SUMMARY
Phosphoinositide 3-kinase (PI3K) and its product phosphatidylinositol(3,4,5)-trisphosphate (PIP3) control cell growth, migration, and other processes by recruiting proteins with pleckstrin homology (PH) domains and possibly other domains to the plasma membrane (PM). However, previous experimental and structural work with PH domains left conflicting evidence about which ones are PIP3 regulated. Here we used live-cell confocal imaging of 130 YFP-conjugated mouse PH domains and found that 20% trans-located to the PM in response to receptor-generated PIP3 production. We developed a recursive-learning algorithm to predict PIP3 regulation of 1200 PH domains from different eukaryotes and validated that it accurately predicts PIP3 regulation. Strikingly, this algorithm showed that PIP3 regulation is specified by amino acids across the PH domain, not just the PIP3-binding pocket, and must have evolved several times independently from PIP3-insensitive ancestral PH domains. Finally, our algorithm and live-cell experiments provide a functional survey of PH domains in different species, showing that PI3K regulation increased from approximately two C. elegans and four Drosophila to 40 vertebrate proteins.
INTRODUCTION
Genetic, biochemical, and pharmacological evidence shows that phosphoinositol 3-kinase (PI3K) is a necessary regulator of processes that range in vertebrates from the control of cell size, growth, proliferation, survival, and secretion to chemotaxis and growth cone extension (Engelman et al., 2006; Vanhaesebroeck et al., 2005). The signature of this signaling pathway is the PI3K-mediated production of phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which has the highest known negative charge among plasma membrane (PM) lipids and which can increase in cells up to 40-fold within seconds of receptor stimulation and subsequent activation of PI3K (Stephens et al., 1993). These unique features of PIP3 make it well suited as a specific and dynamic second messenger linking PI3K to cellular effectors. Given the diverse roles of PIP3, the question has been raised how a single second messenger can regulate so many functions. Identifying which proteins in a particular species are regulated by PIP3 would be a significant step forward in understanding this ubiquitous signaling pathway important in cancer, as well as metabolic and cardiac diseases.
The current mechanistic understanding of PIP3 signaling is based on the finding that the cell-growth regulator Akt (PKB) is activated by PI3K-dependent translocation from the cytoplasm to the PM (for review, see Fayard et al., 2005). PM localization of Akt then leads to its dual phosphorylation and activation. The translocation of Akt to the PM was shown to be mediated by the pleckstrin homology (PH) domain of Akt, and this PH domain was shown to bind PIP3 with high affinity. Further studies showed that the PH domains from the Arf GEFs Cytohesin-1 (PSCD1) and GRP1 (PSCD3) also bind PIP3 (Klarlund et al., 1997), arguing that PH domains can serve more generally as membrane adaptors that recruit proteins to the PM in response to receptor-triggered increases in PIP3 (Hemmings, 1997).
Nevertheless, only a small fraction of the over 250 PH domains in the mammalian genome are thought to bind PIP3, and several PH domains have been shown to bind specifically to other phosphoinositides. For example, the PLCδ-PH domain has been shown to selectively bind to PI(4,5)P2 (Lemmon et al., 1995), and, more recently, the FAPP-PH domain has been shown to selectively bind to PI(4)P (Godi et al., 2004). In addition, many PH domains have been shown to bind phosholipids nonspecifically, weakly, or not at all (Yu et al., 2004).
By building on the in vitro binding results, we showed previously that Akt and PLCδ PH domains can be conjugated with GFP to generate biosensors that can measure changes in specific phosphoinositide lipid concentrations in living cells (Kontos et al., 1998; Stauffer et al., 1998). The success of this strategy of expressing GFP-conjugated PH domains and testing for receptor-triggered PM translocation in the context of a cell was the incentive for the study presented here. Our goals were to obtain a data set of PH domains verified to be PIP3 regulated in a physiological, live-cell context and to then use this data set to develop an algorithm that could predict all PIP3-regulated PH domains in different model organisms.
To achieve these goals, we first expressed 130 YFP-conjugated mouse PH domains and found that 27 of them translocated to the PM following receptor-mediated PIP3 production. Markedly, there was only minimal overall sequence homology among the PIP3-regulated PH domains. We used a recursive-learning approach to develop an algorithm that could predict which PH domains were PIP3 regulated and validated the algorithm by cloning and imaging additional PH domain constructs from human, C. elegans, and Drosophila melanogaster, as well as from S. pombe and S. cerevisiae. We found that the algorithm predicted the experimental results in nearly all cases. Our analysis further suggested that these different PIP3-regulated PH domains evolved in parallel from non-PIP3-regulated PH domains and that the sequence information that best predicts PIP3 regulation is not confined to the structurally known PIP3-binding pocket but is distributed broadly across the PH domain structure. According to the algorithm and our experimental results, there are ~40 PIP3-regulated, PH domain-containing genes in the vertebrate genomes, while Drosophila and C. elegans have only four and two, respectively. Mammalian PIP3-regulated PH domains include signaling adaptor proteins, regulators of small GTPases, kinases, phosphatases, and a motor protein that are predicted to regulate a wide range of cellular processes, suggesting that PIP3 functions as a hub in the cellular signaling network.
RESULTS
Live-Cell Translocation Experiments to Identify PIP3-Regulated PH Domains
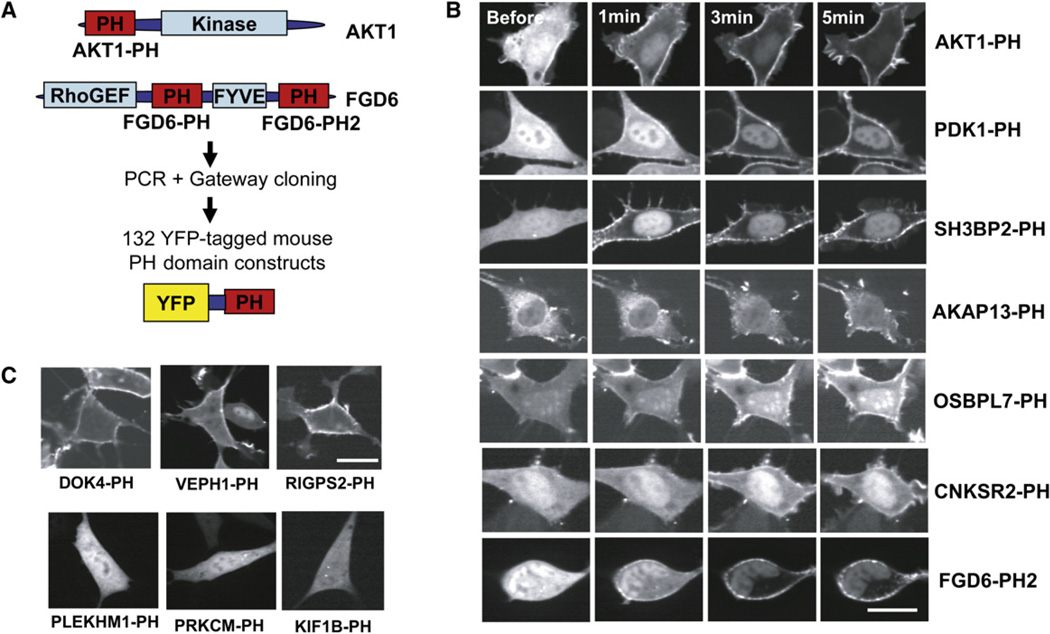
Using confocal imaging, we carried out live-cell localization and translocation studies on 130 YFP-tagged mouse PH domain constructs and two tandem PH domain constructs expressed in NIH 3T3 fibroblasts (Figure 1A; ~40% of predicted mouse PH domains). We stimulated the cells with PDGF to induce robust production of PIP3 at the PM and found that 27 of the single PH domain constructs and both of the tandem PH domain constructs translocated to the PM (see Figure S1 and Table S1 available online). Translocation was assessed by visual inspection and by measuring the receptor-triggered change in cytosolic over PM fluorescence intensity in multiple cells (Table S7).
