Abstract
Purpose: Accurate patient registration is crucial for effective image-guidance in open cranial surgery. Typically, it is accomplished by matching skin-affixed fiducials manually identified in the operating room (OR) with their counterparts in the preoperative images, which not only consumes OR time and personnel resources but also relies on the presence (and subsequent fixation) of the fiducials during the preoperative scans (until the procedure begins). In this study, the authors present a completely automatic, volumetric image-based patient registration technique that does not rely on fiducials by registering tracked (true) 3D ultrasound (3DUS) directly with preoperative magnetic resonance (MR) images.
Methods: Multistart registrations between binary 3DUS and MR volumes were first executed to generate an initial starting point without incorporating prior information on the US transducer contact point location or orientation for subsequent registration between grayscale 3DUS and MR via maximization of either mutual information (MI) or correlation ratio (CR). Patient registration was then computed through concatenation of spatial transformations.
Results: In ten (N = 10) patient cases, an average fiducial (marker) distance error (FDE) of 5.0 mm and 4.3 mm was achieved using MI or CR registration (FDE was smaller with CR vs MI in eight of ten cases), which are comparable to values reported for typical fiducial- or surface-based patient registrations. The translational and rotational capture ranges were found to be 24.0 mm and 27.0° for binary registrations (up to 32.8 mm and 36.4°), 12.2 mm and 25.6° for MI registrations (up to 18.3 mm and 34.4°), and 22.6 mm and 40.8° for CR registrations (up to 48.5 mm and 65.6°), respectively. The execution time to complete a patient registration was 12–15 min with parallel processing, which can be significantly reduced by confining the 3DUS transducer location to the center of craniotomy in MR before registration (an execution time of 5 min is achievable).
Conclusions: Because common features deep in the brain and throughout the surgical volume of interest are used, intraoperative fiducial-less patient registration is possible on-demand, which is attractive in cases where preoperative patient registration is compromised (e.g., from loss/movement of skin-affixed fiducials) or not possible (e.g., in cases of emergency when external fiducials were not placed in time). CR registration was more robust than MI (capture range about twice as big) and appears to be more accurate, although both methods are comparable to or better than fiducial-based registration in the patient cases evaluated. The results presented here suggest that 3DUS image-based patient registration holds promise for clinical application in the future.
Keywords: registration, volumetric true 3D ultrasound, US-MR registration, correlation ratio, mutual information, capture range, image-guidance
INTRODUCTION
Intraoperative navigation based on preoperative images has become the standard-of-care in most open cranial procedures because it enables more effective neurosurgical interventions. An accurate image-to-patient transformation (commonly referred to as “patient registration”) is crucial to the overall performance of intraoperative neuronavigation because it defines the degree of alignment between structures of interest in the operating room (OR) and their counterparts in preoperative images (most often preoperative magnetic resonance, MR, and computed tomography, CT). Seamless integration of the registration procedure with surgical workflow is becoming increasingly important as pressures mount to improve efficiencies in all phases of OR patient management in order to increase throughput and/or reduce health care costs.
Patient registration is typically accomplished by matching a set of homologous fiducial markers implanted in skull or affixed to skin with their corresponding locations identified in MR. Matching skull-implanted fiducials yields the highest registration accuracy [∼1 mm (Ref. 1)], and is often regarded as the “gold standard”; however, registration based on skin-affixed fiducials is more common in practice due to their noninvasiveness, despite the relatively inferior registration accuracy [typically 2–5 mm (Refs. 2 and 3) in terms of fiducial registration error, FRE (Ref. 4)]. Fiducial-based registration (FBR) usually requires manual identification of the markers in the OR (typically with a digitizing stylus) as well as in image space, which not only consumes personnel time but also involves communication and coordination between individuals with sufficient expertise to ensure proper anatomical/image pairing. FBR may also require special imaging sequences in addition to typical diagnostic scans, which can add time and associated costs. Because fiducials must remain on the patient's head from the start of preoperative image acquisition (usually on the day of surgery) until registration is completed in the OR, risks of marker movement, loss, or removal (in which case patient registration may be compromised or even become infeasible) always exist (typically FBR is no longer possible once surgery begins).
As a result, alternative patient registration techniques that do not rely on fiducials have been of considerable interest. Various approaches have been proposed and their performances examined. Although methods based on discrete anatomical landmarks are simple to implement, they have not gained acceptance in practice because their registration accuracy has been limited.5 Most fiducial-less techniques rely on surface-based registration (SBR) in which the natural curvatures/features of the face, forehead (e.g., Refs. 6, 7, 8, 9, 10), or exposed cortical surface (e.g., Refs. 11 and 12) are recovered with laser range scanning6, 7, 8, 9, 11 or stereovision.10 Registration is then performed by matching the reconstructed surface in the OR with its counterpart derived from MR or CT using algorithms based on the iterative closest point (ICP; Ref. 13) technique. When texture information is available, image intensity has also been employed to enhance registration (e.g., “surface mutual-information” registration11).
Registration accuracy with SBR has also been evaluated. Although comparing the registration accuracies between studies is imperfect because of the multiplicity of error metrics and techniques which have been used, these quantitative assessments provide important insights into the relative performance of the various registration techniques that have been investigated. For example, an accuracy of <3 mm in target registration error (TRE; Ref. 4) can be achieved in the frontal region using natural facial contours for registration,10 but TRE degrades significantly in regions farther away from the surface (e.g., Refs. 6 and 7). Mascott et al.14 statistically compared the performances of FBR and SBR and confirmed that skull-implanted fiducials yield statistically superior registration accuracy (1.7 ± 0.7 mm). However, no significant difference in accuracy was found in other skin-based methods by matching either fiducials or surfaces (4.0 ± 1.7 mm), except when discrete anatomical landmarks were used instead (4.8 ± 1.9 mm). Similarly, a more recent comparison between different registration techniques suggests that extracranial methods using skin fiducials or face-based registrations are statistically equivalent in terms of TRE (5.3 mm) based on distinct vessel bifurcations and gyral points taken as targets from the exposed cortical surface. However, the accuracy was improved significantly (to approximately 2 mm) with intracranial reregistration using discrete features, vessels, or intensity in an effort to compensate for brain shift.12
In addition to having similar registration accuracy, SBR and FBR are also both performed before the start of surgery (or at the very beginning of surgery in the case of intracranial reregistration12). Once retraction or tumor resection occurs which prevent access to or distort features used for establishing correspondence, patient registration is no longer possible. Further, because same-sided superficial features are typically involved in SBR techniques, degradation of TRE away from these surfaces (e.g., Refs. 6 and 7) and deeper in the brain may occur which could lead to poor and/or erratic registration accuracy when the tumor or other structures of interest are located well below the surface.
In this paper, we present an image-based patient registration technique that directly establishes coordinate system correspondence between tracked 3D volumetric ultrasound (3DUS) and the MR image volume automatically and without fiducials or other information on the location and orientation of the US scan-head. Similar to SBR methods, the approach does not rely on fiducials or require manual image processing operations; however, it differs significantly from other techniques because volumetric image data including features deep in the brain (as opposed to on the surface) are used for registration, which reduces TRE within the surgical volume of interest (e.g., around the tumor) when located deeper in the brain. In addition, our registration technique allows intraoperative patient registration on-demand even after surgery begins or in emergency cases when preoperative patient registration is not always possible (because fiducials were not in place in time) as long as features deep in the brain are visible in 3DUS. Although US has been successfully applied to improve fiducial-based patient registration at the start of surgery by reregistering with MR images to compensate for initial brain deformation (e.g., Refs. 17 and 30), the technique reported in this paper represents a significant advance because it achieves the same advantage of volumetric image-based patient registration but does so automatically and without fiducials or other prior information on location/orientation of the US transducer.
