Abstract
Extracellular adenosine triphosphate (ATP) activates P2X receptors, which are involved in diverse physiological functions. Using a proteomic approach, we identified the neuronal calcium sensor VILIP1 as interacting with P2X2 receptors. We found that VILIP1 forms a signaling complex in vitro and in vivo with P2X2 receptors and regulates P2X2 receptor sensitivity to ATP, peak response, surface expression, and diffusion. VILIP1 constitutively binds to P2X2 receptors and displays enhanced interactions in an activation- and calcium-dependent manner owing to exposure of its binding segment in P2X2 receptors. VILIP1-P2X2 interactions are also enhanced in hippocampal neurons during conditions of action potential firing known to trigger P2X2 receptor activation. Our data thus reveal a previously unrecognized function for the neuronal calcium sensor protein VILIP1 and a mechanism for regulation of ATP-dependent P2X receptor signaling by neuronal calcium sensors.
INTRODUCTION
Purinergic signaling has emerged as a widespread cell sensing mechanism in species from several phyla (1). Released ATP activates P2X receptor ligand-gated cation channels (1). Pharmacology, gene knockout, and knockdown approaches establish that P2X receptor signaling is involved in numerous pathological and physiological processes (1) and is a fundamental feature of cell-cell communication in several species. In particular, P2X2, P2X3, P2X4, and P2X7 subunits are widely expressed and the evidence for a role of these subunits in normal sensory transduction and pain is overwhelming (2–15). P2X receptors define a distinct structural family of ion channels unlike other ionotropic receptors (16). P2X receptor subunits range from 379 (P2X6) to 595 (P2X7) amino acids in length. Each P2X receptor subunit is thought to have two membrane-spanning segments (TM1 and TM2) separated by an extracellular region containing 10 conserved cysteine residues (17, 18). TM2 lines the pore (19–21), and the N and C termini comprising the cytosolic domain contain putative protein interaction–rich sequences that suggest the existence of interacting proteins (22). This topology is the simplest among ionotropic receptors (16, 23). Individual P2X receptors contain only three subunits (24–27). P2X receptor topology and stoichiometry are thus similar to those of acid-sensing channels, although they do not share sequence homology (28). P2X receptor signaling is thought to start when the pore opens. This leads to membrane potential depolarization and calcium entry. The pores of P2X receptors select for calcium ions (29), with the family as a whole showing high calcium flux relative to other transmitter-gated channels (29). Moreover, calcium flux is key to the role of P2X receptors in presynaptic facilitation (30–32), postsynaptic potentiation (33), and muscle contraction (34, 35). However, the possibility that P2X receptors may be regulated by calcium-sensitive proteins has not been fully explored.
Calcium ions serve numerous roles (36, 37). Signaling specificity is achieved through sensor proteins that convert calcium signals into specific effects on cellular proteins and processes (38). Neuronal calcium sensors are an emerging class of calcium-binding proteins (38). These include frequenin, hippocalcin, recoverin, the guanylate cyclase–activating proteins (GCAPs), the Kv channel–interacting proteins (KChiPs), and the visinin-like proteins (VILIPs), which comprise three family members (VILIP1 to 3). VILIP proteins interact with signaling molecules (39–42) and thus are thought to exert physiological and modulatory roles.
We have identified VILIP1 as part of a signaling complex associated with the P2X2 receptor. We show that VILIP1 affects P2X2 function, expression, and mobility and that its interactions increase following receptor activation.
RESULTS
Identification of VILIP1 as part of a P2X2 receptor signaling complex
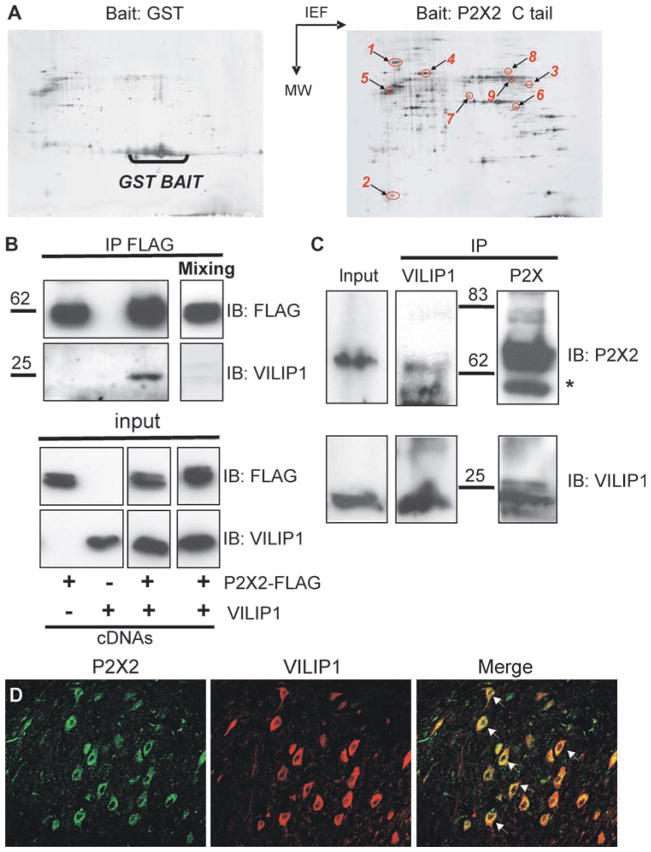
We used the P2X2 receptor C terminal tail domain as bait in glutathione-S-transferase (GST) “pull-down” assays (43) and identified heat shock protein 90 (HSP90), vacuolar-type H+–adenosine triphosphatase (V-ATPase), N-ethylmaleimide–sensitive factor (NSF), tubulin1α, vesicle amine transport protein 1 homolog (VAT1), glutamic acid decarboxylase (GAD), synapsin IIb, glutamine synthase, and VILIP1 as brain proteins interacting with P2X2 receptors (Fig. 1A and table S1). Of these, VILIP1 has overlapping distribution with P2X2 receptors in many areas of the brain (44, 45); moreover, it is regulated by Ca2+ through an EF hand motif (1, 38, 46, 47). P2X2 receptors and VILIP1 coimmunoprecipitated from human embryonic kidney (HEK) cells (Fig. 1B) in a calcium-independent manner (fig. S1) when they were coexpressed, but not when the two protein samples were mixed (Fig. 1B). We interpret this to indicate that the interaction requires coassembly, specific protein conformations that are not rendered by mixing alone, or both. Moreover, native P2X2 receptors and VI-LIP1 coimmunoprecipitated from whole brain (Fig. 1C), and immunocytochemical analysis indicated that native P2X2 receptors and VILIP1 also colocalized in some parts of the brain, such as the deep cerebellar nuclei (Fig. 1D) and dentate gyrus (fig. S2A), but not in others (for instance, cerebellar Purkinje and granule cells; fig. S2B).
Fig. 1.
Identification of P2X2 receptor–interacting proteins. (A) Typical 2D gels are shown. Left: As a control, GST alone was used as bait in the pull-down assay. Right: P2X2 receptor used as bait. Identified proteins indicated are as follows: 1, HSP90; 2, VILIP1; 3, synapsin 2b; 4, vacuolar type H+-ATPase; 5, tubulin α1; 6, glutamine synthase; 7, vesicular amine transporter 1; 8, vesicular fusion protein NSF; 9, glutamate decarboxylase, 65-kDa isoform. (B) VILIP1 coimmunoprecipitation with P2X2 in HEK cells. P2X2-FLAG and VILIP1 plasmids were transfected either alone or in combination in HEK cells and P2X2 was immunoprecipitated with FLAG. The top panel shows immunoblot (IB) of P2X2 and VILIP1 after FLAG immunoprecipitation (IP). The bottom panel show immunoblots of a fraction (10%) of protein extracts before immunoprecipitation (input). “Mixing” indicates that detergent-solubilized extracts from HEK expressing either P2X2 or VILIP1 were mixed before immunoprecipitation. (C) Coimmunoprecipitation of VILIP1 with P2X2 receptor from mouse brain membrane extracts. Solubilized proteins from brain membrane were immunoprecipitated with either anti-P2X2 receptor or anti-VILIP1 antibodies. The star indicates immunoglobulin heavy chains; “Input” represents a fraction (10%) of soluble membrane protein extract that was used in immunoprecipitation. (D) Immunocytochemical analysis of deep cerebellar nuclei. The P2X2 receptor and VILIP1 colocalized in neurons of the deep cerebellar nuclei with 47 ± 6% (n = 12) overlap in area per neuron.
