identified (Probe 3) |
H/L Ratio (Probe 4) |
# of Peptides Identified (Probe 4 |
Kinase Group |
|---|---|---|---|---|---|---|
| BRK | 3 + 4 | 7.7 | 2 | > 28 | 2 | Tyrosine |
| CSK | 3 + 4 | 2.0 | 8 | 8.8 | 3 | Tyrosine |
| EPHA2 | 3 + 4 | 15 | 12 | 5.0 | 27 | Tyrosine |
| EPHB2 | 3 + 4 | 15 | 6 | 35 | 12 | Tyrosine |
| EPHB4 | 3 + 4 | 35 | 6 | 71 | 5 | Tyrosine |
| FAK | 3 + 4 | 2.5 | 1 | 37 | 4 | Tyrosine |
| FRK | 3 + 4 | > 100 | 1 | > 110 | 1 | Tyrosine |
| JNK1 | 3 + 4 | 1.9 | 12 | 6.3 | 2 | CMGC |
| JNK3 | 3 + 4 | 2.5 | 7 | 15 | 4 | CMGC |
| LOK | 3 + 4 | 2.7 | 12 | 11 | 6 | STE |
| PYK2 | 3 + 4 | 2.7 | 59 | 3.9 | 28 | Tyrosine |
| SLK | 3 + 4 | 1.9 | 3 | 27 | 2 | STE |
| smMLCK | 3 + 4 | 1.9 | 6 | 9.2 | 2 | CAMK |
| SRC | 3 + 4 | 14 | 18 | > 32 | 7 | Tyrosine |
| SRM | 3 + 4 | 9.9 | 2 | > 67 | 1 | Tyrosine |
| CDC2 | 3 | 2.1 | 2 | CMGC | ||
| ERK2 | 3 | 19 | 3 | CMGC | ||
| GSK3β | 3 | 3.6 | 4 | CMGC | ||
| p38α | 3 | 5.1 | 4 | CMGC | ||
| PIP4K2C | 3 | 3.9 | 8 | PIP5K2 | ||
| TAO1 | 3 | 2.0 | 3 | STE | ||
| EGFR | 4 | 6.1 | 1 | Tyrosine | ||
| EPHB3 | 4 | > 22 | 1 | Tyrosine | ||
| ERBB4 | 4 | 11 | 3 | Tyrosine | ||
| HGK | 4 | > 17 | 2 | STE | ||
| KHS1 | 4 | 12 | 20 | STE | ||
| MST1 | 4 | 4.6 | 1 | STE | ||
| MST3 | 4 | 2.4 | 1 | STE | ||
| MST4 | 4 | 3.8 | 7 | STE | ||
| PAK2 | 4 | 2.3 | 2 | STE | ||
| PITSLRE | 4 | 15 | 1 | CMGC | ||
| RIPK1 | 4 | 11 | 6 | TKL | ||
| SYK | 4 | > 14 | 1 | Tyrosine | ||
| TNIK | 4 | 3.2 | 1 | STE |
LOK adopts the DFG-out conformation when bound to type II inhibitor 1
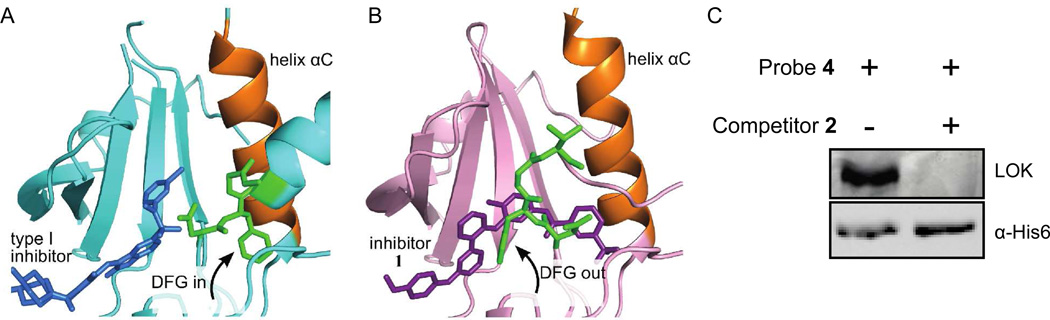
Of the targets that were selectively enriched by our photo-probes, we were particularly interested in the STE kinase LOK [probe 3 (H/L ratio = 2.7); probe 4 (H/L ratio = 11)] because it is distantly related to kinases that have already been characterized to adopt the DFG-out conformation. We have also previously demonstrated that this kinase binds to affinity reagents based on the type II triazolo-pyridine inhibitor scaffold.38 As confirmation of our proteomics results, labeling experiments with the purified catalytic domain of LOK were performed (Figure 4C). As expected, probe 4 was found to efficiently label the active site of LOK. Furthermore, no crosslinking was observed in the presence of an active site competitor. Next, we obtained a crystal structure of a type II ligand (inhibitor 1) bound to the catalytic domain of LOK. Analysis of