Intensity-based registration between US and MR is challenging because of differences in their underlying physics and image characteristics.15 Successes with both mutual information (MI) and correlation ratio (CR) (along with their variants) as image similarity measures have been reported (e.g., for brain,16, 17 liver,18 and phantoms19 based on MI; and for brain20, 21 based on CR). For example, Rasmussen et al.16 developed a coregistration method for automatic brain-shift correction by rigidly registering reconstructed 3D US with MR using MI-based image similarity metrics (either between anatomical US and MR images or between power-Doppler US and MR angiography). More recently, we have also employed normalized mutual information (nMI) as an image similarity metric to reregister 2D US images with MR using an initial starting point generated from FBR. In these studies, we achieved millimeter-level registration accuracy around the tumor boundary (reduced from an initial misalignment of 2.5 mm on average with a maximum of 4.9 mm) with translational and rotational capture ranges of 5.9 mm and 5.2°, respectively.17 In parallel, Roche et al.26 developed CR as a new image similarity measure based on standard theoretical statistics. Its applications to MR, CT, and PET images suggest that CR provides a favorable trade-off between accuracy and robustness. Subsequently, the authors extended the measure to include both MR intensity and gradient information (i.e., bivariate-CR) to register MR with US and achieved accuracy comparable to MR voxel resolution using phantom, pediatric, and adult brain datasets. Comparison of the original CR (i.e., monovariate) and MI suggested that the relative robustness of these different image similarity measures largely depends on the image data and whether MR intensity or gradient images were used. Similarly, a more recent comparison21 of registration between reconstructed US and MR of the liver using nMI, CR, and their variants found that accuracy was predominantly a function of the quality and sampling volume of the US images. However, CR (with a MR-gradient-norm image volume) was significantly more robust, although with longer computation times (about twice that of nMI).
Given these prior investigations, we investigated both MI and (monovariate) CR image similarity measures to rigidly register 3DUS with gradient-encoded MR as the basis for patient registration without using fiducials or introducing prior information on the US transducer contact point location and orientation, and compared their relative performances. The details of our image-based registration technique and its outcomes with MI and CR applied to MR and 3DUS image data from ten patient cases are presented in the following sections.
METHODS
Establishing the spatial transformation between the 3DUS and MR image volumes directly is key to successful patient registration without using fiducials. For operational acceptance, a fully automated procedure which requires as little prior information as possible, but is sufficiently accurate (comparable or superior to FBR) and robust, is desired. To this end, a two-step registration scheme was developed in which binary image volumes generated from 3DUS and MR were first registered. The resulting registration provided an initial starting point for a second registration between grayscale 3DUS and MR volumes (details appear in Secs. 2C, 2D, respectively).
Image acquisition
Ten (N = 10) patients undergoing open cranial procedures (nine tumor resections and one epilepsy surgery) where intraoperative 3DUS was deployed at Dartmouth-Hitchcock Medical Center were retrospectively evaluated in this study (Table 1). Patient recruitment and image analyses were approved by the Institutional Review Board at Dartmouth College. The only criterion for inclusion in the analysis was the presence of parenchymal boundary features contralateral to the craniotomy in 3DUS. All patients had T1-weighted, gadolinium-enhanced axial MR scans from a 1.5 T GE scanner prior to surgery (256 × 256 × 124; voxel size in dx × dy × dz: 0.94 × 0.94 × 1.5 mm3; 16-bit grayscale). The brain was automatically segmented with a level-set technique22 to generate a triangular brain surface. For each patient, a set of 3DUS volumes was acquired (3–9 volumes) before dural opening using a broadband matrix array transducer (X3-1) connected to a dedicated ultrasound system (iU22, Philips Healthcare, N.A.; Bothell, WA). For consistency across patients, we used the first 3DUS image volume with sufficient scan-depth (typically 14–16 cm) to capture the parenchymal boundary contralateral to the craniotomy for patient registration. All image acquisitions were configured to cover the maximum angular ranges allowed by the scanner (−42.9° to 44.4° in θ, and −36.6° to 36.6° in φ, respectively; see Ref. 23 for details of 3DUS image format and coordinate system).
Table 1.
Summary of patient age, gender, type, and location of brain lesion, and size of craniotomy.
| Patient | Age/gender | Type of lesion | Location of lesion | Size of craniotomy |
|---|---|---|---|---|
| 1 | 46/M | Low grade glioma | Left frontal | 10 × 6 cm2 |
| 2 | 27/F | Epilepsy | Right parietal | 5 × 5 cm2 |
| 3 | 51/M | Low grade glioma | Right frontotemporal insula | 6 × 7 cm2 |
| 4 | 60/F | Meningioma | Supratentorial | 6 × 6 cm2 |
| 5 | 74/F | Meningioma | Right frontal | 6.5 × 6.2 cm2 |
| 6 | 50/F | Meningioma | Right temporal | 3.5 × 3.5 cm2 |
| 7 | 48/F | Low grade glioma | Left fronal | 7.4 × 4.5 cm2 |
| 8 | 70/F | Glioblastomamultiforme | Right temporal | 6 × 6 cm2 |
| 9 | 56/M | Glioblastomamultiforme | Left frontal | 7 × 6.2 cm2 |
| 10 | 55/M | Low grade glioma | Left frontal | 10 × 7 cm2 |
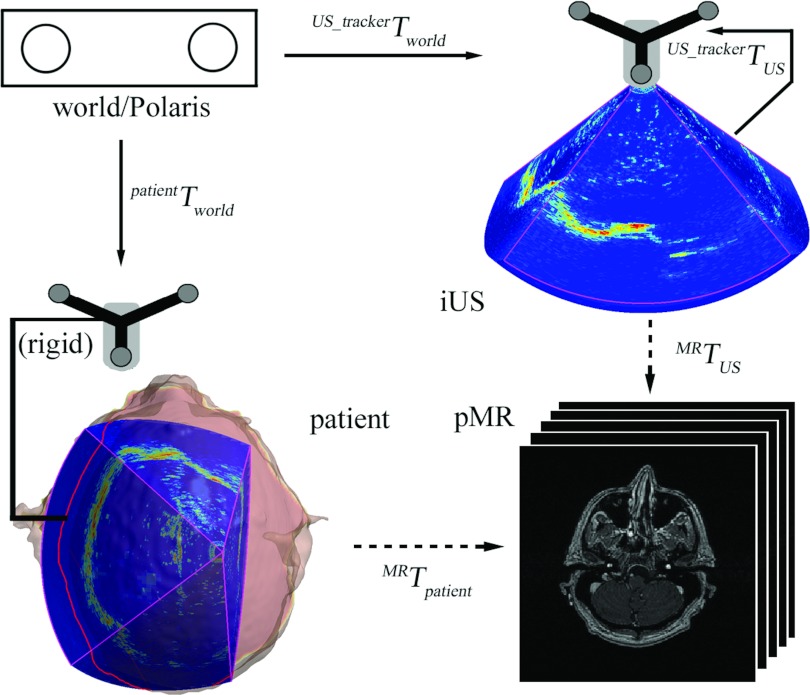
Coordinate systems involved in patient registration
Because image-to-patient registration was computed by concatenating spatial transformations, a “world” coordinate system was necessary to provide a common reference and was defined by an optical tracking system (Polaris; The Northern Digital Inc., Canada) that determines the spatial positions and orientations of the trackers rigidly fixed to the patient's head clamp and 3DUS transducer (i.e., patientTworld and , respectively; Fig. 1). In order to transform the 3DUS image volume from its image coordinate system into the tracker space (i.e., ), the transducer was calibrated prior to surgery using phantom images (with an accuracy of approximately 1 mm; Ref. 24). When the spatial transformation between 3DUS and MR image volumes (i.e., MRTUS) is available, the transformation between the patient in the OR and MR (i.e., “patient registration” or MRTpatient) can be readily computed from
| (1) |
which completes the volumetric image-based patient registration technique described this work (Fig. 1).