VILIP1 affects P2X2 receptor function
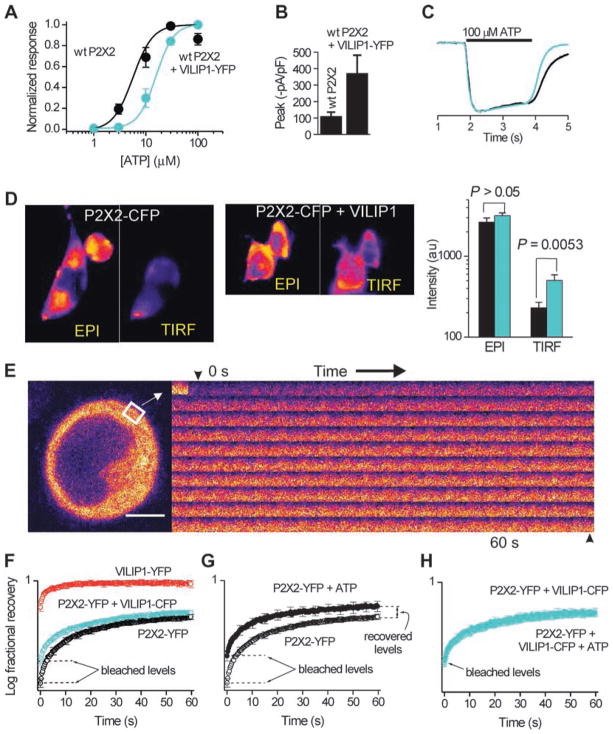
In HEK cells expressing P2X2 receptors and a VILIP1–yellow fluorescent protein (YFP) fusion protein (VILIP1-YFP), the ability of ATP to evoke inward currents measured as the effective concentration (EC50) was significantly different at 15 ± 2 versus 6 ± 1 μM for P2X2 alone (n = 5, the Hill slopes were ~2; Fig. 2A; P = 0.005 by unpaired t test). The effect of VILIP1 on the ATP responses was profound at concentrations of ATP less than 10 μM, similar to those measured on neurons during action potential firing (48) (Fig. 2A). The effect of VILIP1 on P2X2 receptor ATP sensitivity was not abolished by intracellular dialysis with 10 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) to chelate Ca2+ ions (EC50 19 ± 4 μM; n = 7; fig. S3). Similarly, coimmunoprecipitation experiments indicated that the basal interaction of VILIP1 with P2X2 receptors is calcium independent (fig. S1). Coexpression of VILIP1-YFP with P2X2 also increased peak ATP-evoked currents more than 300% (Fig. 2B; P < 0.05; n = 5). However, coexpression of VILIP1 did not affect P2X2 receptor activation kinetics (Fig. 1C), nor did it affect responses evoked by repetitive (fig. S4) or prolonged ATP applications or recovery from desensitization (fig. S5A). The effect of VILIP1 on P2X2 receptors was specific to the extent that other members of the NSC family did not affect P2X2 receptor ATP–evoked inward current responses (EC50 or peak current density) and VILIP1 did not affect ATP-evoked currents at two other P2X receptors (fig. S5, A and B).
Fig. 2.
VILIP1 affects P2X2 receptor function. (A) ATP-evoked inward current dose–response curves for P2X2 receptors (P2X2) with and without VILIP1. Black represents the wild-type (wt) P2X2 receptor alone, and aqua represents the P2X2 receptor expressed concurrently with VILIP1. (B) Peak currents recorded from HEK cells (100 μM ATP) expressing P2X2, with and without VILIP1. (C) Similar time course of normalized current waveforms from cells expressing P2X2 with and without VILIP1. Black represents the wt P2X2 receptor alone, and aqua represents the P2X2 receptor expressed concurrently with VILIP1. (D) Epifluorescence and TIRF images of HEK cells expressing P2X2-CFP with and without VILIP1, as well as average data showing that VILIP1 preferentially increases the P2X2-CFP TIRF signal (n = 14). (E) Representative FRAP images for a small region of interest over time. (F) FRAP curves for cells expressing the indicated constructs. Note that VILIP is bleached less and recovers faster than P2X2-YFP. Moreover, the arrows indicate that VILIP1 reduces the amount of P2X2-YFP receptors that can be bleached and speeds the FRAP recovery time. (G) FRAP curves for P2X2-YFP before and during application of 100 μM ATP. (H) As in (F) but for cells expressing P2X2-YFP and VILIP-CFP. In (F) to (H), the arrows point to the bleached levels.
Using noise analysis (fig. S5, E and F), we found the conductances of P2X2 receptors and P2X2 receptors + VILIP1 were 18 ± 3 and 18 ± 2 pS, respectively (n = 9 and 14), close to the published 19-pS value (49). The fluorescence intensity of cells expressing P2X2 receptor–cyan fluorescent protein (CFP) fusion proteins (P2X2-CFP receptors) and P2X2-CFP receptors + VILIP1 was identical when assayed by epifluorescence microscopy (Fig. 2D) to measure total cellular P2X2-CFP receptor proteins (50). However, plasma membrane expression of P2X2-CFP measured by total internal reflection (TIRF) microscopy was three times higher for cells expressing P2X2-CFP receptors + VILIP1 compared to those expressing P2X2-CFP receptors alone (Fig. 2D). This increase was in the same range as that observed for the change of current densities measured by electrophysiology (Fig. 2B).
We next measured the mobility of P2X2-YFP receptors in the plasma membrane by fluorescence recovery after photobleaching (FRAP; Fig. 2E) (51). The FRAP time constant for P2X2-YFP (τ = 20 ± 1 s; n = 66) was slower than for VILIP1-YFP (τ < 5 s; n = 25), as expected for a cytosol-dominant protein pool compared to one localized mainly to the plasma membrane (51). Upon coexpression of P2X2-YFP with VILIP1-CFP, we found that P2X2-YFP bleached markedly less than in cells expressing P2X2-YFP alone (see arrows in Fig. 2F; τ = 20 ± 1 s; n = 29). Diffusion of P2X2-YFP receptors increased during activation by ATP. Thus, the extent of bleaching was reduced (arrow in Fig. 2G), the τ was faster (16 ± 1 s; n = 35; P < 0.05, t test), and the total recovery was greater (Fig. 2G). When VILIP1 was coexpressed, ATP failed to modulate P2X2-YFP mobility (the graphs overlap in Fig. 2H; P > 0.05), implying that VILIP1 is associated with recruitment of the P2X2 receptor to the plasma membrane and enhanced mobility in this compartment.
Dynamic interactions between P2X2 receptors and VILIP1
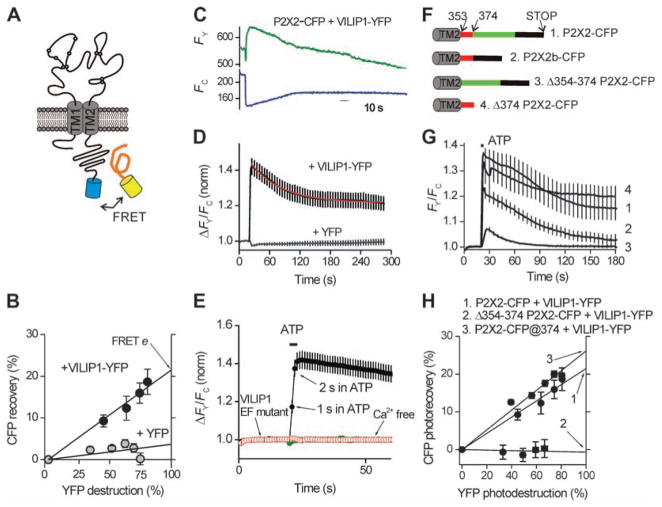
We next used fluorescence resonance energy transfer (FRET) to study real-time interactions between P2X2 receptors and VILIP1 in living cells. We determined FRET efficiency (e) by donor dequenching (52) between P2X2-CFP receptors and VILIP1-YFP (Fig. 3A). P2X2 receptors tolerate CFP at the C tail without altering function (53). We measured robust FRET between P2X2-CFP and VILIP1-YFP at ~22% (Fig. 3B). This was not attenuated in cells loaded with BAPTA-acetoxymethyl (AM) ester and bathed in low–Ca2+ concentration solutions (0.1 mM; fig. S6), implying that VILIP1 binding to P2X2 receptors exists even when intracellular Ca2+ concentrations are buffered, as also shown by coimmunoprecipitation experiments (fig. S1). In contrast, we measured negligible FRET between P2X2-CFP receptors and YFP at ~3% (Fig. 3B), ruling out random nonspecific fluorophore interactions. FRET e falls as ~1/R6 as a function of distance between fluorophores. Thus, the distance between P2X2 receptors and VILIP1 is ~60 Å, i.e., approaching the size of a single P2X2 receptor (54).
Fig. 3.