the LOK-1 complex clearly shows that the ATP-binding site of LOK is in the DFG-out conformation (Figure 4B). The conformation of LOK when bound to inhibitor 1 is strikingly similar to that of the tyrosine kinase SRC bound to a similar triazolo-pyridine inhibitor.39 The molecule sits deep within the catalytic cleft, spanning the adenine and DFG-out pockets. In addition to the two hydrogen bonds that 1 forms with the hinge region of LOK, this inhibitor makes two hydrogen bond contacts that are characteristic of type II ligands. These two hydrogen bonds are both between the amide group of the 3-trifluoromethylbenzamide moiety and LOK: the nitrogen of 1 with Glu-81 in helix-αC (equivalent to Glu-310 in Figure 1A) and the carbonyl of 1 with the backbone of the DFG motif. These results demonstrate that if a kinase is selectively labeled by probes 3 or 4 it is most likely able to adopt the DFG-out conformation.
Figure 4.
The STE kinase LOK adopts the DFG-out conformation. (A) Crystal structure of the catalytic domain of LOK bound to the type I inhibitor SU11274 (PBD 2j7t). The DGF-motif of LOK (shown in green) is in the active conformation. (B) Crystal structure of the LOK-inhibitor 1 complex. The DFG-motif of LOK (shown in green) adopts the DFG-out conformation when bound to inhibitor 1. Inhibitor 1 makes all of the characteristic contacts of type II inhibitors. (B) In vitro labeling of the purified catalytic domain of LOK with probe 4.
In situ activities of probes 3 and 4
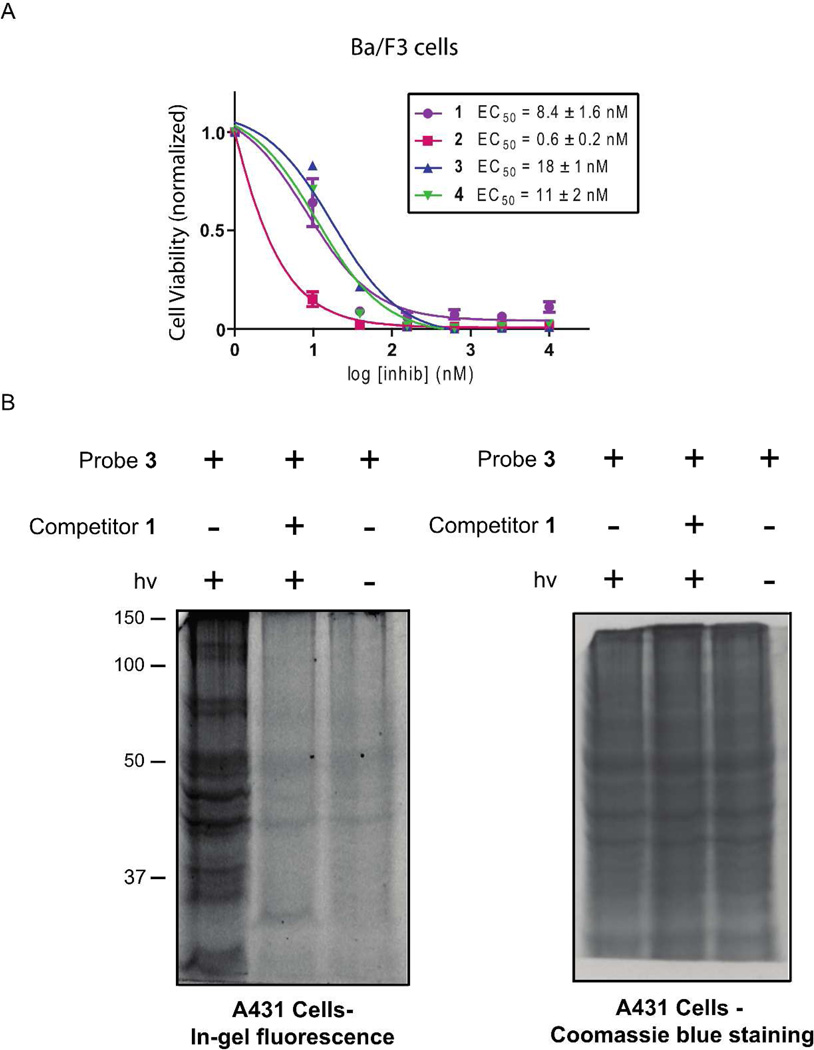
An advantage of using small molecule probes to profile protein kinases is they have the potential to label their targets in situ. To test the cell permeability of photo-crosslinkers 3 and 4, their ability to block the proliferation of Ba/F3 cells stably expressing BCR-ABL was determined with a cell viability assay (Figure 5A). Ba/F3 cells are dependent on the kinase activity of BCR-ABL for survival when grown in the absence of cytokines. Probes 3 and 4 are potent inhibitors of ABL kinase activity in vitro (Figure 1C), and should be able to block the growth of BCR-ABL-expressing Ba/F3 cells if they possess sufficient cell permeability. Indeed, probes 3 (EC50 = 18 nM) and 4 (EC50 = 11 nM) are able to block the proliferation of BCR-ABL-expressing Ba/F3 cells, indicating that they are able to access intra-cellular protein kinase targets (Figure 5A).
Figure 5.