Figure 1. Genome-wide Live-Cell Imaging Identifies a Subset of Mouse PH Domains Regulated by Receptor Stimulation.
(A) In the initial set of experiments, 132 YFP-tagged mouse PH domain constructs were transfected into NIH 3T3 cells and imaged.
(B) Examples of PH domains that translocate to the PM in response to the addition of PDGF (5 nM final concentration).
(C) Examples of PH domains that are constitutively localized to the PM (top row) and that remain cytosolic even after PDGF receptor stimulation (bottom row). Scale bars are 20 µm.
Figure 1B shows translocation time courses of PH domains from AKT, a well-known PIP3-sensitive growth regulator; from PDK1, which was often thought to be persistently at the PM, but which we now show can translocate in living cells; and from five translocators we identified: the PH domains from the adaptor proteins SH3BP2, CNSKR2, and AKAP13, a RhoGEF FGD6, and a steroid-related protein OSBPL7.
We also found 24 PH domains that were prelocalized to the PM and remained PM localized in response to PDGF stimulation (Figure 1C, top panel). We did not see significant reductions of the PLCδ—or of any other constitutively PM localized—PH domain upon PDGF stimulation, indicating a lower relative activation of PLCγ compared to that of PI3K.
The majority of PH domains were cytosolic or vesicular localized and were not regulated by receptor stimulation (Figure 1C, bottom panel). Basal localization images of all tested mouse PH domains are shown Figure S2, and phenotypes are summarized in Tables S2 and S7.
PM Translocation of PH Domains Is Regulated by PI3K
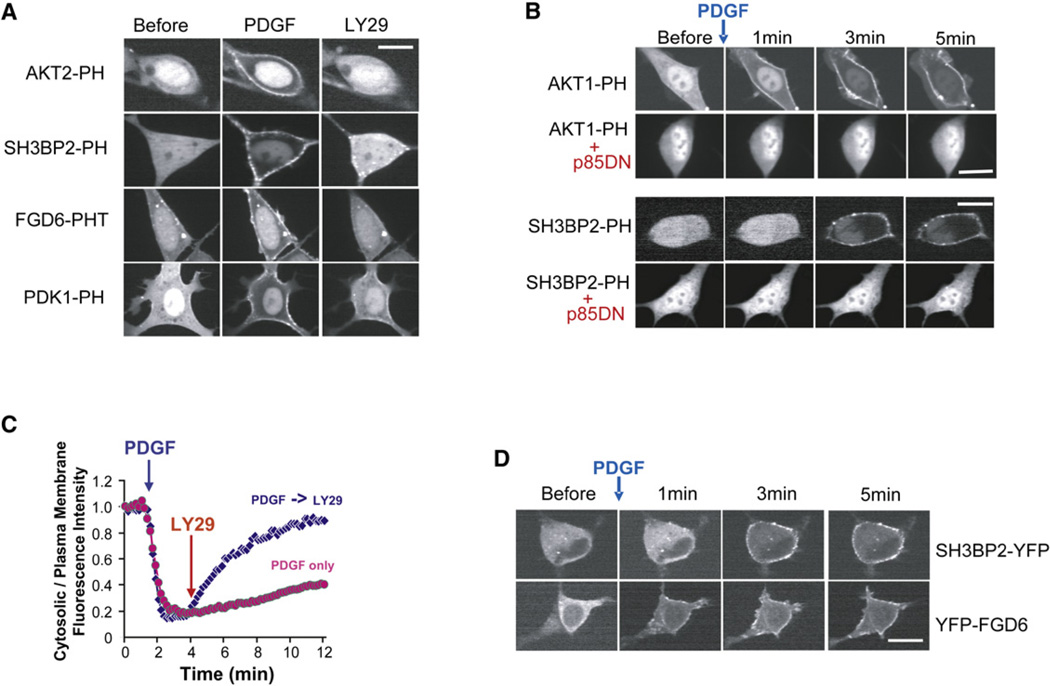
To confirm that the translocation of the PH domains to the PM after PDGF stimulation was due to PI3K activation, we carried out three sets of control experiments. First, we verified that adding the PI3K inhibitor LY294002 dissociated the translocated PH domains from the PM (Figure 2A shows four examples). Second, we showed that pretreatment of cells with 50 µM LY294002 for 3 min before stimulation prevented PDGF-induced PM translocation of the previously translocating PH domains (see Figures S9C and S9F). Third, we coexpressed a version of the p85 regulatory subunit of PI3K that acts as a dominant-negative construct (Dhand et al., 1994) and showed it prevented PDGF-induced PM translocation (see Figure 2B for two examples).
Figure 2. Control Experiments and Characterization of Two Translocators: SH3BP2-PH and FGD6-PH.
(A) Inhibition of PI3K using 50 µM LY294002 reversed the PDGF receptor-triggered PM translocation of PH domains in NIH 3T3 cells. Cells were treated with PDGF for 3 min prior to LY294002 treatment. Images were taken 5 min after the treatment with LY294002. See also Figures S9A and S9B.
(B) Inhibition of PI3K using a dominant-negative PI3K construct (DN-p85).
(C) Time course of SH3BP2-PH domain translocation to the PM in response to sequential addition of PDGF and LY294002. The y-axis is the ratio of cytosolic over PM fluorescence intensity.
(D) PDGF receptor-triggered PM translocation of the PH domain in a protein resulted in translocation of the full-length protein. FGD6 and SH3BP2 full-length proteins were conjugated with YFP. Scale bars, 20 µm.
The results of these three sets of control experiments are consistent with 26 of the translocating PH domains being PIP3 regulated. The one exception was AKAP13-PH, which translocated to the PM as an indirect effect of PIP3-induced peripheral actin polymerization (Figure S3).
For all translocating PH domain constructs tested, the half-maximal dissociation time from the PM was ~100 s following LY294002 addition (Figure 2C shows a time course of the SH3BP2-PH domain). We also confirmed for two of the PIP3-regulated PH domains we identified that translocation of the PH domain translates into translocation of the full-length protein as has been shown previously for AKT and PSCD. Figure 2D shows the receptor-triggered translocation of full-length FGD6 and SH3BP2, which suggests that the translocation of the PH domains can confer their translocation capacity to the full-length proteins.
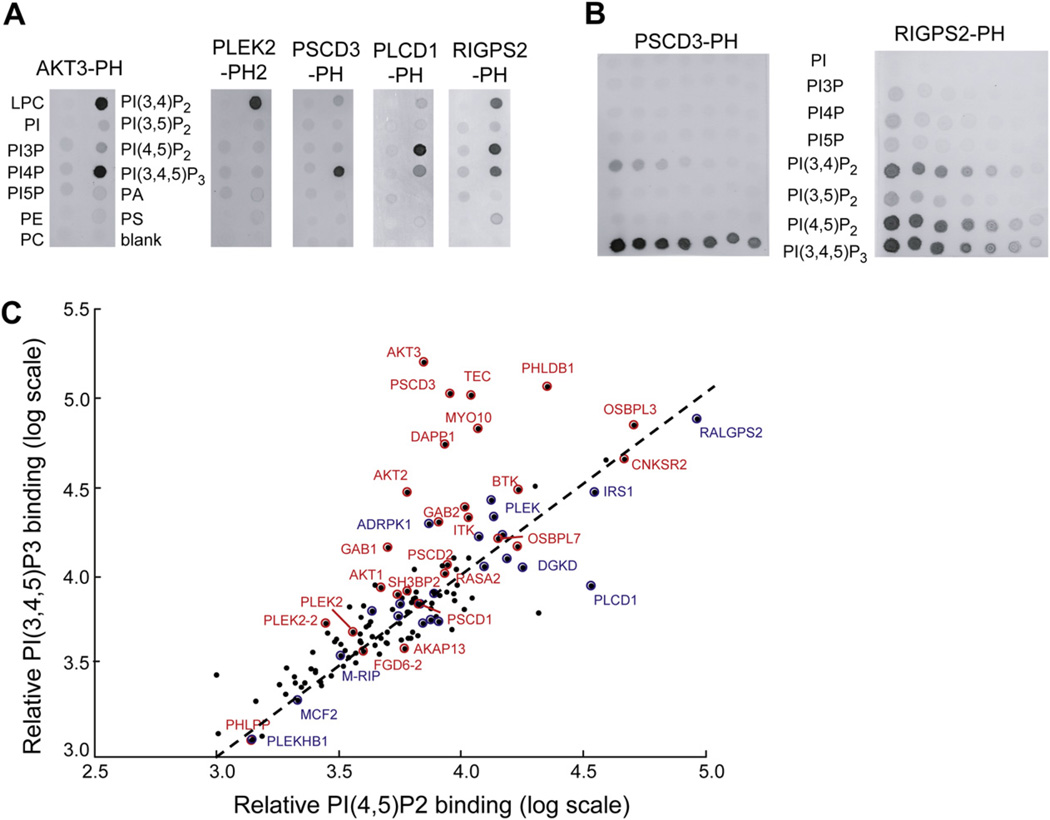
In Vitro PI(4,5)P2 versus PI(3,4,5)P3 Binding Correlated with PM Localization and Translocation
We used lipid blot assays to compare the live-cell translocation and localization data to in vitro binding to different phosphoinositol lipids (Figure 3). Table S2 contains a summary of all measured lipid blot binding interactions. Figure S2 contains bar graphs showing selected lipid blot binding values for all mouse PH domain constructs.