VILIP1 undergoes FRET with P2X2 receptors and is recruited during receptor activation. (A) The cartoon shows P2X2 receptors with a CFP tag and YFP-tagged VILIP; if the proteins interact, FRET occurs. (B) Progressive photodestruction of YFP (x axis) plotted against photorecovery of CFP (y axis). FRET efficiency is equal to the right-hand y-axis intercept. (C) Representative traces for ATP-evoked ratio-metric changes in fluorescence intensity of VILIP-YFP and P2X2–CFP. (D) Average FRET data from experiments such as those in (B), with cells expressing P2X2-CFP and cytosolic YFP serving as controls (n > 10). Note the ATP-evoked increase in FRET was biphasic. (E) A zoomed-in view of ATP-evoked responses showing that increased FRET occurred within ~2 s of P2X2 receptor activation, was abolished in Ca2+-free buffers, and was not observed with VILIP1 with EF hand mutants that abolish Ca2+ binding. (F) P2X2-CFP deletion constructs. (G) Plots of ATP-evoked increases in FRET for the wild-type (wt) P2X2-CFP and VILIP-YFP and similar measurements for the indicated P2X2-CFP deletion constructs. (H) FRET efficiency determinations for the indicated constructs. The highest FRET efficiency was seen when CFP was inserted near residues K374 of P2X2, and no FRET was seen when the juxtamembrane region between 354 and 374 was deleted.
We next analyzed whether the VILIP1–P2X2 receptor interaction was dynamically modulated by activity and calcium—two key requirements for feedback regulation by a calcium sensor protein. Using dual-emission FRET imaging (48) of VILIP1-YFP and P2X2-CFP, we found that ATP activation of P2X2 receptors (100 μM for 3 s) resulted in increased FRET (measured as FY/FC, the ratio of YFP emission over CFP emission; Fig. 3C) between P2X2-CFP and VILIP1-YFP, as would be expected if VILIP1 bound more tightly to P2X2 receptors. The average data showed that the increase in FRET displayed two kinetic phases, with ~46% of the FRET increase decaying with a τ of 56 s (rate, ~0.02 s−1) and ~54% persisting for >5 min after ATP application (Fig. 3D). The ATP-evoked increase in FRET between P2X2-YFP and VILIP1-CFP was complete in 2 s (rate, ~0.8 s−1; Fig. 3E). Furthermore, the ATP-evoked increase in FRET between P2X2-CFP and VILIP1-YFP required the EF hand motif of VILIP1 and was abolished in Ca2+-free buffers (Fig. 3E), as expected for a calcium sensor (38, 47). However, selectively activating coexpressed P2X4 receptors (48, 55, 56) to increase intracellular Ca2+ did not result in increased FRET between ATP binding–deficient K69A P2X2-CFP receptors (48, 57) and VILIP1-YFP (fig. S7). This suggests that the C tail of P2X2-CFP receptors may undergo a conformational change to allow increased FRETwith VILIP1-YFP. We report data on this possibility in the following sections.
The VILIP1-binding segment in the P2X2 receptor undergoes an ATP-evoked conformational change
We determined where VILIP1 binds to the P2X2 receptor by making a series of P2X2 receptor-CFP deletion constructs and assaying for the ability of ATP to evoke increased FRET between P2X2-CFP and VILIP1-YFP (Fig. 3F). All of the P2X2-CFP receptor deletion constructs shown in Fig. 4F were functionally expressed in the plasma membrane, as assessed by CFP fluorescence, allowing us to measure their FRET with VILIP1-YFP and ATP-evoked changes in FRET (Fig 3G). Thus, the P2X2b splice variant, which lacks a stretch of residues in the middle of the C-terminal tail, underwent robust ATP-evoked FRET with VILIP1-YFP (Fig. 3G). We next truncated the C-terminal tail of P2X2 after K374 (58) to generate P2X2-ΔK374-CFP. This mutant underwent ATP-evoked increases in FRET (Fig. 3F) similar to those of the full-length P2X2 receptor (Fig. 3D), suggesting that most of the C-terminal tail is unnecessary for VILIP1 interactions. We then deleted the region between residue 353 (just intracellular to TM2) and position K374 of the C-terminal tail to generate P2X2-Δ353-374-CFP and found significantly (P < 0.05) reduced ATP-evoked FRET changes (Fig. 3G). Moreover, in this construct we measured negligible FRET e between this deletion and VILIP1-YFP (Fig. 3H). These data suggest that a major locus for the interaction site between the P2X2 receptor and VILIP1 is between residues 353 and 374, in a juxtamembrane region of the P2X2 receptor C-terminal tail. To further explore this possibility, we generated a P2X2 construct in which we left the C-terminal tail intact and inserted CFP just after position 374 (P2X2-CFP@374; Fig. 3F); this receptor was functional with an ATP EC50 of 12 ± 2 μM (n = 7). For a specific set of comparative experiments, FRETe between CFP at position 374 in a full-length P2X2 C-terminal tail and VILIP1-YFP was ~26%. The higher FRET e for P2X2-CFP@374 as compared to that of the C-terminal tail tip (22%; Fig. 3H) suggests that VILIP1 is ~3 to 5 Å closer to the juxtamembrane region than the C-terminal tail tip.
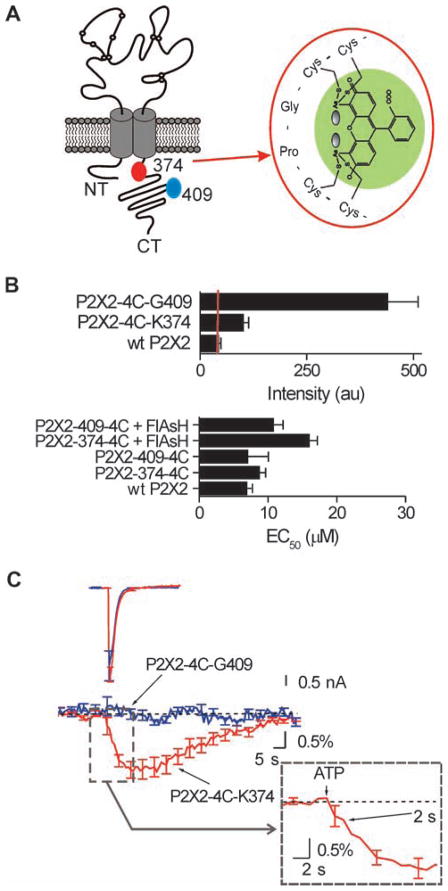
Fig. 4.
The VILIP1 binding segment on P2X2 receptors is exposed during channel opening. (A) The cartoon shows a P2X2 receptor subunit with the location of the sites where 4C tags were inserted. The expanded blow out shows how a FlAsH fluorophore is held in place by the 4C tag. (B) Fluorescence intensity of the FlAsH-labeled constructs shown in (A), compared to that of wt P2X2. We observed meaningful FlAsH staining only in constructs carrying 4C tags. The lower graph shows that 4C-tagged and -labeled P2X2 constructs display ATP EC50 values similar to those of wt P2X2 (P > 0.05) (C) Superimposed ATP-evoked currents (upper left) and FlAsH fluorescence (middle), before during, and after ATP exposure for the indicated constructs. Only the P2X2-4C-K374 construct showed optical signals in response to ATP. The blow out shows the rising phase of the optical signal; a significant change was detected within 2 s of applying ATP.
How does ATP evoke closer interactions between VILIP1 and P2X2 receptors? One possibility is that the VILIP1-binding segment in the juxtamembrane region of the P2X2 receptor undergoes ATP-evoked conformational changes resulting in a closer interaction between VILIP1 and the channel. We tested this idea by using combined patch-clamp and spectroscopy (55). We generated P2X2 constructs carrying tetracysteine (4C) tags in the juxtamembrane region (at K374), as well as at a more distal site in a region not required for VILIP1 binding (at position G409; Fig. 4A). These 4C-tagged P2X2 receptors were expressed alone and labeled with the biarsenical fluorophore FlAsH (Fig. 4B) without altering EC50 values for ATP-evoked inward currents. We next recorded ATP-evoked currents during brief ATP applications (1 s, 100 μM ATP) while recording the emission intensity of FlAsH, which reports conformational rearrangements of the region to which it is bound (55). For P2X2 receptors labeled in the juxtamembrane region with FlAsH we measured decreases in intensity (τ = 5 ± 1 s), whereas for FlAsH in the distal region, there were no changes (Fig. 4C). This is in contrast to the finding that ATP-evoked currents for both constructs were equal in peak amplitude and time course (Fig. 4C), providing direct evidence that neither optical signal is a result of flow of calcium or sodium ions. This is in accord with experiments showing that FlAsH signals are not affected by ion flow (55). These results indicate that ATP evoked a conformational change of the juxtamembrane region of P2X2 receptor. The change in FlAsH intensity was apparent within ~2 s of ATP activation, recalling the time course of the increased FRET between VILIP1-YFP and P2X2-CFP receptors (Fig. 4C, inset) and further supporting the idea that an ATP-evoked conformational change in the P2X2 receptor leads to its closer interaction with VILIP1.