Probes 3 and 4 are cell permeable. (A) The abilities of inhibitors 1–4 (1–10,000 nM) to block the growth of BCR-ABL-dependent Ba/F3 cells were determined with a cell viability assay. The number of viable cells was normalized relative to a DMSO control. (B) In situ labeling of A431 cells with probe 3. A431 cells were incubated with probe 3 (1 µM) in the presence (lane 2) or absence (lane 1) of an active site competitor (10 µM) for 30 minutes and then irradiated with UV light for 7 minutes. Lysate obtained form these cells was tagged with rhodamine-azide, resolved by SDS-PAGE, and labeled proteins were detected with in-gel fluorescence scanning. Lane 3 shows the results of an in situ labeling experiment performed without UV irradiation. A coommassie-blue stained gel showing all of the proteins loaded is shown on the right.
We next determined the abilities of probes 3 and 4 to label protein kinases in situ. To do this, cultured mammalian cells were incubated with probes 3 or 4 for 30 minutes, irradiated with UV light for seven minutes, washed with PBS, and lysed. Labeled proteins in the cell lysate were then conjugated to rhodamine-azide, separated by SDS-PAGE, and visualized using a fluorescent gel scanner. Fluorescent gels for photo-crosslinking experiments performed with probe 3 in A431 cells and K562 cells are shown in Figure 5B and Figure S4, respectively. Consistent with probe 3 possessing the ability to robustly label multiple kinases in situ, a large number of fluorescent bands are observed. This labeling appears to be specific as most of these bands are absent when photo-crosslinking is performed in the presence of competitor 1 (10 µM) or the absence of UV light. Probe 3 was found to consistently provide more specific labeling than probe 4 in situ (data not shown) and was selected for further characterization.
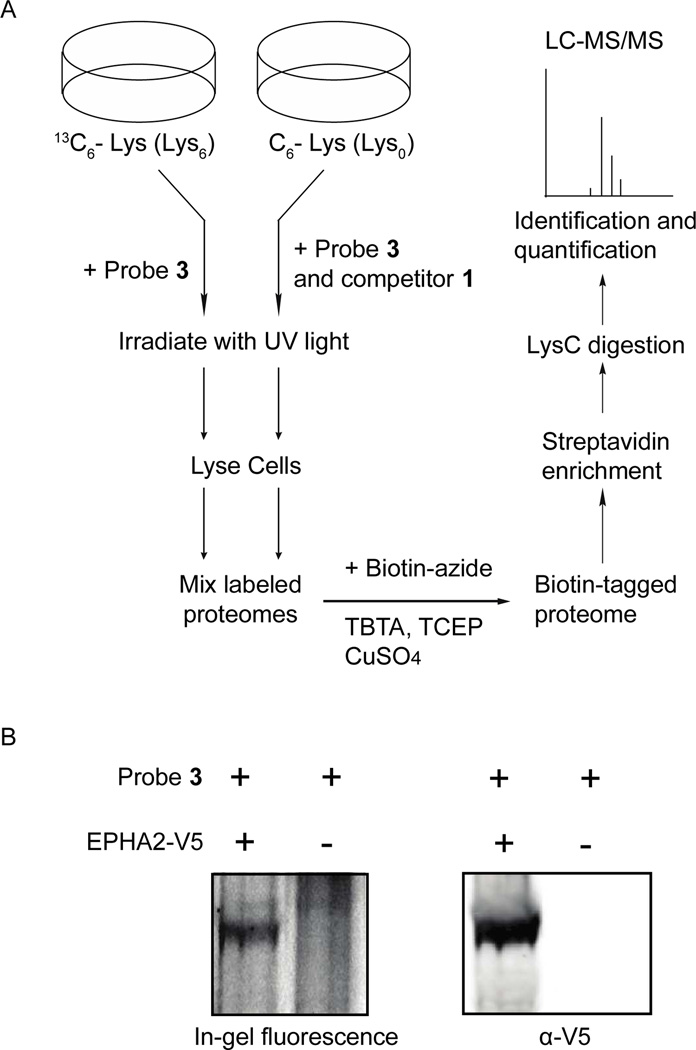
To initially characterize the intra-cellular targets of our photo-crosslinkers, an in situ photo-crosslinking experiment was performed with probe 3 (1 µM) in A431 cells metabolically labeled with heavy Lys (Figure 6A). A parallel experiment was carried out with probe 3 (1 µM) in the presence of competitor 1 (10 µM) with A431 cells grown in light Lys. After photo-crosslinking and cell lysis, equal amounts of heavy and light sample protein were mixed, conjugated to biotin-azide, and enriched with streptavidin-conjugated beads. The beads were subjected to extensive