Figure 3. In Vitro Binding Selectivity of PH Domains to PI Lipids and Lipid Controls.
In vitro PI-lipid binding specificity of PH domains. Extracts of cells with overexpressed PH domains were added to PIP strips (A) and lipid arrays (B). (C) Constitutive PM targeting and receptor-triggered PM targeting partially correlate with in vitro binding to PI(4,5)P2 and PI(3,4,5)P3, respectively. PH domains with observed receptor-triggered PM translocation are shown in red. PH domains with constitutive, unregulated PM localization are shown in blue, while PH domains that remained constitutively cytosolic are marked in black without labels. A summary of all measured lipid blot binding interactions can be found in Table S2, and Figure S2 contains bar graphs showing selected lipid blot binding values.
We applied different clustering approaches to the measured binding, localization, and translocation data. Increased PI(4,5)P2 binding correlated with increased PIP3 binding (Figure 3C) and with increased PI(3,4)P2 binding (Figure S10).
In the few cases where the PH domain bound significantly more to PIP3 or PI(3,4)P2 than to PI(4,5)P2 in vitro, the PH domain did translocate (for example, AKT3 and DAPP1). Also, the PH domain from PLCδ1, which is widely used as a biosensor for PI(4,5)P2, stands out as a PI(4,5)P2-specific binder. However, other than these cases, there was no apparent correlation between the in vitro lipid binding and in vivo localization studies, and most of the PM-translocating and persistently PM-localized PH domains bound PIP3 and PI(4,5)P2 to a similar degree.
A few of the PH domains, such as the second PH domain in PLEK2 (PLEK2-PH2), showed PI(3,4)P2- over PI(3,4,5)P3-specific binding. However, we did not observe that the PLEK2-PH2 domain translocation time to the PM was significantly slower than that of more PIP3-specific lipid binders as has been reported in lymphocytes (Allam and Marshall, 2005), suggesting that PI(3,4)P2 and PIP3 serve a similar functional role in inducing translocation for the receptor stimuli and cell type used in our study. As a technical comment, it should also be noted that some of the domains that were reported in the literature to bind PIP3 in vitro or that failed to translocate in our live-cell experiments might be a result of different flanking sequences and other construct properties or may reflect insufficiently strong receptor stimulation to identify weak interactors.
Search for a PIP3-Binding Motif that Predicts the Translocation Data
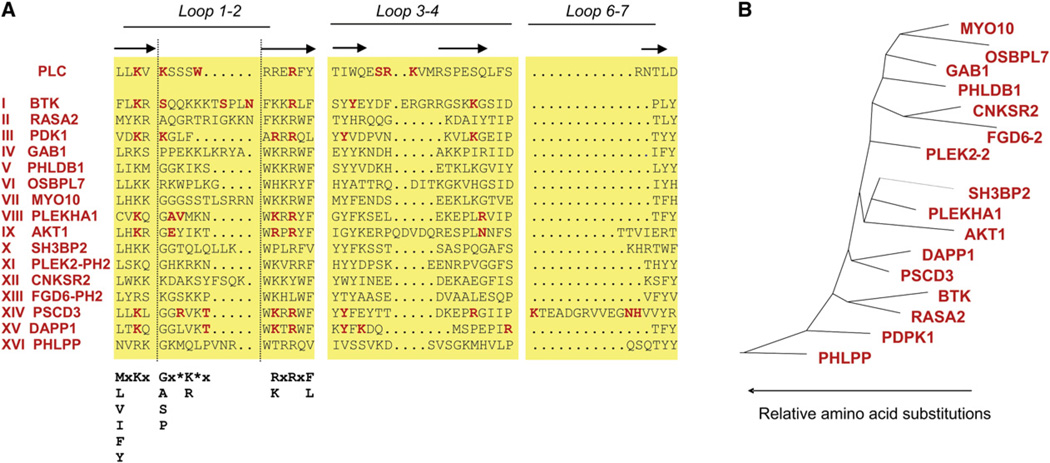
To find a sequence motif for PIP3 regulation, the PH domain sequences first needed to be correctly aligned. Using ClustalW and other multisequence alignment methods that we tried did not result in an alignment consistent with the known structural data, presumably due to the low sequence homology among PH domains (~7%–30%). We used instead a Hidden Markov Model (HMM) for PH domains (Pfam PF00169) and verified that the resulting alignment was consistent with the locations of the variable loops and ends of β strands by comparison to known structures: PDK1 (Komander et al., 2004), PLEKHA1 (Thomas et al., 2001), Akt (Thomas et al., 2002), BTK (Hyvonen and Saraste, 1997), PSCD3 (Ferguson et al., 2000; Lietzke et al., 2000), and DAPP1 (Ferguson et al., 2000).
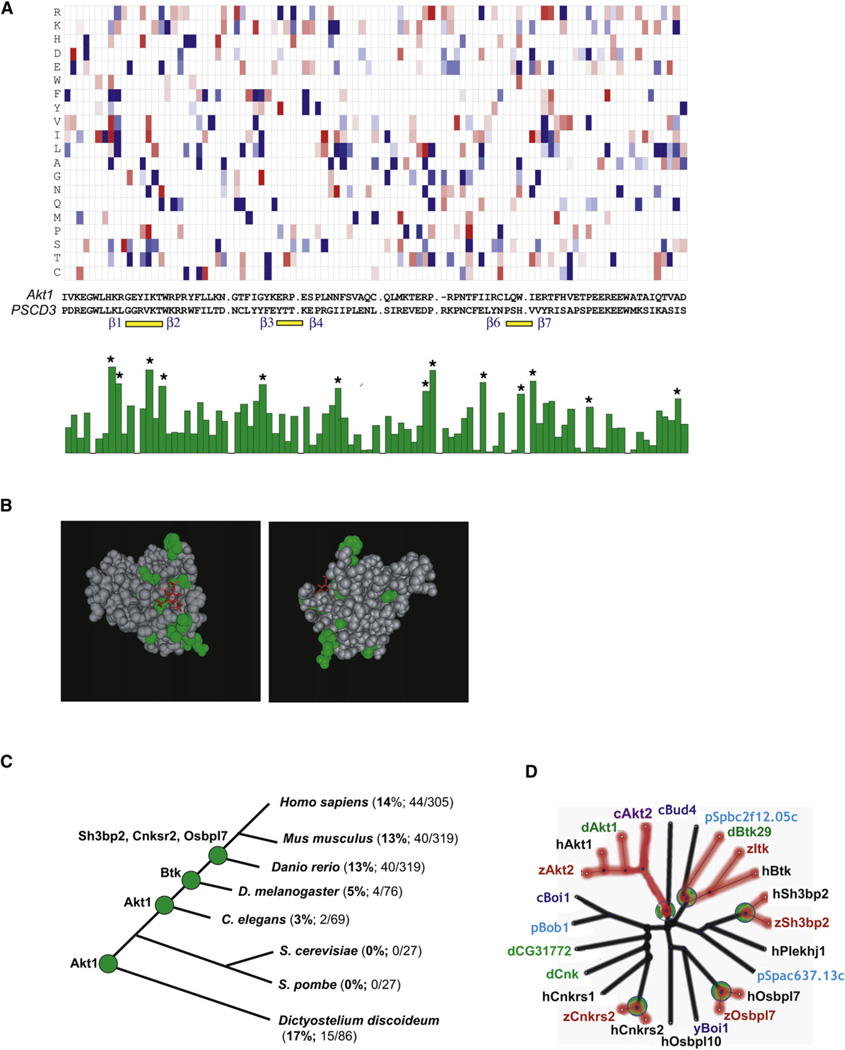
These known PH domain structures show that the loop region between the first and second β strands forms the core of the interaction with the phosphates of the inositol headgroup. For several of the PH domains, secondary interactions occur in the loop region between β strands 3 and 4, as well as in the loop region between β strands 6 and 7. Figure 4A shows an alignment of the three loop regions for selected PIP3-regulated PH domain constructs identified in our study. We highlighted in red the amino acids that have been shown to directly interact with the phosphates of the inositol headgroups. When aligning the 26 identified PIP3-regulated PH domains, we found that they belonged to 16 different subclasses with little sequence homology between them (Figure 4B).