Increased FRET between VILIP1 and P2X2 receptors during endogenous ATP release
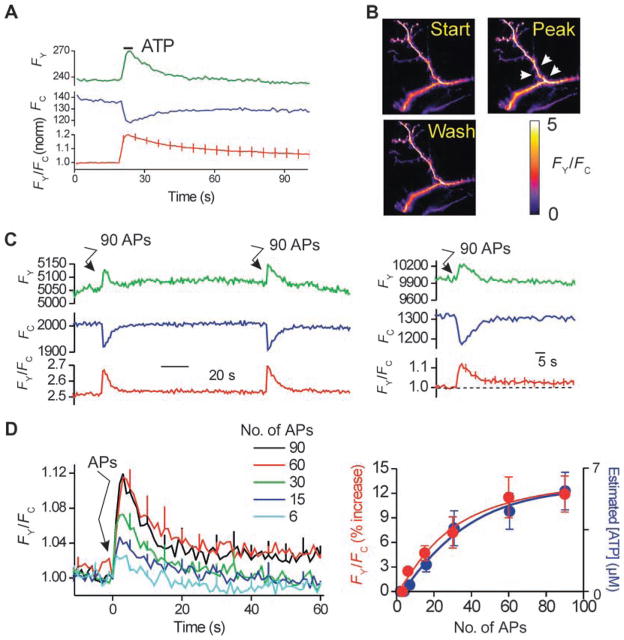
We determined whether VILIP1 undergoes increased FRET with P2X2 receptors when the latter are activated in hippocampal neurons known to express both proteins in vivo (59, 60) and whether VILIP1–P2X2 receptor interactions could be triggered by electrical activity. To this end, we used FRET microscopy of hippocampal dendrites expressing P2X2-CFP and VILIP1-YFP. We locally applied ATP to dendrites and recorded robust increases in FRET between VILIP1 and P2X2 receptors (Fig. 5, A and B). We next used electrical field stimulation under conditions that cause vesicular ATP release and P2X2 receptor activation in hippocampal neurons (48). We found that 90 action potentials (APs; in 3 s) evoked a robust increase in FRET between P2X2-CFP receptors and VILIP1-YFP (Fig. 5C). The action potential–mediated increase in FRET depended on the number of APs in the 3-s period (Fig. 5D). Furthermore, the dependency of AP-evoked VILIP1–P2X2 receptor FRET was similar to AP-evoked ATP release (superimposed in blue in Fig 5D, right panel) (48). These data suggest that action potential firing causes ATP release onto P2X2 receptors and triggers closer VILIP1 interactions in neurons under physiological settings.
Fig. 5.
Increased FRET between VILIP1 and P2X2 receptors during action potential firing in neurons. (A) P2X2-CFP (FC) and VILIP-YFP (FY) intensity over time, as well as the ratio FY/FC from multiple experiments. Note that local ATP application caused increased FRET between P2X2-CFP and VILIP-YFP. (B) Representative FY/FC images of dendrites expressing P2X2-CFP and VILIP-YFP receptors before, during, and after ATP (100 μM; 3 s). (C) Representative (left-hand graph) and average FRET data (right-hand graph) for experiments similar to those in (A), but with 90 APs triggered at the time point indicated. (D) Left-hand graph shows the dependence of FRET increases between P2X2-CFP and VILIP-YFP and action potential number. The right-hand graph shows the increase in FRET between P2X2-CFP and VILIP-YFP versus action potential number, and the amount of ATP released into the buffer as measured in our recent study (48).
DISCUSSION
The main findings of the present study are the identification of components of the P2X2 receptor signaling complex that interact with the C tail of the receptor. Of the nine interacting proteins identified, we studied VILIP1 in-depth and report its effects on several aspects of P2X2 receptor function. Our data reveal the molecular events linking P2X2 receptor activation by exogenous ATP, and during ATP released by action potential firing, to closer interactions between P2X2 receptors and VILIP1. Our data suggest that VILIP1 may modulate P2X2 receptor function through two mechanisms. Through a constitutive interaction, VILIP1 modifies P2X2 receptor sensitivity to ATP, enhances its membrane expression and peak response, and increases its local diffusion in the plasma membrane. In addition, our results demonstrate that VILIP1 interactions with P2X2 receptors increase in an activity-and calcium-dependent manner. Our data suggest a molecular explanation for this observation; on activation, the P2X2 receptor undergoes a conformational change in the proximal C tail region, which carries the VILIP1 binding site. This results in a closer interaction between P2X2 receptors and VILIP1. In addition, we show that calcium entry through P2X2 receptors is also necessary for this interaction, suggesting that after calcium binding in an EF domain-dependent manner, VILIP1 may also undergo a conformational change (38). Altogether, these data indicate that distinct molecular events coordinated by activity are necessary to promote closer VILIP1 interactions with P2X2 receptors. Moreover, because ATP-evoked VILIP1–P2X2 receptor interactions greatly exceeded the duration of the ATP pulse, these data provide evidence for activation-dependent interactions between an accessory protein and a P2X receptor and satisfy a key requirement for a feedback mechanism.
Our experiments do not reveal whether a single trimeric P2X2 receptor binds one, two, or three VILIP1 molecules. Our imaging experiments cannot distinguish between the possibilities that VILIP1 is constitutively bound, with receptor activation leading to conformational changes in both proteins and thereby tighter binding, and the alternative that activation stimulates the binding of more VILIP1 molecules. Indeed, our biochemical data suggest that receptor activation does not likely recruit more VILIP1 to P2X2 receptors because we did not see any change in the coimmunoprecipitation experiments when P2X2 receptors were activated with ATP (fig. S1). We plan to investigate this possibility in the future with molecular and spectroscopic methods pioneered for the interactions between calcium channels and calmodulin (61, 62) and by quantal bleaching analysis of VILIP1-YFP bound to P2X2-CFP receptors (63).
This study provides substantial advances in our understanding of the molecular mechanisms that finely tune ion channel functions. Plasma membrane ion channels are increasingly found to be components of larger signaling complexes consisting of associated molecules (64), including kinases, phosphatases, scaffolding proteins, and regulatory proteins. Such associated proteins may be constitutively bound and thus directly regulate ion-channel function or subcellular localization, or the associations may be enhanced by a stimulus that gates the channel, offering a way to fine-tune its activity. This latter type of regulation is of physiological importance in the case of channels with significant calcium permeability. Calcium entering the cell can trigger conformational change of calcium-binding proteins, which in turn can modulate the channel’s function. Our work shows that the neuronal calcium sensor protein VILIP1 forms a complex with neurotransmitter-gated P2X2 receptors, functions as a channel regulator, and likely binds more tightly to P2X2 receptors in an activation- and calcium-dependent manner.
What might be the pathological and physiological roles for the interaction we have reported? Recent data suggest that VILIP1 is widely expressed and involved in physiological and disease processes throughout the body, including tumor formation (38, 65). In light of the ubiquitous expression of P2X2 receptors (1), these data point to widespread functional interactions between the pair. Furthermore, our experiments provide the framework to probe these interactions in different organs and tissues. Of the areas we have examined to date, the strongest co-localization between P2X2 receptors and VILIP1 was for deep cerebellar neurons. It would thus be interesting to study physiological ATP responses in these neurons under conditions of differing extracellular calcium concentrations and intracellular calcium buffering. Our FRAP data imply that VILIP1 may regulate pre- or postsynaptic responses mediated by P2X2 receptors, for example, by restricting the mobility of P2X2 receptors to and from sites of activation. In future work, it will be interesting to test this hypothesis during ATP synaptic transmission in hippocampal (33, 48) and myenteric synapses (66). Conversely, neuronal calcium sensor proteins modulate G protein–coupled receptor kinase activities in a calcium-dependent manner (67) and VILIP1 modulates cyclic adenosine 3′,5′-monophosphate concentrations. It is thus conceivable that P2X2 receptor interactions with VILIP1 could underlie cross talk between ATP-gated ion channel and G protein–coupled receptor signaling pathways.
This study also defines the real-time molecular physiology of a neuronal P2X2 receptor signaling complex by identifying VILIP1 as a P2X2 receptor-interacting protein in vitro and in vivo. Functional experiments suggest VILIP1 regulates P2X2 receptor ATP sensitivity (Fig. 2A), peak currents (Fig. 2B), surface expression (Fig. 2D), and basal and ATP-evoked mobility (Fig. 2, F to H). Imaging experiments indicate that VILIP1 interactions with P2X2 receptors increase by ATP activation in ~2 s (Fig. 3) and that VILIP1 binds to the juxtamembrane region of the C tail (Fig. 3, F to H), which undergoes an ATP-evoked conformational change. Finally, we provide evidence for enhanced VILIP1–P2X2 interactions during action potential firing (Fig 5).