Figure 1.
Coordinate systems involved in patient registration. Solid/dashed arrows indicate transformations determined from calibration/registration. A transformation reversing the arrow direction is obtained by matrix inversion.
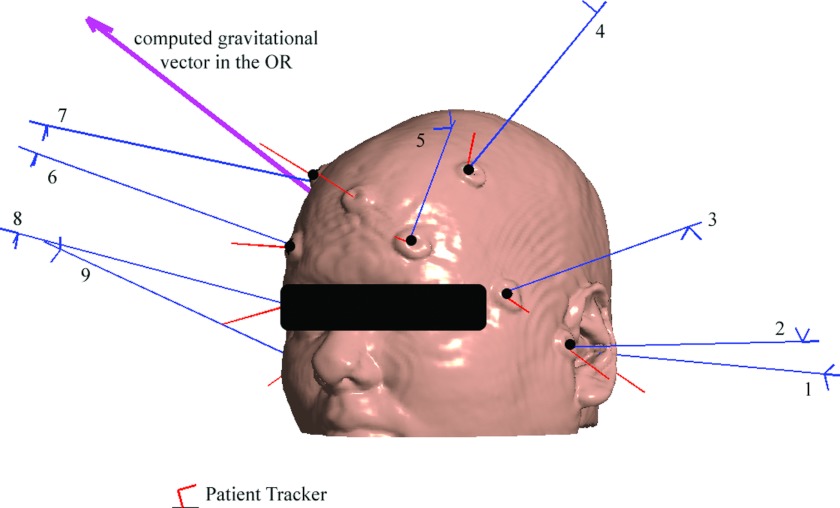
The FBR was computed by matching two sets of homologous markers separately identified in the OR using a digitizing stylus (typically 8–10 fiducials were successfully identified) and in the MR image space using a custom interactive software tool. These markers were not paired (no special effort was made to record their acquisition orders with either the digitizing stylus or the software interface to simplify surgical workflow at the start of a case), and a genetic algorithm25 was used to establish the transformation between the patient in the OR and MR [i.e., MRTpatient in Eq. 1]. Figure 2 illustrates a representative fiducial marker layout used for patient registration in a typical surgery.
Figure 2.
Illustration of fiducial markers identified in the OR (long thin lines represent the sequence of markers located with a digitizing stylus; nine markers in total) and in the MR image space (short thin lines; ten markers in total) for patient 6. Six fiducial markers (black dots) that produced the best FRE were used to determine patient registration in this case. The computed gravitational vector in the OR is also shown for reference (patient was lying on the right).
Binary image volume registration
Instead of registering grayscale 3DUS and MR directly, binary registrations were performed to generate an initial starting point without imposing any prior information on the point of contact of orientation of the US transducer with respect to the segmented brain surface. Essentially, binary image volumes highlighting parenchymal boundaries opposite to the craniotomy from both 3DUS and MR were produced via automated image preprocessing (see Sec. 2D for details). The sum of squared intensity differences (SSD; Ref. 15) was chosen as the image similarity measure and a gradient descent optimization was applied.
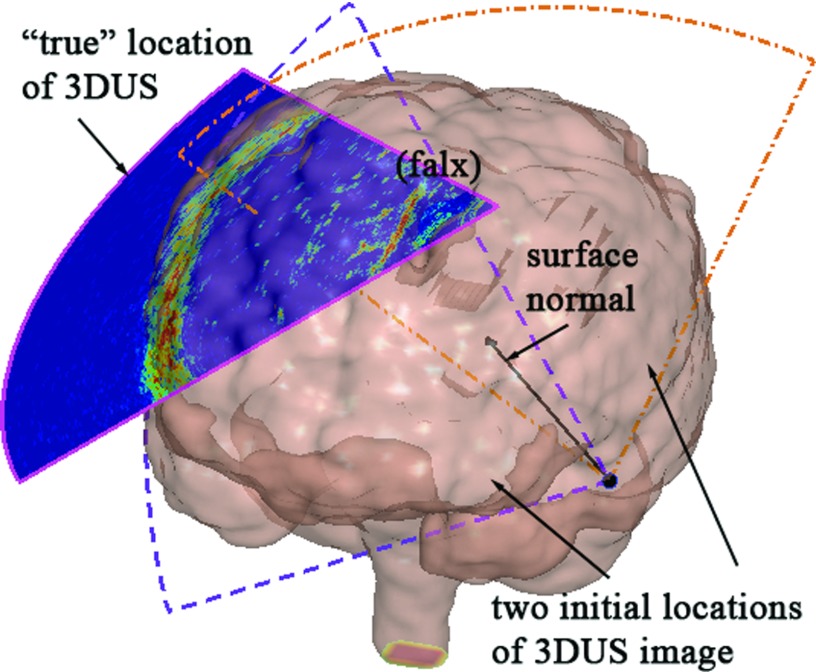
Because an initial alignment between the binary image volumes was purposely not introduced, a multistart approach was implemented using a set of random initial positions. To generate these initial starting points, the 3DUS transducer was assumed to be in contact with and pointing towards the parenchyma during image acquisition. Conceptually, when the location of craniotomy is known, the transducer tip location relative to the brain can be further constrained (e.g., via manual positioning of the transducer), which would reduce the number of initial starting points necessary for successful registration convergence. In this work, however, we minimized the use of any prior knowledge about the initial starting point relative to patient in order to explore the clinical feasibility and potential utility of the technique. Therefore, a subset of triangulated brain surface nodes (50 in total) was randomly chosen as potential transducer tip positions, and the central axis of the 3DUS image (i.e., both θ and φ are zeroed) was positioned along the corresponding surface normal. The only remaining degree-of-freedom (DOF) was the transducer's orientation with respect to the central axis (0°–360°), and ten equally spaced orientations were generated for each transducer position. In total, 500 (50 × 10) initial starting positions were evaluated [see Fig. 3f or illustration of the initial starting points].
Figure 3.
Generation of random initial starting points for registering binary 3DUS and MR image volumes. A typical transducer location was generated to coincide with a brain surface node and was directed along the inward (towards the parenchyma) normal. Equally spaced angular orientations of the transducer were subsequently created (dashed fan-shapes showing two representative orientations). The “true” location of the 3DUS relative to the triangular brain surface is also shown for comparison.