Figure 4. Applying a Previously Published PIP3-Binding Motif to Explain the Translocation Data.
(A) Sequence comparison of the three variable loops between β strands 1–2, 3–4, and 6–7 of the PIP3-regulated PH domains shows only minimal sequence similarity. Amino acids that have been shown in known structures to interact with the 1, 3, 4, and 5 phosphates of PIP3 are highlighted in red. The motif from Isakoff et al. (1998) is shown in bold black type.
(B) Phylogenetic tree with scaled branches showing distinct subclasses of PIP3-binding PH domains.
By building on available structural data, a PIP3-binding motif based on eight criteria was proposed (Isakoff et al., 1998). The main criteria in this motif were the presence of a Lys residue two amino acid positions before the end of the first β strand and in the second β strand a Lys or Arg at the second amino acid position and an Arg in the fourth amino acid position.. When applied to the sequences of the tested mouse PH domains, we found that 10 of the 26 translocating PH domains did not meet the eight criteria (summarized in Table S3). This prompted us to search for a method that is better suited to predict PIP3 regulation.
Development of a Recursive-Learning Strategy to Predict PIP3 Regulation
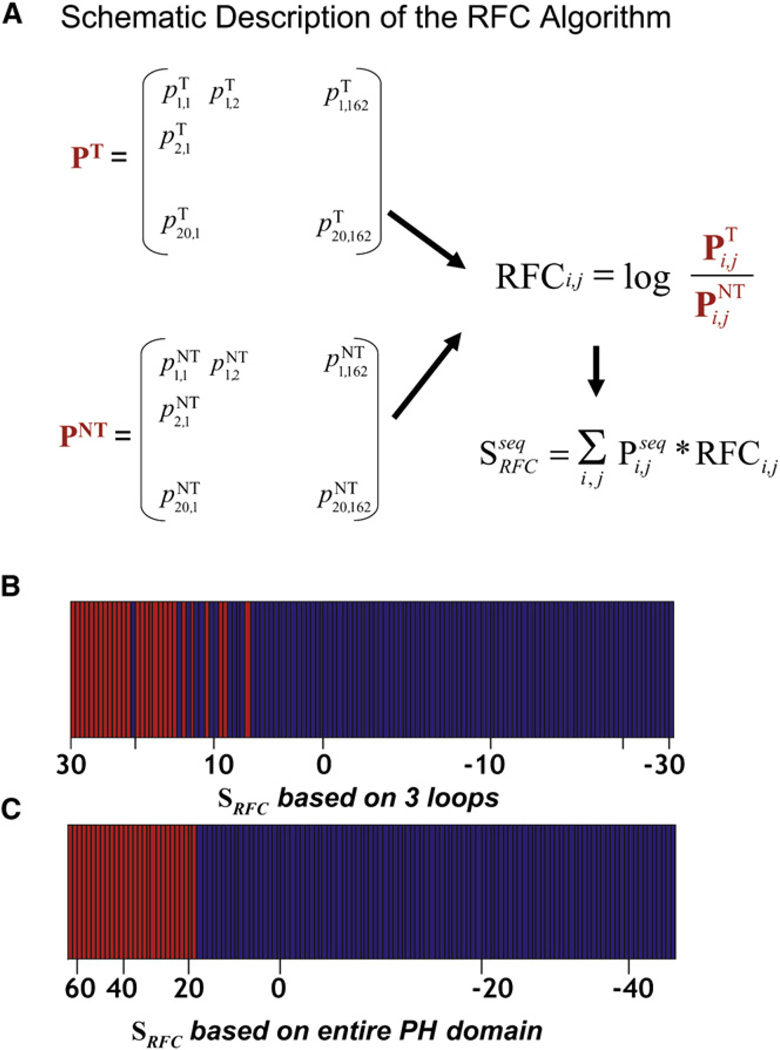
We tested the idea that a probabilistic sequence comparison can distinguish between PIP3-regulated and nonregulated PH domains. The approach that we developed was based on separating the aligned sequences of the tested PH domains into two groups: PIP3-regulated PH domains and non-PIP3-regulated PH domains. For the group of PIP3-regulated PH domains, we calculated a two-dimensional sequence profile matrix, PT. This 162 × 20 matrix (Figure 5A) contained for each of the 162 positions in the aligned sequences the probabilities to find one of the 20 amino acids. The column values at a position summed to 1, unless there was a gap at that position in one or more of the sequences, in which case the column values summed to less than 1. Analogously, we used the aligned sequences of the non-PIP3-regulated PH domains and derived a second sequence profile matrix, PNT, with the same 162 × 20 dimensions.
Figure 5. Development of a Recursive-Learning Strategy for Predicting PIP3 Regulation.
(A) Schematic description of the RFC algorithm.
(B) Restricting the RFC algorithm to the three variable loop regions between the 1–2 β strands, the 3–4 β strands, and the 6–7 β strands showed only partial predictive capabilities. Rank-ordered values are shown (x-axis) for the RFC score of the 130 initially tested mouse PH domains. Translocating PH domains are shown in red, and non-translocating ones are shown in blue. All tested mouse PH domains are used in the bar diagram.
(C) Allowing all amino acid positions in the PH domain to contribute to the RFC score enables exact separation of PIP3-regulated from non-PIP3-regulated PH domains. Same color bar representation as in (B).
We then made the assumption that each sequence position contributes independently to PIP3 regulation. This allowed us to use logarithms in our calculation of an overall score. We defined a recursive functional classification matrix (RFC-matrix) by ratioing the elements in the PT and PNT matrices and taking the logarithm:
In the resulting RFC-matrix, values > 0 indicate positive correlation with PIP3 regulation and values < 0 indicate negative correlation.
This RFC-matrix facilitates the calculation of a predicted PIP3-regulation score, SRFC, for a particular PH domain. SRFC was calculated by first converting the sequence of the PH domain to be scored into a binary sequence profile matrix, Pseq, with the same dimensions as the RFC-matrix. All the values in each column of Pseq are equal to 0, except for the value corresponding to the amino acid in the sequence, which is equal to 1. The PIP3-regulation score for a particular PH domain can then be calculated by element-by-element multiplication of the two matrices, Pseq and RFC, and then summation (Figure 5A). The resulting RFC score was used to predict PIP3 regulation.