Overall, the data reveal a previously unknown function for VILIP1 as well as a paradigm of P2X receptor signaling—regulation by neuronal calcium sensors. This substantially adds to the signaling repertoire of this class of ligand-gated ion channels. VILIP1 is known to interact with ligand-gated α4β2 nicotinic (39) and GluR6 kainate receptors (68); however, the mechanisms have remained elusive. This study provides a functional characterization of how VILIP1 modifies an ion channel, provides direct evidence that the interaction is dynamic, and suggests that VILIP1 may function as a general regulator of ligand-gated ion channels.
MATERIALS AND METHODS
Plasmid construction
Plasmid construction methods are included in the Supplementary Materials.
GST pull-down from brain membrane proteins
Synthesis of the recombinant proteins [GST–tobacco etch virus (TEV)–P2X2b] in BL21 cells (GE Healthcare) was induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside for 3 hours at 37°C. Cells were sonicated in a buffer containing 2% sarkosyl, 15 mg lysozyme, 150 mM NaCl, 50 mM tris (pH 7.5), 5 mM dithiothreitol (DTT), and Complete Protease Inhibitor Cocktail (Roche Molecular Biomedical). After sonication, 4% Triton X-100, 10 mM MgSO4, and 2 mM ATP were added to the cell lysate. After sonication, the cell lysate was centrifuged at 8500 rpm for 30 min. Soluble GST-fusion proteins in the supernatant were purified on bulk glutathione-Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions, with the exception of the use of buffer containing 500 mM tris (pH 7.5), 125 mM NaCl, and 5 mM EDTA. After fixation, resin beads were washed in a buffer containing 50 mM tris (pH 7.5), 250 mM NaCl, and 5 mM EDTA. Brain membrane protein extraction and GST pull-down assay was essentially performed as described in (43) except that brain membrane proteins retained on the affinity column were eluted with TEV proteolytic cleavage of the bait (P2X2b C-terminal tail) from the GST protein. Following binding of membrane protein to the GST-Tev-P2X2b bait, recombinant AcTev (Invitrogen) carrying a C-terminal 6xHis tag was added to the bead slurry and incubated for 4 hours at room temperature according to the manufacturer’s condition. Released proteins were collected. TEV protease was removed by affinity purification with QIAexpress Ni-NTA protein purification system (Qiagen) according to the manufacturer’s instruction in the supernatant after centrifugation, precipitated by trichloroacetic acid, and stored at −80°C. Proteins were subsequently identified by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry.
Two-dimensional gel electrophoresis and protein identification by MALDI-TOF mass spectrometry
Proteins were first separated according to their isoelectric point along linear immobilized pH-gradient (IPG) strips (pH 3 to 10, 18 cm long; Amersham Biosciences). Sample loading for the first dimension was performed by passive in-gel reswelling. The IPG strips were then equilibrated for 10 min in a buffer containing 6 M urea, 50 mM tris–HCl (pH 6.8), 30% glycerol, 2% SDS, 10 mg/ml DTT, and bromphenol blue and then equilibrated for 15 min in the same buffer containing 15 mg/ml iodoacetamide instead of the DTT. For the second dimension, the strips were loaded onto vertical 12.5% SDS polyacrylamide gels. Gels were silver stained. For comparison purposes, gels (GST versus P2X2-GST) were always processed and stained in parallel. Gels were scanned with a computer-assisted densitometer (Amersham). Spot detection, gel alignment, background subtraction, and spot quantification were performed with the Melanie 5 software (Amersham); three to four gels were run and analyzed. Protein spots that were not detected in control gels were excised and digested in gel with the use of trypsin (Promega). Digest products were loaded onto the target of an Ultraflex MALDI-TOF mass spectrometer (Bruker-Franzen Analytik) and mixed with the same volume of α-cyano-4-hydroxy-trans-cinnamic acid (10 mg/ml in acetonitrile–trifluoroacetic acid, 50% to 0.1%; Sigma). Analysis was performed in reflectron mode with an accelerating voltage of 20 kV and a delayed extraction of 400 ns. Spectra were analyzed with/by the XTOF software (Bruker-Franzen Analytik) and auto-proteolysis products of trypsin [relative molecular mass (Mr): 842.51, 1045.56, 2211.10] were used as internal calibrates. Identification of proteins was performed with/by the Mascot software previously described. All identified proteins were covered above 30% of their total sequence.
Coimmunoprecipitation of recombinant VILIP1 and P2X receptors
HEK cells were transfected with plasmids encoding VILIP1 and P2X2-FLAG proteins with the use of Lipofectamine2000 according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were scraped in 500 μl of lysis buffer [20 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM EDTA, 1% NP-40 and Complete Protease Inhibitor Cocktail set (Roche Molecular Biomedical)]. After 30 min of solubilization at 4°C under agitation, lysates were centrifuged (16,000g, 10 min, 4°C) and the supernatant was collected. Equal amounts of protein (500 μg) were incubated with 40 μl of anti–FLAG-conjugated agarose beads (Sigma) for 1 hour at 4°C on a rotating wheel. Beads were washed three times (5 min at 4°C) in lysis buffer (as described above except that NaCl was adjusted to 500 mM NaCl) and once with normal lysis buffer. Proteins were eluted by incubation with Laemmli sample buffer and boiled for 5 minutes. Proteins were resolved by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes and visualized with either anti–flag horseradish peroxidase (HRP) or anti VILIP (1/5000) followed by an anti–rabbit HRP antibody and SuperSignal West Pico substrate (Pierce). Control experiments (mixing) were performed by mixing lysate from HEK cells transfected with either VILIP1 or P2X2-FLAG plasmids.
Coimmunoprecipitation of native VILIP1 and P2X2 receptors
Mouse brain was homogenized in ice-cold homogenization buffer [0.32 M sucrose, 10 mM Hepes (pH 7.4), 2 mM EDTA, Complete Proteases Inhibitor Cocktail] with the use of a glass–Teflon Potter homogenizer. Nuclear fraction was removed by centrifugation at 300g for 20 min and the pellet was discarded. The supernatant was centrifuged at 200,000g for 15 min and the resulting membrane pellet was resuspended in Hepes lysis buffer [50 mM Hepes (pH 7.4), 100 mM NaCl, 2 mM EDTA, protease inhibitor, 1% NP-40] and incubated on a rotating wheel for 2 hours. Insoluble proteins were pelleted by centrifugation at 13,000g for 10 min and the protein content of the solubilized membranes was determined with a protein assay kit (Bio-Rad). Solubilized membrane proteins were precleared in solubilization buffer and 25 μl protein A-Sepharose beads for 1 hour on a rotating wheel at 4°C. Beads were pelleted by centrifugation and supernatants were then incubated overnight at 4°C with either rabbit anti-VILIP1 or guinea-pig anti-P2X2 antibodies. Protein A–Sepharose beads (20 μl) were added and the mixtures were incubated for 1 hour at 4°C. Beads were washed three times for 5 min with solubilization buffer containing 500 mM NaCl followed by a final wash in 100 mM NaCl solubilization buffer. Proteins were eluted with Laemmli sample buffer and boiled for 5 min. Immunoprecipitates were detected by Western blot analysis as described above.
Immunocytochemistry
Immunocytochemistry was performed as described (60). Briefly, 3-week-old rat brains were fixed by intracardiac perfusion of freshly made paraformaldehyde (4%). Brains were then transferred into phosphate-buffered saline (PBS) + 30% sucrose solution overnight and kept in PBS+10% sucrose until used. Brains were mounted in optimal cutting temperature medium and snap-frozen on dry ice. A Leica cryostat was used to cut 50-μm sections. Sections were collected in cold PBS. The sections were incubated in blocking solution [10% normal goat serum (NGS, Vector Laboratories), 1% Triton X-100 in PBS] for 1 hour at room temperature (in a 24-well plate, two to three sections per well). Sections were incubated in primary antibodies [anti-P2X2, 1:500 (Neuromics), anti-VILIP1 1:1000 (obtained from D. Lavant)] in PBS containing 5% NGS and 0.1% TX-100 and incubated overnight at 4°C. A control with no primary antibody was made for every experiment. For P2X2 receptor control experiments, blocking peptide of the target epitope was pre-incubated with the antibody for 1 hour at a concentration of 2 mg/ml before use. Sections were washed three times in PBS at room temperature (10, 15, and 30 min) and then transferred into secondary antibody (anti–guinea pig Alexa 488 to detect the anti-P2X2 primary and anti-rabbit Alexa 594 to detect VILIP-1; 1:200, Molecular Probes) in PBS for 4 hours. Sections were washed three times in PBS at room temperature (10, 15, and 30 minutes). Alternatively, peroxidase labeling was performed with a vectastain ABC kit (rabbit immunoglobulin G or guinea pig immunoglobulin G; Vector Laboratories) following the manufacturer’s protocol. Revelation was done with the DAB substrate kit (Vector Laboratories). Sections were then mounted with the use of Vectashield mounting media (Vector Laboratories) and sealed with clear nail polish.