Registration between grayscale image volumes
For each patient, the converged transformation corresponding to the minimum SSD among all binary image registrations was selected as the initial starting point for the subsequent volumetric grayscale image registration. Two image similarity measures, mutual information (MI; Ref. 15) and correlation ratio (CR; Ref. 26) were used to register preprocessed grayscale 3DUS and MR image volumes (see Sec. 2D for details on image preprocessing) in order to compare their registration performances. Given two image sets, X and Y, their MI (I(X,Y)), is defined as
| (2) |
where H(X) and H(Y) are the marginal entropy of the respective images, and H(X,Y) is their joint entropy. A gradient descent optimization scheme in the Insight Segmentation and Registration Toolkit (ITK; version 3.10; Ref. 27) was utilized to maximize MI between the 3DUS and MR image volumes.15, 17
CR-based registration of two images, X and Y, finds a spatial transformation, T, that maximizes the functional dependence between the two images defined as
| (3) |
where η(Y|X) is the correlation ratio, E(Y|X) is the conditional expectation of Y given X, and Var(Y) is the variance. Implementation Eq. 3 was based on a discrete approach.26 Specifically, for an overlapping image region, Ω, the total number of voxels is defined as its cardinal, N = Card(Ω). Similarly, for iso-sets of X, Ωi = {ω ∈ Ω, X(ω) = i}, their cardinals, Ni = Card(Ωi), also represent the total number of voxels in each set. The total (σ) and conditional (σi) variance and the mean of Y for the overlapping region as well as for iso-sets of X(m and mi, respectively) are determined from
| (4) |
and,
| (5) |
respectively, in which case the expression for CR is finally obtained as26
| (6) |
We implemented CR-based registration through the Insight Segmentation and Registration Toolkit (ITK; version 3.10; Ref. 27) with multithreading to compute the similarity metric.
Preprocessing of MR and 3DUS image volumes
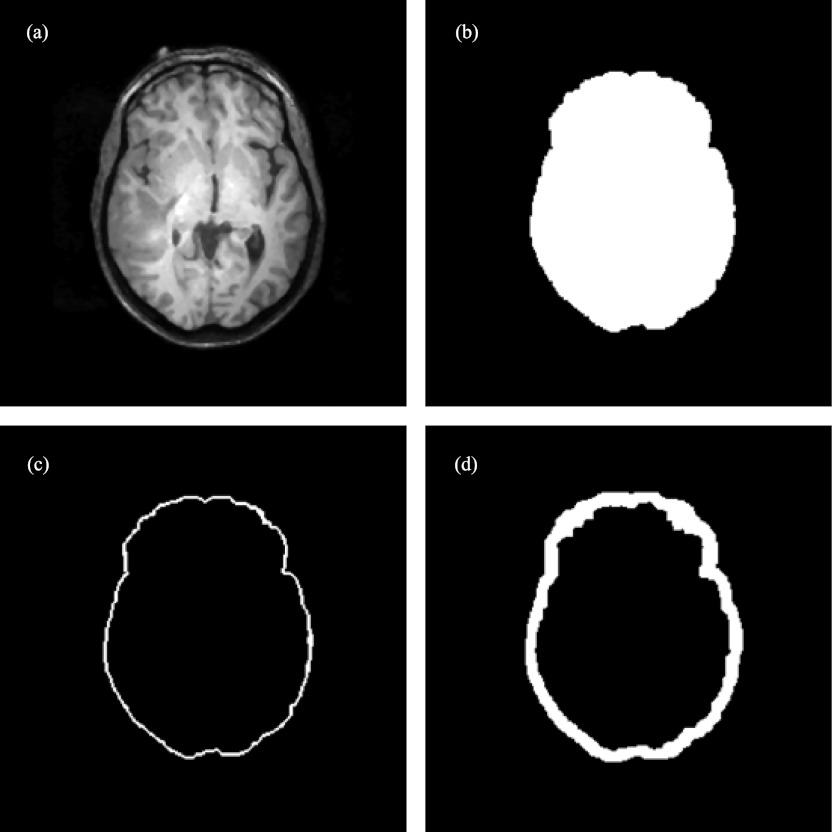
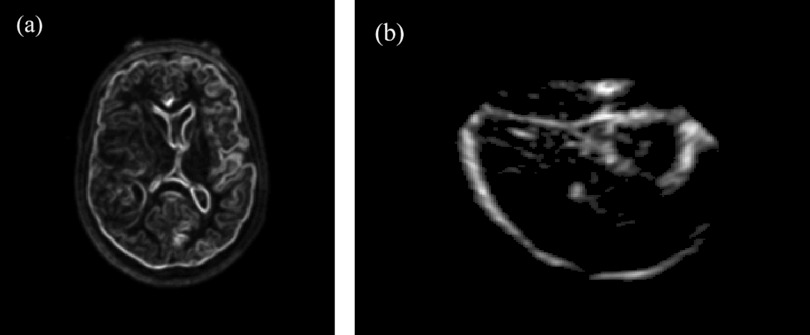
Generation of the binary image volumes was achieved through automatic preprocessing with computation time of less than 20 s on average. Specifically, for each MR volume, a gradient image of the segmented brain was produced. The resulting gradient image volume was then thresholded29 to highlight parenchymal features, and further dilated (with a kernel of 5 × 5 × 5) to emulate its appearance in the corresponding 3DUS image volume as illustrated in Fig. 4.
Figure 4.
Image preprocessing of MR to generate its binary image volume (patient 8): (a) axial MR image; (b) binary image of the segmented brain; (c) thresholded gradient image; (d) dilated parenchymal surface.
For 3DUS, the acquisition was rasterized into a 200 × 200 × 200 Cartesian grid [Ref. 28; Fig. 5a], Gaussian smoothed (with a kernel of 5 × 5 × 5) to reduce speckle noise [Fig. 5b], and thresholded29 to produce a binary image volume [Fig. 5c]. The result was eroded (with a kernel of 3 × 3 × 3) to create the largest connected region in 3D and dilated [with a kernel of 3 × 3 × 3; Fig. 5d]. Erosion and dilation maximized the presence of parenchymal boundary features while minimizing the appearance of internal structures.
Figure 5.
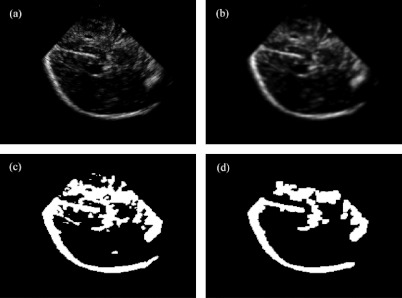
Image preprocessing of 3DUS to generate its binary image volume (patient 8). (a) rasterized 3DUS image; (b) 3DUS after Gaussian smoothing and thresholding; (c) corresponding binary image; and (d) binary image after erosion, selection of largest connected region in 3D, and dilation.
Prior to grayscale registration, the MR (grayscale) gradient image was Gaussian smoothed (with a kernel of 5 × 5 × 5) to emulate the feature appearance in 3DUS [Fig. 6a]. Each gray-level 3DUS volume was also Gaussian smoothed (with a kernel of 5 × 5 × 5) to reduce speckle noise and only voxels with intensities larger than the global threshold value were retained for registration [effectively, this result in Fig. 6b was generated by multiplying images corresponding to Figs. 5b, 5c at the voxel level].
Figure 6.
Illustration of typical preprocessed grayscale (a) MR and (b) 3DUS image volumes for CR reregistration.