We first used this approach to score just the positions in the three variable-loop and flanking β strand regions that have been shown to interact with PIP3 (Figure 5A). While scoring these three loop regions showed an improved predictive value over the motif above for separating the non-PIP3-regulated (blue) from the PIP3-regulated (red) PH domains, there was still not a clean separation (Figure 5B). We then calculated an overall RFC score by allowing all amino acid positions to contribute to the score. Markedly, the RFC score based on the entire PH domain accurately separated PIP3-regulated PH domains from the non-PIP3-regulated ones (Figure 5C).
Validating the RFC Algorithm by Testing PH Domains from Other Species
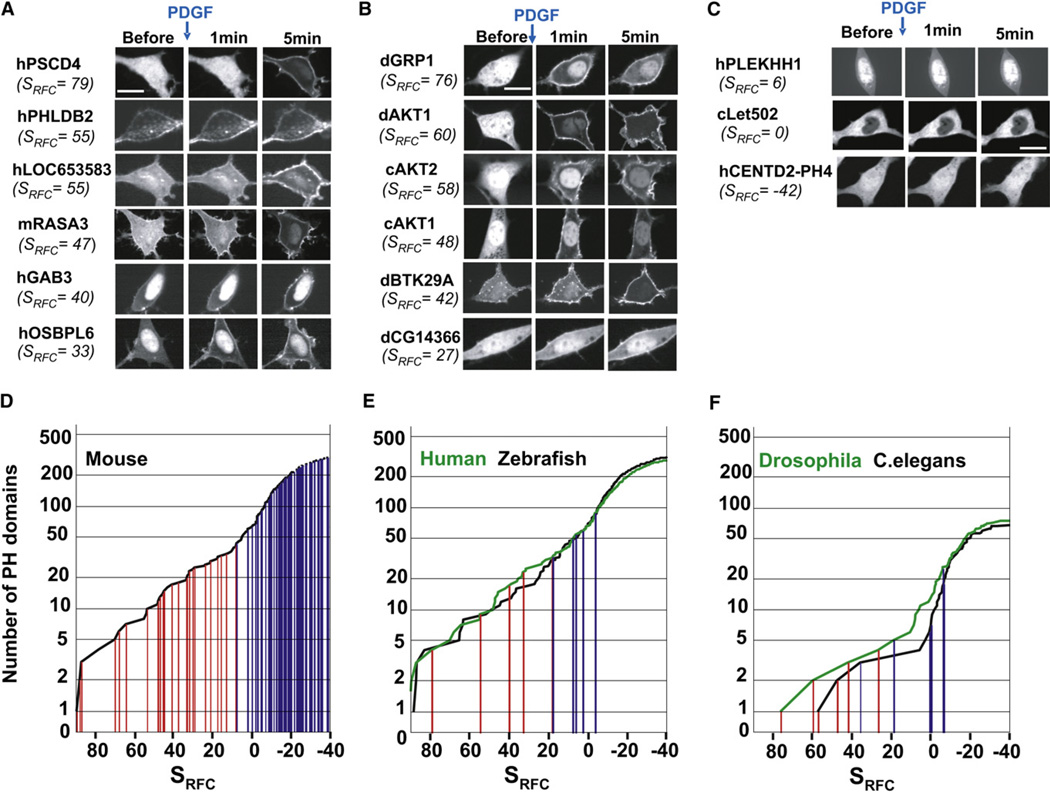
While the RFC algorithm accurately predicted the PIP3 responses of the 130 tested mouse PH domains, a validation of the RFC algorithm required applying it to an independent set of untested PH domains. We used the algorithm to score all PH domains from human, C. elegans, D. melanogaster, S. pombe, and S. cerevisiae, as well as the mouse PH domains we had not tested in our first set of experiments. In a first test of the predictions, we cloned and tested mammalian PH domains that were not homologous to PH domains we had already tested. We successfully cloned 11 human PH domains and two additional mouse domains from the 26 sequences we targeted by PCR. Out of the seven PH domains that had high SRFC scores and were thus predicted to be PIP3 regulated, all seven translocated (Figure 6A and Figure S8). The human PH domains with scores at or below borderline either weakly translocated (Sbf1, SRFC =8) or did not translocate at all (Figure 6C and Figure S8).
Figure 6. Prediction of PIP3-Regulated PH Domains in Other Species.
(A) High-scoring predicted human and mouse PH domains translocated to the PM. (B) High-scoring C. elegans and Drosophila PH domains translocated to the PM. (C) Example of PH domains that scored near or below the cutoff score and did not translocate. Distribution of PIP3-regulated PH domains in mouse (D), human and zebrafish (E), and C. elegans and Drosophila (F). The cumulative representations in (D) and (E) show the PIP3 score of each PH domain in a particular species on the x-axis and the number of PH domains that have a specific PIP3 score or higher on the y-axis. This representation enables one to visualize the distribution of the PIP3 scores and number of PH domains in a particular species. The tested PH domains that were PIP3 regulated are marked in red, and those that were not PIP3 regulated are marked in blue. Table S6 lists all SRFC scores for the different species. Figure S7 shows images of all tested PH-domains used for validation of the RFC algorithm. Figure S8 shows time courses of mammalian translocating PH domains.
PI3K signaling has been studied extensively in Drosopohila and C. elegans, where it has been shown to control cell size, cell number, and aging. Based on the prediction of the RFC algorithm, there were five potential PIP3-regulated PH domains in Drosophila, and three in C. elegans. We tested seven Drosophila PH domains. Out of the four that the RFC algorithm predicted to be PIP3 regulated, all four of them translocated (dAkt1, dGrp1, dBtk29a, and dCG14366; Figure 6B). dCG5004-PH and dCG12467-PH, which had RFC scores in the border region (SRFC = 19 and 8, respectively), and dVap7-PH, which had a very low score (SRFC = −6), all did not translocate.
In C. elegans, we tested six PH domains. Among the three PH domains predicted to be PIP3 regulated, we identified two translocators (cAkt1-PH and cAkt2-PH; Figure 6B) but found that cSec7-PH, which had a high RFC score, did not translocate. The failure of the cSec7 to be PIP3 regulated could be due to missing positive charges in the first loop that are present in the other translocating Sec7 homologs and in translocating PH domains from other species. The low-scoring C. elegans PH domains we tested, cF59A6.5, cObr1, and cLet502, did not translocate. Figure 6C shows examples of low-scoring PH domains that did not translocate, and Table S6 lists all SRFC scores for the different species.
We then used the RFC algorithm to search the S. cerevisiae and S. pombe genomes for PIP3-regulated PH domains. While both yeast species do not have PI3K isoforms, there is a PTEN-related putative PIP3 phosphatase in S. pombe, and it has been suggested that PIP3 might be generated via other pathways (Mitra et al., 2004). Almost all the yeast PH domains scored below the cutoff value for PIP3 regulation, but a few had RFC scores in the boundary region (Figure S4 and Table S6). We cloned and tested 12 of the highest scoring S. pombe and S. cerevisiae PH domains but did not find evidence for PIP3-mediated translocation.
These translocation results from other species were then used to recursively update the RFC-matrix. This allowed for an even more accurate prediction of the PI3K-regulated proteome in the different species. Figures 6D–6F show surveys of the predicted and the tested PIP3-regulated PH domains from C. elegans to mammals (data for S. pombe, S. cerevisiae, and Dictyostelium are shown in Figures S4 and S5). These plots show the cumulative number of PH domains (y-axis) with values higher than a given RFC score (x-axis). The cumulative representation was chosen because it shows the distribution of the RFC scores, as well as the corresponding number of PH domains present in a particular species. Figure 6D shows the numbers of PH domains predicted for mouse, with tested PH domains marked by colored bars: red for PIP3 regulated, and blue for not regulated. As a control, we compared the distribution of SRFC scores for scrambled versions of the tested mouse PH domains to the actual scores (Figure S6). The average SRFC score of the scrambled sequences was −5 with a standard deviation of 9. SRFC scores > 22 become trustworthy because they are three standard deviations from the median of this random noise. This scrambled sequence analysis also defined a boundary region where the PIP3 prediction is ambiguous one to three standard deviations from the median corresponding toSRFC scores between 4 and 22.