HEK 293 cell culture and transfection
HEK 293 cells (obtained from American Type Culture Collection) were maintained in 75-cm2 cell culture flasks in Dulbecco’s modified Eagle’s medium/F12 media with Glutamax (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Cells were grown in a humidified atmosphere of 95% air/5% CO2 at 37°C in a cell culture incubator. The cells were split 1 in 10 when confluence reached 60% to 90%, which was generally every 3 to 4 days. Cells were prepared for transfection by plating onto six-well plates at the time of splitting, 3 to 4 days before transfection. They were transfected at 60 to 90% confluence. For transient expression in HEK-293, we used 0.5 to 1 μg of plasmid complementary DNA and the Effectene transfection reagent (Qiagen) for each well of a six-well plate. Where appropriate, 100 ng of CFP was used as a marker of transfected cells. The manufacturer’s instructions were followed; the transfection efficiency was 40 to 60%. Cells were gently dispersed and plated on polylysine-coated coverslips (12-mm diameter). Cell density was 20 to 60% so that isolated single cells could be found for imaging and recording. Cells were left to attach for at least 2 hours before labeling.
FlAsH labeling of HEK cells
The FlAsH was bought from Invitrogen and is sold under the name of LumioGreen. We started by preparing a 25 mM 1,2 ethandithiol (EDT; from EMD) solution by mixing 2.1 μl of EDT with 1 ml of dimethyl sulfoxide (Sigma) by gentle vortexing for 1 to 2 s. We then incubated 4 μl of FlAsH with 1 μl of the EDT solution for 10 min at room temperature to ensure there was no unbound FlAsH. We then added 200 μl of 1× Hanks’ balanced salt solution (HBSS)–glucose (bought as a 10× stock from Gibco) to the FlAsH–EDT mixture and waited for another 10 min at room temperature. All of this was done in a fume hood because of the pungent odor of EDT. We transferred the 200 μl of FlAsH labeling solution to 1.8 ml of HBSS-glucose. The solution now contained 4 μM FlAsH and 12.5 μM EDT (final labeling solution). We washed HEK cells twice in 1× HBSS–glucose and added 200 μl of final labeling solution to each coverslip with attached cells, with each coverslip in one well of a 24-well plate. We left this at 37°C for 1 hour. We then washed the cells in HBSS–glucose twice. To lower the background, we incubated the cells in HBSS–glucose + 250 mM EDT for 10 to 30 min. We then washed the cells twice in HBSS–glucose to remove excess EDT and replaced the cells in growth medium until used for recording or imaging. Cells were typically used within 8 hours of labeling. Our labeling protocol is based on one already published (55).
Patch-clamp electrophysiology
HEK 293 cells were used for recordings 24 to 48 hours after transfection as described (53). The extracellular recording solution was composed of (in mM) NaCl 147, KCl 2, MgCl2 1, CaCl2 1, Hepes 10, and glucose 10 (pH 7.4), and the pipette solution contained (in mM) KCl (or CsCl or NaCl) 154, EGTA 11, and Hepes 10. In some cases (as indicated), NaCl was replaced by equimolar amounts of N-methyl-D-glucamine. Whole-cell voltage clamp recordings were made with 3- to 5-megohm borosilicate glass electrodes (WPI), with an Axopatch 200B or 700A amplifier controlled by a computer running pCLAMP 8.1 software via a Digidata 1322A interface (Axon Instruments). Data were filtered at 2 to 5 kHz and digitized at >5 kHz. The chamber housing the glass cover-slip was perfused with extracellular buffer at a rate of 2 to 3 ml/min.
FRET microscopy
For most of the experiments we used an Olympus BX50 microscope equipped with a Peltier cooled (−15°C) Imago charge-coupled device camera (640 × 480 pixels; each pixel 9.9 × 9.9 μm), epifluorescence condenser, hardware control unit and Polychrome IV (from TILL Photonics, Germany). The hardware was controlled by a personal computer, an appropriate frame grabber (TILL Photonics), and macros driven by TILLvisION v3.3 software. The cells were viewed with a 40× water immersion objective lens with a numerical aperture of 0.8 (Olympus). We used the following filters for acquiring CFP or YFP images (from Glen Spectra, Stanmore, UK; order is dichroic, emitter in nanometers): CFP (455DRLP, 480AF30) and YFP (525DRLP, 545AF35). To photo destroy the YFP, we used 525-nm light from the monochromator and a 525 DRLP dichroic. FY/FC changes over time were recorded with a beam splitter (Cairn Optosplit) inserted in the emission path of the microscope.
FRAP microscopy
FRAP was carried out with an Olympus BX61WI and FV300 Fluo-view confocal laser scanning microscope. Bleaching was achieved with 100% laser power for ~1 s followed by examination of recovery every 0.3 s with the laser power at 0.1% of maximum. For these experiments we used the 488-nm laser line of an argon laser and a 40× objective lens with a numerical aperture of 0.8 (Olympus).
Data analysis
For donor dequenching experiments, the FRET efficiency (e) was calculated as e = (1 − [IC-before/IC-after]) × 100, where IC-before is the donor fluorescence intensity before photodestruction and IC-after is the intensity after photo-destruction. The photodestruction of the YFP proceeds with a rate equivalent to the dequenching of the donor, and plotting the photorecovery versus photo destruction yields a linear plot. We used such linear plots and extrapolated to 100% acceptor photodestruction to calculate the maximum donor dequenching for epifluorescence microscopy; e is given by the y-axis intercept. FY/FC traces and movies were made with the use of the Cairn Optosplit Applet in Image J. Concentration response curves were fitted to the Hill equation in Origin 7.5, and charge transfer was measured by integration of the current traces in Clampfit 10.1 or Origin. Single-exponential fits (Origin 7.5) were used to measure the FRAP time constant (τ).
Software and statistical analysis
Electrophysiological analysis was performed with Clampfit 10.1 (Molecular Devices Axon Instruments) or Origin 6.1 or 7.5 (OriginLab Corp) and all statistical tests were run in GraphPad Instat 3.0 (GraphPad Software). Image analysis was performed with ImageJ (NIH). Data are presented as the mean ± SEM from at least five experiments (indicated in the text as n numbers).
Supplementary Material
Fig. S1. The constitutive interaction between P2X2 receptors and VILIP1 is calcium independent.
Fig. S2. Colocalization between P2X2 receptors and VILIP1.
Fig. S3. ATP concentration–response curve for HEK cells expressing P2X2 receptors and VILIP-YFP, and dialyzed with BAPTA.
Fig. S4. Responses evoked by repetitive ATP application.
Fig. S5. Effects of VILIP1 on P2X2 receptor function.
Fig. S6. FRET efficiency determination between P2X2-CFP receptors and VILIP1-YFP in the absence of calcium
Fig. S7. Calcium entry independently of P2X2 receptors does not lead to elevated FRET between P2X2-CFP and VILIP1-YFP.
Table S1. Proteomic analysis of proteins interacting with the carboxyl-terminal cytoplasmic region of the P2X2 receptor.
Acknowledgments
We thank E. Shigetomi and H. Singh for discussions and K.-H. Braunewell and R. Burgoyne for plasmids. This work was supported by Human Frontier Science Program and Stein/Oppenheimer Awards (B.S.K.). E.R. was supported by a Neural Repair Training Grant Predoctoral Fellowship, and E.T. partly by the Fondation francaise pour la Recherche Medicale. F.R. was supported by the CNRS and a grant from Fondation pour la Recherche Médicale. V.C. was supported by Association pour la Recherche sur le Cancer. Proteomic analysis was made possible thanks to the help of the Plateforme de Proteomic Fonctionnelle of Montpellier. G.H. was supported by a James Cook Fellowship–Royal Society of New Zealand. S.C. was supported by a Human Frontier Science Program Grant to B.S.K.