washing, followed by on-bead digestion with LysC, and the eluted peptides were identified using tandem mass spectrometry. In total, 24 proteins were specifically enriched (H/L ratio > 2) by probe 3, with eight of these being protein kinases (Table S3). Notable in situ targets of probe 3 are kinases that have previously been characterized to adopt the DFG-out conformation, p38α, SRC, and YES, and the receptor tyrosine kinases EPHA2, EPHB2, and EPHB3. Interestingly, the EPH receptor tyrosine kinases were labeled with particularly high efficiency and specificity (H/L ratio between 9 and 67), which demonstrates that probe 3 is able to label transmembrane proteins within live cells. To further confirm the specific labeling of EPH receptor tyrosine kinases in situ, cells that have low endogenous levels of these kinases, COS-7, were transiently transfected with a plasmid containing V5-tagged EPHA2. Transfected cells were then subjected to our standard in situ photo-crosslinking conditions. COS-7 cells that were not transfected with V5-tagged EPHA2 were photo-crosslinked using the same conditions. The lysates obtained from both in situ photo-crosslinking experiments were conjugated to rhodamine-azide and separated with SDS-PAGE. A fluorescent band that corresponds to the molecular weight of EPHA2 was observed in COS-7 cells transiently expressing EPHA2, which was absent in the non-transfected cells (Figure 6B and S5 (SI)). The high efficiency in which probes 3 and 4 label receptor tyrosine kinases in situ will facilitate the interrogation of these multi-domain proteins in their native environment. Currently, we are using probe 3 to profile the active site conformational changes that Eph receptor tyrosine kinases undergo upon ligand stimulation.
Figure 6.
Identification of in situ targets of probe 3. (A) A schematic representation of the in situ photo-crosslinking experiments performed with probe 3 in A431 cells. A table listing kinases that were specifically enriched is in the Supporting Information (Table S3, SI). (B) (left panel) EPHA2-V5-transfected and non-transfected COS-7 cells were photo-crosslinked with probe 3 (1 µM). Lysate obtained from both samples was conjugated to rhodamine-azide, resolved by SDS-PAGE, and labeled proteins were detected with in-gel fluorescence scanning. (right panel) Western blot analysis of EPHA2-V5-transfected and non-transfected COS-7 cells (anti-V5 antibody).