Analogously to the survey of the mouse domains in Figure 6D, Figure 6E shows the PH domain predictions for human and zebrafish and Figure 6F shows them for Drosophila and C. elegans. The observed significant match between predicted cutoff scores and experimental results suggests that the algorithm provides a useful framework to predict the PIP3-regulated proteome in different species.
Because PDGF stimulation of NIH 3T3 cells results in robust PIP3 increases at room temperature, most experiments were carried out at room temperature. However, we did confirm that there is no change in translocation phenotype at 37°C (Table S8).
PIP3 Regulation Is Predicted by Amino Acids at Positions across the PH Domain, Not Just the PIP3-Binding Pocket
We next determined which positions in the aligned PH domains contribute most to predicting whether PH domains are PIP3 regulated or not. Figure 7A shows a two-color representation of the RFC-matrix used to score the plots in Figures 6D–6F. The values are positive (red) if a particular amino acid is more frequently found at that position in the PIP3-regulated group of sequences, and negative (blue) if it is found more frequently in the non-PIP3-regulated group. The positions of the three PIP3-interacting loop regions (marked by yellow lines) and the flanking β strands (labeled in blue), as well as the sequences of AKT1 and PSCD3, have been added underneath the plot for orientation.
Figure 7. Structural and Evolutionary Insights into PIP3-Regulated Proteomes Derived from the RFC Algorithm.
(A) Amino acid positions that contribute to PIP3 regulation are distributed across the PH domain, not just the first β strand loop. A two-dimensional plot of the RFC-matrix used to score Figures 6D–6F is plotted at the top. Amino acid positions contributing the most to PIP3 regulation are marked with a star (*) in the bar graph on the bottom.
(B) Amino acid positions marked in (A) with a star were overlaid on the crystal structure of AKT1-PH. The headgroup of P1(3,4,5)P3 is shown in red.
(C) Number and percentage of PIP3-regulated PH domains in different model organisms.
(D) Radial phylogenetic tree with scaled branches showing nonregulated PH domains from different species that are homologs of five PIP3-regulated mammalian PH domains. The first letter before the gene name specifies the species where the gene is from: y, S. cerevisiae; p, S. pombe; c, C. elegans; d, D. melanogaster; z, zebrafish; and h, human. The branches are colored black if the gene is not PIP3 regulated and red if the gene is PIP3 regulated. The green circles mark where PIP3-regulated genes have branched off from a non-PIP3-regulated ancestor.
We determined the relevance of specific amino acid positions for PIP3-regulated PH domains by averaging the contribution from each position. This was done by squaring the RFC-matrix elements and adding the values for the 20 possible amino acids. The resulting averages were plotted in a bar graph (bottom of Figure 7A). The height of each bar is a measure of the relevance of the position for predicting PIP3 responsiveness. The 13 positions that have the maximal effect on PIP3 regulation were marked with stars in Figure 7A and were overlaid on the structure of AKT1 (Figure 7B). These positions are more prominent near the PIP3-binding pocket, but are also scattered inside the structure and at locations far from the interaction site. Together with the lack of sequence homology among the subclasses of PIP3-regulated PH domains (Figures 4A and 4B), this argues that PIP3 regulation of different subclasses is based on amino acids located across the PH domain.
DISCUSSION
Development of an Algorithm to Predict PIP3-Regulated Proteomes
We have developed and validated a recursive functional classification algorithm that we termed RFC, which can be used to predict whether or not a PH domain is regulated by PIP3. This algorithm is based on an experimentally derived probability matrix and the assumption that different amino acid positions in the PH domain independently contribute to PIP3 regulation.
We have shown by live imaging that this strategy leads to accurate predictions about the PIP3 regulation of PH domains in different species, and we have used it to score all PH domains from C. elegans, Drosophila, S. cerevisiae, S. pombe, Dictyostelium, zebrafish, human, and mouse and to define the scope of the PIP3-regulated proteome in these species. Surprisingly, we found Drosophila has, according to our model and experiments, only four PH domains (5%) that are PIP3 sensitive, as opposed to 13% in mammals. Of significance is that Drosophila CG14366 PH domain has no homology to any mammalian PH domain, yet our algorithm was able to identify it as PIP3 regulated. We then confirmed this by cloning and live-cell imaging. Another finding of great interest to the chemotaxis community is that ~15 (17%) of PH domains in Dictyostelium are most likely PIP3 sensitive, which suggests a yet-unexplored collection of PH domains that could be very important in cell motility.
Our study also argues that the accuracy of the predictions can be recursively improved as more live-cell experimental data on PH domains are included in the RFC-matrix. Thus, this RFC approach provides a relatively simple and unbiased probabilistic strategy that can likely be extended to other domains and proteins. The key requirement is a core experimental data set that functionally groups a set of homologous sequences and thereby allows the generation of a predictive RFC-matrix.
Genome-wide Investigation of Phosphoinositide Signaling Based on Live Imaging and In Vitro Lipid Binding
Our study provides a comprehensive investigation of the localization and receptor-triggered translocation of mammalian PH domains and includes a comparison of the binding specificity of PH domains to PI lipids in vitro. A main conclusion from our study is that the lipid blot results could not be used to make definitive predictions about the regulation of PH domains in the cellular context, except in a few cases such as for AKT3 and PSCD3. This argues that the investigation of PIP3-regulated PH domains requires live imaging and measurement of PM translocation in response to physiologically generated PIP3 signals. It should be noted that alternative in vitro assays for phosphoinositide binding could be better suited to determine phosphoinositide binding specificity (for example, Dowler et al., 2002).
In addition to the PH domains that translocate to the PM in a PI3K-dependent manner, we found a similar number of PH domains that were constitutively PM localized and not regulated by growth factors. These are putative PI(4,5)P2 binding PH domains. However, our study found no clear correlation between in vitro binding selectivity for PI(4,5)P2 and membrane localization, as was also observed for PH domains from S. cerevisiae (Yu et al., 2004). Nevertheless, it is likely that improved in vitro binding measurements could further increase the value of direct binding studies.
This raises the question of what the roles are of the remaining ~60% of PH domains that do not translocate and are not constitutively localized to the PM. While they may bind phosphoinositides only weakly in the context of other binding interactions, it is plausible that they have roles in protein-protein interactions as has been suggested for the PH domain from AKAP13 (Figure S3) and several other PH domains (Lemmon, 2007).
Recent in vitro studies show that proteins with polybasic clusters such as K-Ras, MARCKS, and WAVE, as well as a DHR motif present in DOCK180, can also bind PIP3 and trigger at least partial increases in the PM localization of the associated proteins (Cote et al., 2005; Heo et al., 2006). While these might be coregulatory mechanism whereby PIP3 augments a signaling response, it may also point to a broader role of PIP3 in cell signaling beyond the regulation of PH domains. Further studies are needed to understand the physiological importance of interactions between different phosphoinositides and such polybasic regions.
Cell Functions and Processes Controlled by the Mammalian PIP3-Regulated Proteome
Our combined live imaging and prediction strategy shows that ~40 PH domain-containing mammalian gene products are PIP3 regulated. This included isoforms of proteins already known to be PIP3 sensitive, as well as six proteins that we now identified as PIP3 sensitive: FGD6, OSBPL7, SH3BP2, CNKSR2, OSBPL3, and PHLPP. A striking observation was that many of these PIP3-regulated proteins are involved in cytoskeleton restructuring. PHLDB2 has been characterized as a regulator of the actin cytoskeleton that mediates its function by binding filamin (Paranavitane et al., 2007). MYO-10 is a myosin motor that triggers actin polymerization and filopodia formation (Bohil et al., 2006); PLEK2 has been shown to enhance actin polymerization and lamellipodia formation (Hu et al., 1999); while FGD proteins regulate filopodia formation by activating CDC42 (Kim et al., 2004). One of the FGD isoforms has also been linked to a genetic disease, faciogenital dysplasia (Orrico et al., 2000).