REFERENCES AND NOTES
- 1.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87:659–797. doi: 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- 2.Rong W, Gourine AV, Cockayne DA, Xiang Z, Ford AP, Spyer KM, Burnstock G. Pivotal role of nucleotide P2X2 receptor subunit of the ATP-gated ion channel mediating ventilatory responses to hypoxia. J Neurosci. 2003;23:11315–11321. doi: 10.1523/JNEUROSCI.23-36-11315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- 4.Cockayne DA, Hamilton SG, Zhu QM, Dunn PM, Zhong Y, Novakovic S, Malmberg AB, Cain G, Berson A, Kassotakis L, Hedley L, Lachnit WG, Burnstock G, McMahon SB, Ford AP. Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature. 2000;407:1011–1015. doi: 10.1038/35039519. [DOI] [PubMed] [Google Scholar]
- 5.Cockayne DA, Dunn PM, Zhong Y, Rong W, Hamilton SG, Knight GE, Ruan HZ, Ma B, Yip P, Nunn P, McMahon SB, Burnstock G, Ford AP. P2X2 knockout mice and P2X2/P2X3 double knockout mice reveal a role for the P2X2 receptor subunit in mediating multiple sensory effects of ATP. J Physiol. 2005;567:621–639. doi: 10.1113/jphysiol.2005.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souslova V, Cesare P, Ding Y, Akopian AN, Stanfa L, Suzuki R, Carpenter K, Dickenson A, Boyce S, Hill R, Nebenuis-Oosthuizen D, Smith AJ, Kidd EJ, Wood JN. Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature. 2000;407:1015–1017. doi: 10.1038/35039526. [DOI] [PubMed] [Google Scholar]
- 7.Tsuda M, Shigemoto-Mogami Y, Koizumi S, Mizokoshi A, Kohsaka S, Salter MW, Inoue K. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature. 2003;424:778–783. doi: 10.1038/nature01786. [DOI] [PubMed] [Google Scholar]
- 8.Coull JA, Beggs S, Boudreau D, Boivin D, Tsuda M, Inoue K, Gravel C, Salter MW, De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- 9.Chessell I, Hatcher JP, Bountra C, Michel AD, Hughes JP, Green P, Egerton J, Murfin M, Richardson J, Peck WL, Grahames CB, Casula MA, Yiangou Y, Birch R, Anand P, Buell GN. Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain. 2005;114:386–396. doi: 10.1016/j.pain.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis MF, Burgard EC, McGaraughty S, Honore P, Lynch K, Brennan TJ, Subieta A, Van Biesen T, Cartmell J, Bianchi B, Niforatos W, Kage K, Yu H, Mikusa J, Wismer CT, Zhu CZ, Chu K, Lee CH, Stewart AO, Polakowski J, Cox BF, Kowaluk E, Williams M, Sullivan J, Faltynek C. A-317491, a novel potent and selective non-nucleotide antagonist of P2X3 and P2X2/3 receptors, reduces chronic inflammatory and neuropathic pain in the rat. Proc Natl Acad Sci USA. 2002;99:17179–17184. doi: 10.1073/pnas.252537299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson DW, Gregg RJ, Kort ME, Perez-Medrano A, Voight EA, Wang Y, Grayson G, Namovic MT, Donnelly-Roberts DL, Niforatos W, Honore P, Jarvis MF, Faltynek CR, Carroll WA. Structure–activity relationship studies on a series of novel, substituted 1-benzyl-5-phenyltetrazole P2X7 antagonists. J Med Chem. 2006;49:3659–3666. doi: 10.1021/jm051202e. [DOI] [PubMed] [Google Scholar]
- 12.McGaraughty S, Honore P, Wismer CT, Mikusa J, Zhu CZ, McDonald HA, Bianchi B, Faltynek CR, Jarvis MF. Endogenous opioid mechanisms partially mediate P2X3/P2X2/3-related antinociception in rat models of inflammatory and chemogenic pain but not neuropathic pain. Br J Pharmacol. 2005;146:180–188. doi: 10.1038/sj.bjp.0706346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGaraughty S, Wismer CT, Zhu CZ, Mikusa J, Honore P, Chu KL, Lee CH, Faltynek CR, Jarvis MF. Effects of A-317491, a novel and selective P2X3/P2X2/3 receptor antagonist, on neuropathic, inflammatory and chemogenic nociception following intrathecal and intraplantar administration. Br J Pharmacol. 2003;140:1381–1388. doi: 10.1038/sj.bjp.0705574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North RA. P2X3 receptors and peripheral pain mechanisms. J Physiol. 2003;554:301–308. doi: 10.1113/jphysiol.2003.048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE. The origin of spontaneous activity in the developing auditory system. Nature. 2007;450:50–55. doi: 10.1038/nature06233. [DOI] [PubMed] [Google Scholar]
- 16.North RA. P2X receptors: A third major class of ligand-gated ion channels. Ciba Found Symp. 1996;198:91–105. doi: 10.1002/9780470514900.ch5. [DOI] [PubMed] [Google Scholar]
- 17.Clyne JD, Wang L, Hume RI. Mutational analysis of the conserved cysteines of the rat P2X2 purinoceptor. J Neurosci. 2002;22:3873–3880. doi: 10.1523/JNEUROSCI.22-10-03873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ennion SJ, Evans RJ. Conserved cysteine residues in the extracellular loop of the human P2X(1) receptor form disulfide bonds and are involved in receptor trafficking to the cell surface. Mol Pharmacol. 2002;61:303–311. doi: 10.1124/mol.61.2.303. [DOI] [PubMed] [Google Scholar]
- 19.Rassendren F, Buell G, Newbolt A, North RA, Surprenant A. Identification of amino acid residues contributing to the pore of a P2X receptor. EMBO J. 1997;16:3446–3454. doi: 10.1093/emboj/16.12.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan TM, Haines WR, Voigt MM. A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J Neurosci. 1998;18:2350–2359. doi: 10.1523/JNEUROSCI.18-07-02350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samways DS, Egan TM. Acidic amino acids impart enhanced Ca2+ permeability and flux in two members of the ATP-gated P2X receptor family. J Gen Physiol. 2007;129:245–256. doi: 10.1085/jgp.200609677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaumont S, Khakh BS. Regulation of ATP-gated P2X channel functions by their cytosolic domains. In: Arias Hugo., editor. Biological and Biophysical Aspects of Ligand-Gated Ion Channel Receptor Superfamilies. Research Signpost Ltd; Kerala, India: [Google Scholar]
- 23.Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442:527–532. doi: 10.1038/nature04886. [DOI] [PubMed] [Google Scholar]
- 24.Stoop R, Thomas S, Rassendren F, Kawashima E, Buell G, Surprenant A, North R. Contribution of individual subunits to the multimeric P2X2 receptor: Estimates based on methanethiosulfonate block at T336C. Mol Pharmacol. 1999;56:973–981. doi: 10.1124/mol.56.5.973. [DOI] [PubMed] [Google Scholar]
- 25.Nicke A, Baumert HG, Rettinger J, Eichele A, Lambrecht G, Mutschler E, Schmalzing G. P2X1 and P2X3 receptors form stable trimers: A novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. Trimeric architecture of homomeric P2X(2) and heteromeric P2X(1+2) receptor subtypes. J Mol Biol. 2004;342:333–343. doi: 10.1016/j.jmb.2004.06.092. [DOI] [PubMed] [Google Scholar]
- 27.Barrera NP, Ormond SJ, Henderson RM, Murrell-Lagnado RD, Edwardson JM. Atomic force microscopy imaging demonstrates that P2X2 receptors are trimers but that P2X6 receptor subunits do not oligomerize. J Biol Chem. 2005;280:10759–10765. doi: 10.1074/jbc.M412265200. [DOI] [PubMed] [Google Scholar]
- 28.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 29.Egan TM, Khakh BS. Contribution of calcium ions to P2X channel responses. J Neurosci. 2004;24:3413–3420. doi: 10.1523/JNEUROSCI.5429-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu JG, MacDermott AB. Activation of ATP P2X receptors elicits glutamate release from sensory neuron synapses. Nature. 1997;389:749–753. doi: 10.1038/39639. [DOI] [PubMed] [Google Scholar]
- 31.Khakh BS, Gittermann D, Cockayne DA, Jones A. ATP modulation of excitatory synapses onto interneurons. J Neurosci. 2003;23:7426–7437. doi: 10.1523/JNEUROSCI.23-19-07426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigetomi E, Kato F. Action potential-independent release of glutamate by Ca2+ entry through presynaptic P2X receptors elicits postsynaptic firing in the brainstem autonomic network. J Neurosci. 2004;24:3125–3135. doi: 10.1523/JNEUROSCI.0090-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pankratov YV, Lalo UV, Krishtal OA. Role for P2X receptors in long-term potentiation. J Neurosci. 2002;22:8363–8369. doi: 10.1523/JNEUROSCI.22-19-08363.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamont C, Vainorius E, Wier WG. Purinergic and adrenergic Ca2+ transients during neurogenic contractions of rat mesenteric small arteries. J Physiol (London) 2003;549:801–808. doi: 10.1113/jphysiol.2003.043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamont C, Wier WG. Evoked and spontaneous purinergic junctional Ca2+ transients ( jCaTs) in rat small arteries. Circ Res. 2002;91:454–456. doi: 10.1161/01.res.0000035060.98415.4b. [DOI] [PubMed] [Google Scholar]