Registration accuracy
Quantitative evaluation of patient registration accuracy is a practical challenge because homologous internal points are difficult to identify with certainty in 3DUS and MR for true TRE assessment. One approach uses segmented features such as tumor boundaries (e.g., Refs. 17 and 30) or centerlines of segmented vessels (e.g., Refs. 30 and 31) to assess TRE. However, these methods introduce segmentation errors that can be equally difficult to assess. More importantly, our 3DUS acquisitions at the start of surgery were optimized to acquire parenchymal features contralateral to the craniotomy to facilitate initial binary image registration (and not to highlight tumor boundaries per se, which are often located closer to the transducer). We also did not have routine access to MR angiography or power Doppler US images in the patient cases used in the study; hence, vessel features were not always available in the conventional T-1 weighted MR or B-mode 3DUS image volumes for quantitative evaluation of registration accuracy.
Instead, we computed the mean FDEs (analogous to FREs) using spatial transformations generated from both the binary and grayscale image registrations based on either MI or CR maximization. These distance results were then compared with the corresponding FREs from the fiducial-based approach, which effectively provided an objective evaluation of the different registration transformations relative to the clinical standard (i.e., fiducial-based registration). Mathematically, FRE (Ref. 4) is defined as
| (7) |
where Tfid represents the spatial transformation obtained from the fiducial-based approach, qi and pi are the corresponding fiducial point locations identified in the MR image space and in the OR physical space, respectively, and L is the total number of fiducial markers used. Analogously, the FDE is defined as
| (8) |
where T is the spatial transformation under scrutiny obtained from the registrations based on either binary or grayscale images.
To further quantitatively evaluate the spatial similarity (as opposed to the “true” accuracy based on TRE) of the MI- and CR-based registrations, we computed the converged locations of 3DUS image voxels of prominent features (segmented via thresholding) using the two registration transformations. The average distances between corresponding point-pairs were then obtained to assess the spatial similarity in the registration results from the two methods.
Capture ranges
Capture range quantifies the extent to which the initial starting point parameters in an iterative registration algorithm can be perturbed from their converged values, and the registration process returned with a high probability to a converged solution in which voxels in the floating image volume are transformed into the same spatial neighborhood.32 Capture ranges of the binary and grayscale image registrations are of practical significance because they define the robustness of the technique, which is important to understand for clinical application in the OR. In this work, both translational and rotational capture ranges were evaluated. A perturbation-reregistration approach similar to that described in Ref. 17 was employed, in which a converged registration was used to generate perturbations that served as starting locations for additional registrations. A converged registration was considered to be one that corresponded to the minimum sum of squared differences (SSD) for the binary image data and maximized MI or CR for grayscale image volumes, respectively. The floating 3DUS image volume was transformed according to these converged registrations and was systematically perturbed by either a linear translation or a pure rotation with step sizes of 0.25 mm and 0.25°, respectively, where the directionality of the translational/rotational axis passing through the centroid of all floating image voxels was randomly generated in space. The “ground-truth” locations of the floating voxels were determined from the average of all converged locations (i.e., image voxel locations when the registration optimization converged) in which the perturbation magnitude (i.e., the initial translational and rotational perturbations) was less than 1 mm and 1°, respectively (four perturbations of each). Registration of floating image volumes with larger perturbations were then executed via SSD minimization or MI/CR maximization (for binary and grayscale image volumes, respectively), and the converged locations of the floating image voxels were compared to their respective ground truth counterparts. A successful registration was considered to occur when the average distance between the two sets of point locations (i.e., distance error) was no more than 2 mm.17 The translational and rotational capture ranges were then defined by the maximum perturbation at or below which the registration success rate was at least 95%.
Data analysis
For each patient, all binary image registrations were executed independently and in parallel on two Linux clusters (2.8–3.1 GHz; 8–16 G RAM) that had a total of 15 dual-quad nodes, allowing 120 (15 × 8) single-threaded registrations to be performed simultaneously. For MI and CR registration, multithreading with 8 CPUs was employed. A sampling rate of 50% was applied to the floating image volume for all registrations as a reasonable trade-off between sufficient registration robustness in terms of convergence and computation time. All of the registrations performed in this study were rigid as intraoperative 3DUS image volumes were acquired before dural opening at which time minimum brain shift is expected, especially for anatomical features deep in the brain that are used for registration. All registration algorithms were implemented in ITK.
For each patient, accuracy from the binary, MI and CR registrations was quantitatively compared with the fiducial-based approach in terms of the resulting FDEs. These registration accuracies were also compared statistically using Wilcoxon's paired tests (which do not require normally distributed data) to examine whether the average registration accuracy differed significantly with choice of registration approach. If the difference in registration accuracy was found to be statistically significant, Wilcoxon and sign tests were further performed to evaluate whether the difference was clinically relevant. To assess the clinical relevance of any FDEs that were found to be statistically different between the registration methods, we added an increment to the mean FDE of the method with smaller error (up to 2 mm, which is consistent with the threshold used to define a successful registration in Sec. 2G), recomputed the statistical tests and found the largest increment which eliminated the statistical difference in the FDEs of the methods as a way of quantifying the clinical relevance of the statistical differences that could then be judged accordingly (rather than subjectively picking a single threshold for clinical relevance and testing for statistical significance). For all statistical analyses, the significance level was defined at 95%. In addition, the relative similarity of the MI- and CR-based image registrations was evaluated in terms of the average distance between converged voxel pair locations from the resulting spatial transformations. The translational and rotational capture ranges using the binary, MI and CR registrations were reported for each patient. Finally, typical execution times to complete image-based patient registration were also indicated. All data analyses were performed in MATLAB (Release 2010b, Mathworks, Inc.).
RESULTS
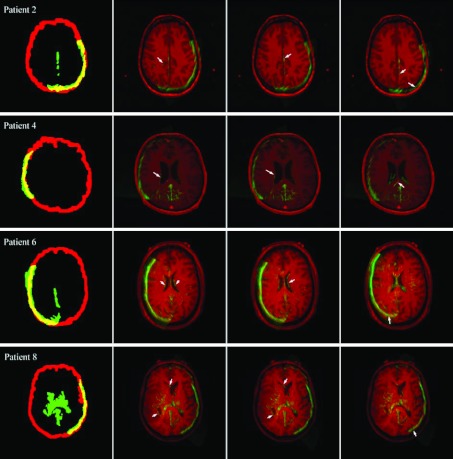
For all patients, the spatial transformation between 3DUS and MR generated from either the binary, MI, CR, or fiducial registrations was used to produce image overlays for qualitative assessment of registration accuracy. Representative results are illustrated in Fig. 7 for four patient cases. The binary image registrations successfully generated an initial transformation with sufficient proximity between features in the two image volumes (see left column in Fig. 7) that the alignment could be further improved with either MI or CR registration (second and third columns in Fig. 7, respectively) using features both at the parenchymal boundary and deep in the brain. The resulting feature alignment was comparable to or even better than the correspondence obtained from the fiducial-based approach (fourth column in Fig. 7).
Figure 7.
Overlays of 3DUS and MR for four patient cases using transformations generated from the binary (first column), MI (second column), CR (third column), and fiducial (fourth column) registrations. Representative feature alignment is noted between MI, CR, and fiducial registrations (arrows).