A number of the identified PIP3-regulated genes have dual roles in regulation of the actin cytoskeleton as well as in gene expression. These include the adaptor protein SH3BP2 that is involved in c-Abl signaling and has been identified to be mutated in some individuals with cherubism disease (Ueki et al., 2001), as well as the adaptor proteins DAPP1 and TAPP1 that enhance Rac and ERK signaling in immune cells (Allam and Marshall, 2005). The PIP3-sensitive TEC, BTK, and ITK tyrosine kinases also have critical roles in immune cells by regulating prolonged Ca2+ signaling, gene expression, and differentiation (Felices et al., 2007). Finally, the adaptor CNKSR2 regulates RhoA and actin cable formation and also has a role in controlling Srcand Ras-mediated gene expression (Jaffe et al., 2004).
A second class of PIP3-regulated gene products regulates cell size, growth, and differentiation. The best known among them are the three Akt protein kinase isoforms, which have in turn a large number of cell growth-related targets, as well as the upstream kinase PDK1 that provides a coactivating phosphorylation for many protein kinases, including PKA, PKC, and Akt itself. PSCD GAP proteins, as well as GAB adaptor proteins, can also have a role upstream of Akt and can act by enhancing growth-factor receptor signaling, possibly by providing a positive feedback that amplifies PI3K signaling (Eswarakumar et al., 2005). A PIP3-regulated negative feedback is provided by the Akt phosphatase PHLPP (Gao et al., 2005), and a similar negative feedback for Ras is provided by the PIP3-sensitive Ras GAP proteins RASA2 and RASA3 (Bottomley et al., 1998).
A third group of PIP3-regulated proteins is involved in vesicular transport processes and possibly in interorganelle contacts and appears to be important in cell metabolism. The group includes again the four PSCD isoforms, as well as three OSBPL isoforms. PSCD proteins have GDP/GTP exchange activity and exert their role by regulating Arf6-mediated vesicular transport and fusion (Kolanus, 2007). OSBPL proteins likely regulate lipid metabolism and have been shown to have a role in regulating contacts between organelles and the PM (Olkkonen, 2004).
Together, these functions of these PIP3-regulated mammalian genes provide a molecular framework for the role of PIP3 as a hub in the cellular signaling system that controls cell migration, cell growth, size, secretion, and possibly other processes.
Amino Acids that Predict PIP3 Regulation Are Not Restricted to the PIP3-Binding Pocket
As shown in Figures 7A and 7B, the RFC-matrix is useful for determining which regions of the PH domain contribute most to PIP3 regulation. The amino acids near the PIP3-binding pocket make a significant contribution, but so do several locations distant from the binding pocket, arguing that PIP3 regulation stems from multiple sites across the PH domain that can differ in subclasses of PH domains. Such a distributed model is also consistent with our empirical finding that a comparison of the entire PH domain sequence is best suited to predict the PIP3-regulated proteome (Figure 5). This shows a strength of the RFC strategy in that it is unbiased and does not require specific knowledge of the relevant region that contains the functional motif.
Different subclasses of PIP3-regulated PH domains are apparent from sequence comparison (Figures 4A and 4B), as well as from structural data. The observation that PSCD-PH, Akt-PH, and other PH domains are binding the PIP3 headgroup in different orientations (Ferguson et al., 2000; Lietzke et al., 2000; Thomas et al., 2002) suggests that they can place the conjugated protein in different orientations in respect to the plane of the PM and the regulated effectors. This provides one possible functional explanation for the existence of PIP3-sensitivity subclasses of PH domains. Together, this argues that PIP3 regulation is defined by positions across the PH domain and that different subclasses of PH domains engage different positions to generate PIP3 selectivity.
Tracking PIP3-Sensitive PH Domains to Eukaryotic Ancestors
Figure 7C shows the number of predicted PIP3-regulated PH domains and the total number of PH domains in selected model organisms from Dictyostelium to human. The sequencing of the genome of the amoeba Dictyostelium confirmed that Dictyostelium split earlier from the animal kingdom branch than the fungi S. pombe and S. cerevisiae. The lack of PI3K in S. pombe and S. cerevisiae, combined with our confirmation that both lack PIP3-regulated PH domains, argues that at least the PIP3-regulated PH domain of Akt must have existed in a common ancestor of yeast and Dictyostelium. Thus, PIP3-regulated PH domains were eliminated in the S. pombe and S. cerevisiae branches.
The ~15 predicted PIP3-sensitive PH domains in Dictyostelium point to a marked expansion of PIP3-sensitive PH domains, possibly due to the role of this pathway in Dictyostelium in migration and stalk formation (Sun and Firtel, 2003). Among the predicted PIP3-regulated PH domains in Dictyostelium, the well-characterized CRAC PH domain and most other proteins have no homologs in C. elegans, Drosophila, or vertebrates. An independent and even larger expansion of PIP3-regulated PH domains also happened with the emergence of vertebrates. This poses the question of whether the different subclasses of PIP3-sensitive PH domains (Figures 4A and B) can be traced back to a single PIP3-sensitive ancestor (such as Akt-PH) or whether PIP3-regulated PH domains may have evolved more than once from distinct non-PIP3-regulated PH domains.
Given that many PH domains, including those in yeast, mediate constitutive PIP- or PIP2-mediated PM targeting, it is likely that an original role of PH domains was simply to target proteins to the PM by binding polyphosphoinositides such as PIPs or PIP2s. The electrostatic nature of much of this binding interaction makes it conceivable that these PH domains had poor specificity between different PIP, PIP2, or PIP3 lipids. This view is consistent with the previously shown broad range of binding specificities for S. cerevisiae PH domains (Yu et al., 2004). PIP3-dependent PM targeting may have evolved more than once by gradual changes in lipid selectivity that enhanced their relative PIP3 binding over PIP2 and PIP binding and thereby allowed these PH domains to be more responsive to PIP3 signaling inputs.
Support for such a hypothesis comes from the observation shown in Figure 7D, where we compared the respective sequence trees for five PIP3-regulated mammalian PH domains and showed that they have closely related homologs that are not PIP3 regulated. These considerations argue for a parallel evolution model based on increases or decreases in the relative sensitivity of different PH domains for PIP3 versus PIP2 and PIP along their evolutionary path. Such a graded parallel evolution model is further supported by the success of the RFC algorithm because this probabilistic approach assumes that the contribution to PIP3 selectivity stems from multiple independent and small contributions from different sites.
Conclusions
Our study introduces an algorithm to identify PIP3-regulated PH domains and shows that 40 mammalian regulatory gene products associated with actin cytoskeleton restructuring, vesicular transport, cell size, and growth are regulated by PI3K. In contrast, PIP3-regulated proteins appear to be rare in C. elegans and Drosophila and absent in S. pombe and S. cerevisiae. This provides a molecular framework for the function of PIP3 as a hub in the vertebrate signaling system, directly regulating multiple effector proteins and cellular processes. Markedly, our analysis showed that this PIP3 regulation is predicted by amino acids across the PH domain, not just the PIP3-binding pocket, and an evolutionary analysis led to the conclusion that PIP3 regulation of PH domains evolved several times in parallel from an ancestral role as a constitutive PM targeting module.
EXPERIMENTAL PROCEDURES
Constructs
The strategy for cloning the PH domain constructs is described in Zavzavadjian et al. (2007) and on the Alliance for Cellular Signaling website (http://www.signaling-gateway.org/data/plasmid/info.html). Briefly, for each PH domain identified, constructs were generated in Gateway vectors with an EF1 promoter and an N-terminal YFP tag. In most cases, each PH domain construct was generated with a 50 amino acid flanking sequence on each end. Exceptions were made when another protein domain or the end of the protein was present within the 50 amino acid length. In those cases, the construct was terminated at the end of the protein or before the next protein domain. In two cases, tandem PH domain constructs were made that contained two adjacent PH domains.
Cell Culture and Transfection
NIH 3T3 cells were purchased from ATCC. Cells were cultured in DMEM containing 10% FBS in 10% CO2 and split every 3 days by a 1/5 dilution at about 90% confluence. For microscopy, 1.5 × 104 cells were plated in each well of a Lab-Tek 4-well chambered coverglass (Nunc; #155383) coated with poly-D-lysine (0.1 mg/ml) and cultured for 24 hr prior to transfection. Cells were transfected with 0.5 ug/well per construct using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol.