- 36.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 37.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Burgoyne RD. Neuronal calcium sensor proteins: Generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin L, Jeanclos EM, Treuil M, Braunewell KH, Gundelfinger ED, Anand R. The calcium sensor protein visinin-like protein-1 modulates the surface expression and agonist sensitivity of the alpha 4beta 2 nicotinic acetylcholine receptor. J Biol Chem. 2002;277:41872–41878. doi: 10.1074/jbc.M206857200. [DOI] [PubMed] [Google Scholar]
- 40.Few AP, Lautermilch NJ, Westenbroek RE, Scheuer T, Catterall WA. Differential regulation of CaV2.1 channels by calcium-binding protein 1 and visinin-like protein-2 requires N-terminal myristoylation. J Neurosci. 2005;25:7071–7080. doi: 10.1523/JNEUROSCI.0452-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lautermilch NJ, Few AP, Scheuer T, Catterall WA. Modulation of CaV2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J Neurosci. 2005;25:7062–7070. doi: 10.1523/JNEUROSCI.0447-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brackmann M, Schuchmann S, Anand R, Braunewell KH. Neuronal Ca2+ sensor protein VILIP-1 affects cGMP signalling of guanylyl cyclase B by regulating clathrin-dependent receptor recycling in hippocampal neurons. J Cell Sci. 2005;118:2495–2505. doi: 10.1242/jcs.02376. [DOI] [PubMed] [Google Scholar]
- 43.Becamel C, Galeotti N, Poncet J, Jouin P, Dumuis A, Bockaert J, Marin P. A proteomic approach based on peptide affinity chromatography, 2-dimensional electrophoresis and mass spectrometry to identify multiprotein complexes interacting with membrane-bound receptors. Biol Proced Online. 2002;4:94–104. doi: 10.1251/bpo39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, Braunewell KH. Expression of the neuronal calcium sensor visinin-like protein-1 in the rat hippocampus. Neuroscience. 2008;153:1202–1212. doi: 10.1016/j.neuroscience.2007.10.067. [DOI] [PubMed] [Google Scholar]
- 46.Spilker C, Braunewell KH. Calcium-myristoyl switch, subcellular localization, and calcium-dependent translocation of the neuronal calcium sensor protein VILIP-3, and comparison with VILIP-1 in hippocampal neurons. Mol Cell Neurosci. 2003;24:766–778. doi: 10.1016/s1044-7431(03)00242-2. [DOI] [PubMed] [Google Scholar]
- 47.Braunewell KH. The darker side of Ca2+ signaling by neuronal Ca2+-sensor proteins: From Alzheimer’s disease to cancer. Trends Pharmacol Sci. 2005;26:345–351. doi: 10.1016/j.tips.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Richler E, Chaumont S, Shigetomi E, Sagasti A, Khakh BS. An approach to image activation of transmitter-gated P2X receptors in vitro and in vivo. Nat Methods. 2008;5:87–93. doi: 10.1038/nmeth1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding S, Sachs F. Single channel properties of P2X2 purinoceptors. J Gen Physiol. 1999;113:695–720. doi: 10.1085/jgp.113.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaiswal JK, Simon SM. Imaging single events at the cell membrane. Nat Chem Biol. 2007;3:92–98. doi: 10.1038/nchembio855. [DOI] [PubMed] [Google Scholar]
- 51.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 52.Nashmi R, Dickinson ME, McKinney S, Jareb M, Labarca C, Fraser SE, Lester HA. Assembly of alpha4beta2 nicotinic acetylcholine receptors assessed with functional fluorescently labeled subunits: Effects of localization, trafficking, and nicotine-induced upregulation in clonal mammalian cells and in cultured midbrain neurons. J Neurosci. 2003;23:11554–11567. doi: 10.1523/JNEUROSCI.23-37-11554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisher JA, Girdler G, Khakh BS. Time resolved measurement of state specific P2X ion channel cytosolic gating motions. J Neurosci. 2004;24:10475–10487. doi: 10.1523/JNEUROSCI.3250-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mio K, Kubo Y, Ogura T, Yamamoto T, Sato C. Visualization of the trimeric P2X2 receptor with a crown-capped extracellular domain. Biochem Biophys Res Commun. 2005;337:998–1005. doi: 10.1016/j.bbrc.2005.09.141. [DOI] [PubMed] [Google Scholar]
- 55.Chaumont S, Khakh BS. Patch-clamp coordinated spectroscopy shows P2X2 receptor permeability dynamics require cytosolic domain rearrangements, but not Panx-1 channels. Proc Natl Acad Sci USA. 2008;105:12063–12068. doi: 10.1073/pnas.0803008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres G, Egan T, Voigt M. Hetero-algometric assembly of P2X receptor subunits. Specificities exist with regard to possible partners. J Biol Chem. 1999;274:6653–6659. doi: 10.1074/jbc.274.10.6653. [DOI] [PubMed] [Google Scholar]
- 57.Jiang LH, Rassendren F, Surprenant A, North RA. Identification of amino acid residues contributing to the ATP binding site of a P2X receptor. J Biol Chem. 2000;275:34190–34196. doi: 10.1074/jbc.M005481200. [DOI] [PubMed] [Google Scholar]
- 58.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein HG, Baumann B, Danes P, Deskman S, Bogart’s B, Gundelfinger ED, Braunewell KH. Regional and cellular distribution of neural visinin-like protein immunoreactivities (VILIP-1 and VILIP-3) in human brain. J Neurocytol. 1999;28:655–662. doi: 10.1023/a:1007056731551. [DOI] [PubMed] [Google Scholar]
- 60.Kanjhan R, Housley GD, Burton LD, Christie DL, Kippenberger A, Thorne PR, Luo L, Ryan AF. Distribution of the P2X2 receptor subunit of the ATP-gated ion channels in the rat central nervous system. J Comp Neurol. 1999;407:11–32. [PubMed] [Google Scholar]
- 61.Mori MX, Erickson MG, Yue DT. Functional stoichiometry and local enrichment of calmodulin interacting with Ca2+ channels. Science. 2004;304:432–435. doi: 10.1126/science.1093490. [DOI] [PubMed] [Google Scholar]
- 62.Dick IE, Tadross MR, Liang H, Tay LH, Yang W, Yue DT. A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature. 2008;451:830–834. doi: 10.1038/nature06529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levitan IB. Signaling protein complexes associated with neuronal ion channels. Nat Neurosci. 2006;9:305–310. doi: 10.1038/nn1647. [DOI] [PubMed] [Google Scholar]
- 65.Fu J, Fong K, Bellacosa A, Ross E, Apostolou S, Bassi DE, Jin F, Zhang J, Cairns P, de Caceres, Braunewell KH, Klein-Szanto AJ. VILIP-1 downregulation in non–small cell lung carcinomas: Mechanisms and prediction of survival. PLoS ONE. 2008;3:e1698. doi: 10.1371/journal.pone.0001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren J, Bian X, DeVries M, Schnegelsberg B, Cockayne DC, Ford AP, Galligan JJ. P2X2 subunits contribute to fast synaptic excitation in myenteric neurons of the mouse small intestine. J Physiol. 2003;552:809–21. doi: 10.1113/jphysiol.2003.047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iacovelli L, Sallese M, Mariggiò S, de Blasi A. Regulation of G-protein–coupled receptor kinase subtypes by calcium sensor proteins. FASEB J. 1999;13:1–8. doi: 10.1096/fasebj.13.1.1. [DOI] [PubMed] [Google Scholar]
- 68.Coussen F, Perrais D, Jaskolski F, Sachidhanandam S, Normand E, Bockaert J, Marin P, Mulle C. Co-assembly of two glur6 kainate receptor splice variants within a functional protein complex. Neuron. 2005;47:555–566. doi: 10.1016/j.neuron.2005.06.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The constitutive interaction between P2X2 receptors and VILIP1 is calcium independent.
Fig. S2. Colocalization between P2X2 receptors and VILIP1.
Fig. S3. ATP concentration–response curve for HEK cells expressing P2X2 receptors and VILIP-YFP, and dialyzed with BAPTA.
Fig. S4. Responses evoked by repetitive ATP application.
Fig. S5. Effects of VILIP1 on P2X2 receptor function.
Fig. S6. FRET efficiency determination between P2X2-CFP receptors and VILIP1-YFP in the absence of calcium
Fig. S7. Calcium entry independently of P2X2 receptors does not lead to elevated FRET between P2X2-CFP and VILIP1-YFP.
Table S1. Proteomic analysis of proteins interacting with the carboxyl-terminal cytoplasmic region of the P2X2 receptor.