Conclusions
Despite widespread interest in protein phosphorylation and the enzymes that catalyze this post-translational modification, many aspects of protein kinase function remain unexplored due to a lack of general profiling tools. Kinase-directed reagents can be used to perform inhibitor selectivity screens and to globally profile the functional states of protein kinase active sites during specific signaling events. Here, we describe two probes, 3 and 4, that are able to label the ATP-binding sites of protein kinases in cell lysates and live cells. To allow covalent modification of protein kinase active sites and visualization/enrichment of labeled proteins, these reagents contain a photo-activatable trifluoromethylphenyl diazirine group and an alkyne tag, respectively. The kinase-targeting elements of probes 3 and 4 are general type II inhibitors that are selective for the DFG-out inactive conformation of protein kinases. Conversion of type II inhibitors into photo-probes does not affect their abilities to interact with kinases that adopt the DFG-out conformation. Furthermore, probes 3 and 4 are able to efficiently photo-label the active sites of the protein kinases that they are inhibitors of. Probes 3 and 4 were used to profile protein kinases in A431 cell lysate. Using these probes in combination with a quantitative mass spectrometry technique, SILAC, allowed the identification of a diverse set of protein kinase targets that were selectively enriched. These targets include several protein kinases that have previously been characterized to adopt the DFG-out conformation including, SRC, p38α, Pyk2, and CSK. Furthermore, a number of protein kinases, particularly in the STE group, that have not been previously characterized to adopt the DFG-out conformation were identified. We obtained a crystal structure of one of these kinases, LOK, bound to a type II inhibitor and found that its ATP-binding site is indeed in the DFG-out conformation. While selective labeling of a particular kinase by one of our probes is not proof that its ATP-binding site can adopt the DFG-out conformation, the assembled list of kinases targets that have been identified are interesting candidates for further characterization. We are in the process of performing structural studies with several of the other novel kinases that are selectively labeled by probes 3 or 4. Beyond lysate profiling experiments, we found that both probes 3 and 4 are cell permeable and able to label a number of protein targets in A431 and K562 cells. Probe 3 was found to selectively label a number of kinases, including p38α, SRC, YES, EPHA2, EPHB2, and EPHB3, in a preliminary quantitative proteomic analysis of its intracellular targets. Current efforts are underway to obtain a full in situ labeling profile of probes 3 and 4. This will not only allow a greater understanding of the conformational state of protein kinase active sites but will also provide information on which kinases can effectively be targeted with type II inhibitors. In addition, by comparing the cell lysate and in situ labeling profiles of probes 3 and 4, a greater understanding of how the local environment of a protein kinase affects its active site conformation will be obtained. Finally, the conformation-selective properties of probes 3 and 4 are being used to monitor the active site conformational changes that specific kinases undergo during signaling events. This will provide a greater understanding of the intra-cellular regulation of this important enzyme family.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the NIH: R01GM086858 (D.J.M) and R01AI089441 (E.A.M.); and the Alfred P. Sloan Foundation (D.J.M.)
Footnotes
Supporting Information Supplementary figures, supplementary tables, experimental methods, and characterization of compounds 1–4. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Cohen P. Nat. Rev. Cell Biol. 2002;4:e127–e130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 2.Brognard J, Hunter T. Curr. Opin. Genet. Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 5.Brehmer D, Greff Z, Godl K, Blencke S, Kurtenbach A, Weber M, Muller S, Klebl B, Cotten M, Keri G, Wissing J, Daub H. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 6.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, Bennett KL, Gao J, Song L, Eschrich S, Superti-Furga G, Koomen J, Haura EB. Nat. Chem. Biol. 2010;l6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach S, Knockaert M, Reinhardt J, Lozach O, Schmitt S, Baratte B, Koken M, Coburn SP, Tang L, Jiang T, Liang DC, Galons H, Dierick JF, Pinna LA, Meggio F, Totzke F, Schachtele C, Lerman AS, Carnero A, Wan Y, Gray N, Meijer L. J. Biol. Chem. 2005;280:31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 9.Rix U, Remsing Rix LL, Terker AS, Fernbach NV, Hantschel O, Planyavsky M, Breitwieser FP, Herrmann H, Colinge J, Bennett KL, Augustin M, Till JH, Heinrich MC, Valent P, Superti-Furga G. Leukemia. 2010;24:44–50. doi: 10.1038/leu.2009.228. [DOI] [PubMed] [Google Scholar]