Registration accuracy was quantitatively evaluated using FDEs for the ten patients and these results are reported in Table 2, together with the average FDEs for the pooled population. Wilcoxon and sign tests indicate that both MI and CR registrations significantly reduced the FDE relative to binary registration, and that the thresholds for preserving the statistical significance in mean FDE between MI and binary registration and CR and binary registration were added-increments of 0.2 mm and 0.8 mm, respectively. These results suggest that MI or CR registration based on the starting point provided by binary registration is important and necessary to further improve patient registration accuracy. However, no significant difference in FDE was found between outcomes from the MI and CR registrations, or when comparing with FRE from fiducial registration (p > 0.05), indicating that the accuracy obtained from either the MI or CR registrations as well as from the fiducial-based approach was statistically comparable in these surgical cases (although FDE in the CR image registrations was lower than that of MI in eight of the ten cases and lower in six of ten cases relative to fiducial registration).
Table 2.
Summary of fiducial distance errors (mean ± std) between markers for the ten patients using transformations generated from the binary (FDEbin), MI (FDEMI), and CR(FDECR) registrations, along with the fiducial-based FRE (FREfid). Units are in mm.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Avrg. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FDEbin | 6.4 ± 2.9 | 3.6 ± 1.0 | 7.5 ± 0.9 | 4.7 ± 2.1 | 7.8 ± 1.6 | 6.4 ± 2.0 | 5.7 ± 1.7 | 7.9 ± 1.7 | 7.6 ± 3.2 | 5.7 ± 2.6 | 6.3 ± 1.5 |
| FDEMI | 5.3 ± 1.1 | 3.2 ± 0.9 | 7.4 ± 0.8 | 3.7 ± 0.7 | 4.7 ± 2.4 | 4.3 ± 1.2 | 4.2 ± 1.8 | 7.1 ± 2.1 | 4.2 ± 1.2 | 5.7 ± 1.6 | 5.0 ± 1.4 |
| FDECR | 4.7 ± 1.2 | 2.8 ± 0.9 | 3.5 ± 0.9 | 3.6 ± 1.1 | 4.3 ± 1.7 | 3.9 ± 1.8 | 5.1 ± 2.0 | 5.3 ± 1.8 | 5.7 ± 1.6 | 4.5 ± 1.2 | 4.3 ± 0.9 |
| FREfid | 3.4 ± 1.9 | 6.1 ± 2.6 | 3.8 ± 1.9 | 2.9 ± 1.1 | 6.3 ± 0.7 | 4.9 ± 2.4 | 4.2 ± 2.9 | 6.8 ± 2.0 | 5.1 ± 2.4 | 6.3 ± 2.0 | 5.0 ± 1.4 |
The spatial similarity between the two grayscale image registrations using MI or CR for the ten patient cases is reported in Table 3. Overall, the distance between voxel pair locations in the 3DUS volume when transformed by the two registrations averaged 2.6 ± 0.9 mm (ranged from 0.9 to 3.9 mm), which is within the typical FRE from FBR of 2–5 mm, once again suggesting that accuracies from the two registration methods are comparable.
Table 3.
Summary of average distance between 3DUS voxel pair locations in the MI- and CR-based registrations (units in mm).
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Avrg. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dist. | 1.8 ± 0.7 | 3.0 ± 1.5 | 3.4 ± 1.2 | 0.9 ± 0.3 | 3.5 ± 1.2 | 3.9 ± 1.8 | 2.3 ± 0.8 | 2.5 ± 1.0 | 2.7 ± 1.1 | 2.5 ± 0.6 | 2.6 ± 0.9 |
Representative scatter plots of distance errors as a function of translational and rotational perturbations for binary, MI, and CR registrations are shown in Fig. 8 for two patients. Distance errors were tightly clustered near zero (typically ∼0.1 mm, ∼0.1–0.01 mm, and∼0.01 mm for binary, MI, and CR registrations) for successful registrations indicating their consistency in convergence regardless of initial starting point.
Figure 8.
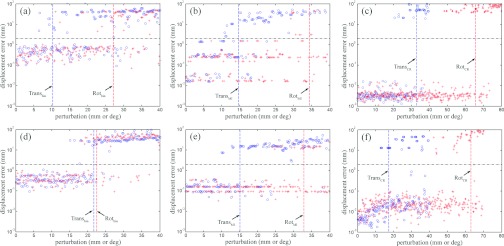
Representative scatter plots of distance error vs translational (open circle) and rotational (cross) perturbations for binary (a) and (d), MI (b) and (e), and CR (c) and (f) registrations for two patients (patient 1: top; patient 8: bottom). Distance errors are shown on a log scale. Vertical dashed lines indicate capture ranges, while the horizontal lines indicate the distance error threshold used to define successful registrations.
For each patient, the translational and rotational capture ranges are summarized in Table 4, together with the averages for the binary, MI, and CR registrations for the group of ten cases. The maximum translational (rotational) capture ranges for the binary, MI, and CR registrations were 32.8 mm, 18.3 mm, and 48.5 mm (36.4°, 34.4°, and 65.6°), respectively.
Table 4.
Summary of translational (mm) and rotational (deg) capture ranges for 3DUS and MR registration using binary, MI, and CR registration for the ten patient cases.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transbin | 10.3 | 21.7 | 26.7 | 24.5 | 29.3 | 23.3 | 28.3 | 21.7 | 21.7 | 32.8 | 24.0 ± 6.1 |
| Rotbin | 27.1 | 22.1 | 32.8 | 33.6 | 19.1 | 30.8 | 36.4 | 22.5 | 20.5 | 25.5 | 27.0 ± 6.1 |
| TransMI | 14.7 | 18.3 | 2.2 | 16.1 | 16.5 | 10.7 | 16.5 | 15.1 | 7.6 | 4.2 | 12.2 ± 5.7 |
| RotMI | 34.4 | 26.7 | 10.3 | 28.5 | 28.5 | 7.4 | 33.4 | 32.8 | 20.9 | 32.7 | 25.6 ± 9.7 |
| TransCR | 32.9 | 12.1 | 20.3 | 16.1 | 26.6 | 48.5 | 30.2 | 17.3 | 10.7 | 11.4 | 22.6 ± 12.0 |
| RotCR | 65.6 | 25.3 | 46.3 | 28.9 | 31.2 | 45.3 | 52.1 | 63.0 | 29.1 | 21.5 | 40.8 ± 15.9 |
The computation time for each independent binary registration was 1–2 min on average resulting in 8–10 min on two Linux clusters to complete all binary registrations. The computation times for MI and CR registrations were similar and were typically 3–4 min using 8 threads. The total execution time was 12–15 min to complete a typical image-based patient registration.
DISCUSSION
Accurate and efficient patient registration is crucial for effective deployment of image-guidance systems that navigate structures of interest based on preoperative images. Although image registration techniques are relatively mature in image-guided neurosurgery, new methods are still emerging that seek to eliminate the dependency on external fiducials without sacrificing registration accuracy (e.g., Refs. 6, 7, 8, 9, 10, 11, 12) and/or improving registration accuracy in targeted areas (e.g., around the tumor boundary17 or on the exposed cortical surface12) through approaches that exploit image features found within the neighborhoods of interest. Efficiency and convenience relative to workflow in the OR environment (e.g., reducing personnel requirements) in order to increase operational throughput is also increasingly important. Most of these techniques reconstruct the natural contours of the face and forehead or intraoperative cortical surface at the craniotomy through which either point or surface registration is performed. The fiducial-less patient registration technique described in this work utilizes a 3DUS image volume to register with its MR counterpart directly via a rigid image-based approach. The resulting interimage registration allows the spatial transformation between the patient's head in the OR and MR to be determined through a concatenation of transformations. Similar to other SBR methods, our patient registration technique is completely automatic and does not require fiducial markers. However, it differs significantly from other FBR or SBR approaches because features deep in the brain are used to establish correspondence enabling intraoperative patient registration on-demand even after surgery begins (e.g., in emergency cases), which is usually not possible with existing SBR or FBR methods that rely on superficial features.