Imaging of PH Domains
NIH 3T3 cells were incubated in serum-free media (DMEM) for 1 hr before imaging. Just prior to imaging, the media was replaced with DPBS (GIBCO #14287). Imaging was carried out at room temperature. For the translocation assays, images were collected every 10 s. An equal volume of 10nMPDGF (PeproTech, Inc.) was added after the fifth image to reach a final concentration of 5 nMPDGF. The NIH 3T3 cell experiments were done on a spinning disc confocal system (PerkinElmer UltraVIEW) attached to an Olympus IX70 inverted microscope with two lasers: a 100mWhelium cadmium laser (442 nm) for visualizing CFP and a 100mWargon laser (514 nm) for visualizing YFP. All images and time courses were taken using an Olympus 40X/1.35 oil objective.
Lipid Binding Assay
Details of the lipid binding assay are given in the Supplemental Data. Briefly, each YFP-PH domain construct was expressed in 293F cells. The cells were lysed, and the cytosolic extract was applied to membrane strips and arrays spotted with different phosphoinositides and control lipids (Echelon Biosciences, Salt Lake City, UT). Antibodies against GFP were used to visualize and quantify the degree of PIP3 domain binding to the respective lipid spots. Spot intensities were measured using Metamorph image processing software (Molecular Devices Corporation, Sunnyvale, CA). For each spot, the local background was subtracted, and the value of lipid binding was measured as the average intensity of the whole spot.
RFC Algorithm
The sequences and information for the PH domains were obtained from the NCBI RefSeq database using 53 PH domain subclasses defined in the NCBI Conserved Domain database. Pfam HMM Model 169 was used to carry out the Hidden Markov Model alignment (PfamWebsite, Sanger, University of Cambridge: http://pfam.sanger.ac.uk/; accession number PF00169; ID, PH).
The sequential steps used to obtain the RFC scores are described in the main text. Adjustments were made to the RFC-matrix to correct for small number fluctuations. Specifically, the sequences were separated into aPIP3-regulated group and a non-PIP3-regulated group. In order for an amino acid to be scored in a position-scoring matrix, it had to be found in at least two sequences in the respective sequence group. Furthermore, due to the limited number of total sequences used, the maximal and minimal values in the RFC-matrix were restricted to −2.5 < log(PT/PNT) < 2.5. This corresponds to an ~12-fold relative enrichment in a particular amino acid between the PIP3-regulated and non-PIP3-regulated group.
The Matlab code for the RFC algorithm can be found in the Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Vitorino, D. Kaplan, and T. Inoue for critical reading of the manuscript. This work was supported by NIH grants GM0653702 and MH064801.
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, ten figures, eight tables, and RFC program and can be found with this article online at http://www.molecule.org/cgi/content/full/30/3/381/DC1/.
REFERENCES
- Allam A, Marshall AJ. Role of the adaptor proteins Bam32, TAPP1 and TAPP2 in lymphocyte activation. Immunol. Lett. 2005;97:7–17. doi: 10.1016/j.imlet.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley JR, Reynolds JS, Lockyer PJ, Cullen PJ. Structural and functional analysis of the putative inositol 1,3,4,5-tetrakisphosphate receptors GAP1(IP4BP) and GAP1(m) Biochem. Biophys. Res. Commun. 1998;250:143–149. doi: 10.1006/bbrc.1998.9179. [DOI] [PubMed] [Google Scholar]
- Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry MJ, Yonezawa K, Kasuga M, Waterfield MD. PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J. 1994;13:511–521. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Kular G, Alessi DR. Protein lipid overlay assay. Sci. STKE. 2002;2002:PL6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphati-dylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fayard E, Tintignac LA, Baudry A, Hemmings BA. Protein kinase B/Akt at a glance. J. Cell Sci. 2005;118:5675–5678. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- Felices M, Falk M, Kosaka Y, Berg LJ. Tec kinases in T cell and mast cell signaling. Adv. Immunol. 2007;93:145–184. doi: 10.1016/S0065-2776(06)93004-1. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Hemmings BA. PtdIns(3,4,5)P3 gets its message across. Science. 1997;277:534. doi: 10.1126/science.277.5325.534. [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MH, Bauman EM, Roll RL, Yeilding N, Abrams CS. Pleckstrin 2, a widely expressed paralog of pleckstrin involved in actin rearrangement. J. Biol. Chem. 1999;274:21515–21518. doi: 10.1074/jbc.274.31.21515. [DOI] [PubMed] [Google Scholar]
- Hyvonen M, Saraste M. Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: molecular explanations for X-linked agamma-globulinaemia. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Aspenstrom P, Hall A. Human CNK1 acts as a scaffold protein, linking Rho and Ras signal transduction pathways. Mol. Cell. Biol. 2004;24:1736–1746. doi: 10.1128/MCB.24.4.1736-1746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Hou P, Gorski JL, Cooper JA. Effect of Fgd1 on cortactin in Arp2/3 complex-mediated actin assembly. Biochemistry. 2004;43:2422–2427. doi: 10.1021/bi036173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling byphosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- Kolanus W. Guanine nucleotide exchange factors of the cytohesin family and their roles in signal transduction. Immunol. Rev. 2007;218:102–113. doi: 10.1111/j.1600-065X.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter Downes C, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos CD, Stauffer TP, Yang WP, York JD, Huang L, Blanar MA, Meyer T, Peters KG. Tyrosine 1101 of Tie2 is the major site of association of p85 and is required for activation of phosphatidylinositol 3-kinase and Akt. Mol. Cell. Biol. 1998;18:4131–4140. doi: 10.1128/mcb.18.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- Mitra P, Zhang Y, Rameh LE, Ivshina MP, McCollum D, Nunnari JJ, Hendricks GM, Kerr ML, Field SJ, Cantley LC, Ross AH. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 2004;166:205–211. doi: 10.1083/jcb.200404150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen VM. Oxysterol binding protein and its homologues: new regulatory factors involved in lipid metabolism. Curr. Opin. Lipidol. 2004;15:321–327. doi: 10.1097/00041433-200406000-00013. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Falciani M, Bracci M, Cavaliere ML, Rinaldi MM, Musacchio A, Sorrentino V. A mutation in the pleckstrin homology (PH) domain of the FGD1 gene in an Italian family with faciogenital dysplasia (Aarskog-Scott syndrome) FEBS Lett. 2000;478:216–220. doi: 10.1016/s0014-5793(00)01857-3. [DOI] [PubMed] [Google Scholar]
- Paranavitane V, Stephens LR, Hawkins PT. Structural determinants of LL5beta subcellular localisation and association with filamin C. Cell. Signal. 2007;19:817–824. doi: 10.1016/j.cellsig.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasmamembrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Jackson TR, Hawkins PT. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim. Biophys. Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- Sun B, Firtel RA. A regulator of G protein signaling-containing kinase is important for chemotaxis and multicellular development in dictyostelium. Mol. Biol. Cell. 2003;14:1727–1743. doi: 10.1091/mbc.E02-08-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Dowler S, Deak M, Alessi DR, van Aalten DM. Crystal structure of the phosphatidylinositol 3,4-bisphosphate-binding pleckstrin homology (PH) domain of tandem PH-domain-containing protein 1 (TAPP1): molecular basis of lipid specificity. Biochem J. 2001;358:287–294. doi: 10.1042/0264-6021:3580287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, Ninomiya C, doAmaral C, Peters H, Habal M, et al .Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism. Nat. Genet. 2001;28:125–126. doi: 10.1038/88832. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zavzavadjian JR, Couture S, Park WS, Whalen J, Lyon S, Lee G, Fung E, Mi Q, Liu J, Wall E, et al. The alliance for cellular signaling plasmid collection: a flexible resource for protein localization studies and signaling pathway analysis. Mol. Cell. Proteomics. 2007;6:413–424. doi: 10.1074/mcp.M600437-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.