- 10.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, Muller A, Baumgartner C, Valent P, Augustin M, Till JH, Superti-Furga G. Leukemia. 2009;23:477–485. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 11.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 12.Hantschel O, Rix U, Schmidt U, Burckstummer T, Kneidinger M, Schutze G, Colinge J, Bennett KL, Ellmeier W, Valent P, Superti-Furga G. Proc. Natl. Acad. Sci. USA. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 14.Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Mol. Cell. Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. Proc. Natl. Acad. Sci. USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knockaert M, Gray N, Damiens E, Chang YT, Grellier P, Grant K, Fergusson D, Mottram J, Soete M, Dubremetz JF, Le Roch K, Doerig C, Schultz P, Meijer L. Chem. Biol. 2000;7:411–422. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 17.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 18.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee JD, Gray NS, Kozarich JW. Chem. Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen MS, Hadjivassiliou H, Taunton J. Nat. Chem. Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Zhang CJ, Chen GY, Yao SQ. J. Am. Chem. Soc. 2012;134:3001–3014. doi: 10.1021/ja208518u. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, Cheng X, Sze SK, Yao SQ. Chem. Comm. 2011;47:11306–11308. doi: 10.1039/c1cc14824a. [DOI] [PubMed] [Google Scholar]
- 22.Fischer JJ, Dalhoff C, Schrey AK, Graebner OY, Michaelis S, Andrich K, Glinski M, Kroll F, Sefkow M, Dreger M, Koester H. J. Proteomics. 2011;75:160–168. doi: 10.1016/j.jprot.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Simon GM, Cravatt BF. J. Biol. Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cravatt BF, Wright AT, Kozarich JW. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 25.Evans MJ, Cravatt BF. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 26.Taylor SS, Bubis J, Toner-Webb J, Saraswat LD, First EA, Buechler JA, Knighton DR, Sowadski J. FASEB J. 1988;2:2677–2685. doi: 10.1096/fasebj.2.11.3294077. [DOI] [PubMed] [Google Scholar]
- 27.Hanks SK, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 28.Wong L, Jennings PA, Adams JA. Acc. Chem. Res. 2004;37:304–311. doi: 10.1021/ar020128g. [DOI] [PubMed] [Google Scholar]
- 29.Huse M, Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 30.Eswaran J, Knapp S. Biochim. Biophys. Acta. 2010;1804:429–432. doi: 10.1016/j.bbapap.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jura N,, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Mol.Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 33.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Gray NS. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 35.Kornev AP, Haste NM, Taylor SS, Eyck LF. Proc. Natl. Acad. Sci. USA. 2006;103:17783–1788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuccotto F, Ardini E, Casale E, Angiolini M. J. Med. Chem. 2010;53:2681–2694. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 37.DiMauro EF, Newcomb J, Nunes JJ, Bemis JE, Boucher C, Buchanan JL, Buckner WH, Cee VJ, Chai L, Deak HL, Epstein LF, Faust T, Gallant P, Geuns-Meyer SD, Gore A, Gu Y, Henkle B, Hodous BL, Hsieh F, Huang X, Kim JL, Lee JH, Martin MW, Masse CE, McGowan DC, Metz D, Mohn D, Morgenstern KA, Oliveira-dos-Santos A, Patel VF, Powers D, Rose PE, Schneider S, Tomlinson SA, Tudor YY, Turci SM, Welcher AA, White RD, Zhao H, Zhu L, Zhu X. J. Med. Chem. 2006;49:5671–5786. doi: 10.1021/jm0605482. [DOI] [PubMed] [Google Scholar]
- 38.Ranjitkar P, Brock AM, Maly DJ. Chem. Biol. 2010;17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunner J, Senn H, Richards FM. J. Biol. Chem. 1980;255:3313–3318. [PubMed] [Google Scholar]
- 41.Choi Y, Syeda F, Walker JR, Finerty PJ, Jr, Cuerrier D, Wojciechowski A, Liu Q, Dhe- Paganon S, Gray NS. Bioorg. Med. Chem. Lett. 2009;19:4467–4470. doi: 10.1016/j.bmcl.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dar AC, Lopez MS, Shokat KM. Chem. Biol. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simard JR, Kluter S, Grutter C, Getlik M, Rabiller M, Rode HB, Rauh D. Nat. Chem. Biol. 2009;5:394–396. doi: 10.1038/nchembio.162. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan JE, Holdgate GA, Campbell D, Timms D, Gerhardt S, Breed J, Breeze AL, Bermingham A, Pauptit RA, Norman RA, Embrey KJ, Read J, VanScyoc WS, Ward WH. Biochemistry. 2005;44:16475–16490. doi: 10.1021/bi051714v. [DOI] [PubMed] [Google Scholar]
- 45.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 46.Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, Schwaller J, Sundstrom M, Knapp S. Proc. Natl. Acad. Sci. USA. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP, Treiber DK. Chem. Biol. 2010;17:1241–1249. doi: 10.1016/j.chembiol.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Ong S, Kratchmarova I, Mann M. J. Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.