The clinical feasibility of image-based, fiducial-less patient registration is largely dependent on its accuracy, robustness, and efficiency. A clinically more important measure of accuracy is perhaps the TRE around the tumor/lesion of surgical interest,33 which is often deep in the brain. Unfortunately, an objective evaluation of TRE is challenging because ground-truth identification of homologous features in 3DUS and MR is not readily available. Although segmenting features in 3DUS manually (e.g., ventricle, falx, gyri, etc.) and comparing them against their counterparts in MR is feasible, the influence of segmentation errors and crossmodality feature correspondence on the TRE assessment is difficult to evaluate. In this paper, we used FDEs as a surrogate measure of registration accuracy, and found that our volumetric image-based, fiducial-less MI or CR patient registration is comparable to typical FBR or SBR approaches (FRE ranged 2–5 mm; e.g., Refs. 2, 3, 12, and 14). Specifically, both MI and CR registrations significantly reduced the FDE in the initial binary image registration, and while not statistically different, the mean FDE of CR image registrations was lower than that of MI in eight of the ten cases (and in six of ten cases for FBR), likely due to its smoother feature space17 as indicated by the much smaller residual displacement error recovered from perturbation (Fig. 8).
Our technique offers some potential advantages over traditional FBR because it could be used to compensate for registration errors when external fiducial markers are lost have moved subsequent to preprocedure imaging or are not available all together. Further, it is likely to be an improvement over FBR or SBR when more significant brain deformation has occurred after the dura has been opened (prior to dural opening brain shift is expected to be small) or when the area of surgical interest is deeper in the brain. In this study, we applied image-based rigid registration, which may become less attractive for maintaining registration later in a case because of the nonrigid brain movement that often occurs in the operative field as surgery progresses. In principle, nonrigid image-based registration methods could replace the rigid transformations we applied here, but nonrigid intermodality registration is more challenging to compute and is less robust.15 Thus, better strategies may exist such as those that incorporate intramodality image-based registration as displacement mapping methods which provide data for model-based image compensation approaches.34
The relative spatial similarity in the registration transformations generated with MI and CR was also investigated. In the ten patients evaluated, we found that the average distance between converged 3DUS voxel locations produced by the two methods was 2.6 mm. Qualitative assessment of internal feature alignment (Fig. 7) also confirmed that the volumetric image-based, fiducial-less patient registration achieved similar or even improved performance relative to FBR, especially deeper in the brain or far away from the surface markers.
Translational and rotational capture ranges for the binary, MI and CR image registrations were compared to evaluate their relative robustness. When data from the ten patient cases were pooled, the average translational and rotational capture ranges were 24.0 mm and 27.0° for the binary registrations, 12.2 mm and 25.6° for MI registrations, and 22.6 mm and 40.8° for the CR registrations, respectively. These values are much larger than the ranges achieved between 2D US and MR using nMI (Ref. 17) because of the much larger and more regularly sampled image volumes available through 3DUS (relative to reconstructed image volumes from 2DUS). Interestingly, the CR capture ranges were much larger than (nearly twice) the MI equivalents, similar to the findings reported in Ref. 21 and are consistent with those reported for pairs of 3DUS image volumes (35.2 mm and 35.8° for translational and rotational capture ranges, respectively;28 32.5 mm for translational capture range35), and between reconstructed 3D power Doppler US and MR angiography (40° for rotational capture range36). These comparisons of the robustness of different registrations between US and MR images provide important quantitative evaluations of their relative performances. In addition, they suggest that automatic “fiducial-less” patient registration may not be possible with hand-swept 2D US images alone. Fully automatic, direct registration of volumetric grayscale 3DUS and MR may also not be feasible because of the significant computation time involved, especially when no prior knowledge on the location of the US transducer (relative to MR) is provided. In this case (i.e., when no prior information on US transducer location is given), a two-step approach that initially registers binary image volumes of 3DUS and MR to obtain a starting point for volumetric grayscale image registration is effective.
The total execution time to complete an volumetric image-based fiducial-less patient registration was 12–15 min on average for each patient using either MI or CR, which was comparable to the personnel time involved in a typical FBR where manual localization of markers is necessary (especially, if the manual effort has to be repeated due to poor registration accuracy). Unlike FBR, however, image-based fiducial-less registration requires no personnel time (beyond 3DUS acquisition which is typically <10 s). The computational efficiency of our volumetric image-based fiducial-less patient registration can be improved by limiting the number of random starting points visited for binary registration (to generate a sufficiently well-defined initial registration for further improvement). In fact, the overall computational efficiency can be improved significantly by restricting the 3DUS transducer tip location to the center of the craniotomy (whose size and approximate position is already known at the time of 3DUS acquisition). Alternatively, the brain surface can be subdivided into a finite number of representative zones to standardize the possible transducer tip locations in order to limit the search range. Because of the large capture ranges available for CR registration between 3DUS and gradient-encoded MR (up to 48.5 mm and 65.6° in the cases presented here), multistart registrations between grayscale 3DUS and MR may be launched directly without the need for binary registration as a starting location. In addition, a reduced image sampling rate may be possible without significantly sacrificing registration accuracy (a 50% sampling rate was used in all registrations in this work). With these enhancements, we estimate that a total execution time of approximately 5 min is readily attainable in the future, and believe that computation time, alone, will not be a barrier to clinical acceptance of the approach. As illustrated in this study, we have successfully applied the image-based technique in ten patient cases involving four types of brain diseases and have achieved patient registration accuracies comparable to FBR. Nonetheless, more prospective studies are warranted to evaluate the approach in a much larger number and range of clinical cases and to understand and further improve the performance of the volumetric image-based”fiducial-less” patient registration technique described here before it can be considered as a possible replacement for and/or (more likely) adjunct to FBR because no fiducials will be available after craniotomy to perform patient registration, which may result in a loss of image-guidance if the fiducial-less registration fails. On the other hand, this fiducial-less registration approach is already applicable in emergency surgical operations where image-guidance is desired but no fiducials are typically used to perform conventional fiducial-based patient registration.
CONCLUSION
In summary, we have developed a completely automatic patient registration technique that does not rely on fiducial markers by directly registering tracked 3DUS with MR. The performance of the method was evaluated in ten neurosurgical patient cases and found to be statistically comparable to fiducial-based registration regardless of whether mutual information or correlation ratio similarity measures were used, although the lowest fiducial distance error (4.3 ± 0.9 mm) was achieved with correlation ratio on average and in eight out of the ten cases, individually. CR capture ranges were also the largest and were about twice the size of those achieved with MI (22.6 mm and 40.8° for CR, vs 12.2 mm and 25.6° for MI, respectively). Computational efficiency was comparable, about 10–15 min, independent of whether CR or MI was used, but can be reduced considerably through several modifications that are not expected to degrade registration performance (i.e., accuracy, robustness, and convenience). Thus, computational cost is not anticipated to be a barrier to clinical acceptance. Because parenchymal features deep in the brain are utilized, the technique also has the potential for intraoperative patient registration that is important for neurosurgical operations where conventional FBR is not possible or impractical (e.g., fiducial loss/movement or absence in emergency cases). However, the current approach does rely on 3DUS features and may not be successful in every surgical case. Registration after craniotomy is potentially very attractive because of its convenience and efficiency, but also carries some risk because an alternative registration may no longer be possible if the method fails. Continued evaluation of the technique in a much larger number and range of surgical cases will be critical to establishing its long-term clinical potential.
ACKNOWLEDGMENTS
Funding from the National Institutes of Health Grant No. R01 CA159324-01 and support from Philips Healthcare for the iU22 3D ultrasound system are acknowledged. The authors are also grateful to Mr. Bennet Vance from the Neukom Institute at Dartmouth College for help on CR implementation in ITK.
References
- Maurer C. R., Fitzpatrick J. M., Wang M. Y., Galloway R. L., Maciunas R. J., and Allen G. S., “Registration of head volume images using implantable fiducial markers,” IEEE Trans. Med. Imaging 16(4), 447–462 (1997). 10.1109/42.611354 [DOI] [PubMed] [Google Scholar]
- Helm P. A. and Eckel T. S., “Accuracy of registration methods in frameless stereotaxis,” Comput. Aided Surg. 3, 51–56 (1998). 10.3109/10929089809148129 [DOI] [PubMed] [Google Scholar]
- Ammirati M., Gross J. D., Ammirati G., and Dugan S., “Comparison of registration accuracy of skin-and bone-implanted fiducials for frameless stereotaxis of the brain: A prospective study,” Skull Base 129(3), 125–130 (2002). 10.1055/s-2002-33458-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. et al. , “Comparison and evaluation of retrospective intermodality brain image registration techniques,” J. Comput. Assist. Tomogr. 21(4), 554–566 (1997). 10.1097/00004728-199707000-00007 [DOI] [PubMed] [Google Scholar]
- Eggers G., Kress B., and Mühling J., “Fully automated registration of intraoperative computed tomography image data for image-guided craniofacial surgery,” J. Oral Maxillofac Surg. 66(8), 1754–1760 (2008). 10.1016/j.joms.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Shamir R. R., Freiman M., Joskowicz L., Spektor S., and Shoshan Y., “Surface-based facial scan registration in neuronavigation procedures: A clinical study,” J. Neurosurg. 111(6), 1201–1206 2009. 10.3171/2009.3.JNS081457 [DOI] [PubMed] [Google Scholar]
- Raabe A., Krishnan R., Wolff R., Hermann E., Zimmermann M., and Seifert V., “Laser surface scanning for patient registration in intracranial image-guided surgery,” Neurosurgery 50(4), 797–803 (2002). 10.1097/00006123-200204000-00021 [DOI] [PubMed] [Google Scholar]
- Marmulla R., Mühling J., Wirtz C. R., and Hassfeld S., “High-resolution laser surface scanning for patient registration in cranial computer-assisted surgery,” Minim Invasive Neurosurg. 47, 72–78 (2004). 10.1055/s-2004-818471 [DOI] [PubMed] [Google Scholar]
- Schlaier J., Warnat J., and Brawanski A., “Registration accuracy and practicability of laser-directed surface matching,” Comput. Aided Surg. 7, 284–290 (2002). 10.3109/10929080209146037 [DOI] [PubMed] [Google Scholar]
- Lee J. D, Huang C. H, Wang S. T, Lin C. W, and Lee S. T, “Fast-MICP for frameless image-guided surgery,” Med. Phys. 37(9), 4551–4559 (2010). 10.1118/1.3470097 [DOI] [PubMed] [Google Scholar]
- Miga M. I., Sinha T. K., Cash D. M., Galloway R. L., and Weil R. J., “Cortical surface registration for image-guided neurosurgery using laser-range scanning,” IEEE Trans. Med. Imaging 22(8), 973–985 (2003). 10.1109/TMI.2003.815868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A., Thompson R. C., Dumpuri P., Dawant B. M., Galloway R. L., Ding S., and Miga M. I., “Laser range scanning for image-guided neurosurgery: Investigation of image-to-physical space registrations,” Med. Phys. 35(4), 1593–1605 (2008). 10.1118/1.2870216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besl P. J. and McKay N. D., “A method for registration of 3-d shapes,” IEEE Trans. Pattern Anal. Mach. Intell. 14(2), 239–256 (1992). 10.1109/34.121791 [DOI] [Google Scholar]
- Mascott C. R., Sol J. C., Bousquet P., Lagarrigue J., Lazorthes Y., and Lauwers-Cances V., “Quantification of true in vivo (application) accuracy in cranial image-guided surgery: Influence of mode of patient registration,” Neurosurgery 59(1), ONS-146–ONS-156 (2006). 10.1227/01.NEU.0000220089.39533.4E [DOI] [PubMed] [Google Scholar]
- Pluim J. P. W., Maintz J. B. A., and Viergever M. A., “Mutualinformation-based registration of medical images: A survery,” IEEE Trans. Med. Imaging 22(8), 986–1004 (2003). 10.1109/TMI.2003.815867 [DOI] [PubMed] [Google Scholar]
- I. A.RasmussenJr., Lindseth F., Rygh O. M., Berntsen E. M., Selbekk T., Xu J., Hernes T. A. N., Harg E., Haberg A., and Unsgaard G., “Functional neuronavigation combined with intra-operative 3D ultrasound: Initial experiences during surgical resections close to eloquent brain areas and future directions in automatic brain shift compensation of preoperative data,” Acta Neurochir. 149, 365–378 (2007). 10.1007/s00701-006-1110-0 [DOI] [PubMed] [Google Scholar]
- Ji S., Wu Z., Hartov A., Roberts D. W., and Paulsen K. D., “Mutual-information-based patient registration using intraoperative ultrasound in image-guided neurosurgery,” Med. Phys. 35(10), 4612–4624 (2008). 10.1118/1.2977728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney G. P., Blackall J. M., Hamady M. S., Sabharval T., Adam A., and Hawkes D. J., “Registration of freehand 3D ultrasound and magnetic resonance liver images,” Med. Image Anal. 8, 81–91 (2004). 10.1016/j.media.2003.07.003 [DOI] [PubMed] [Google Scholar]
- Blackall J. M., Rueckert D., C. R.MaurerJr., Penney G. P., Hill D. L. G., and Hawkes D. J., “An image registration approach to automated calibration for freehand 3D ultrasound,” Medical Image Computing and Computer-Assisted Intervention, edited by Delp S. L., DiGioia A. M., and Jaramaz B., Lecture Notes in Computer Science Vol. 1935 (Springer-Verlag, Berlin Heidelberg, 2000), pp. 462–471. [Google Scholar]
- Roche A., Pennec X., Malandain G., and Ayache N., “Rigid registration of 3D ultrasound with mr images: A new approach combining intensity and gradient information,” IEEE Trans. Med. Imaging 20(10), 1038–1049 (2001). 10.1109/42.959301 [DOI] [PubMed] [Google Scholar]
- Milko S., Melvær E. L., Samset E., and Kadir T., “Evaluation of bivariate correlation ratio similarity metric for rigid registration of US/MR images of the liver,” Int. J. Comput. Assist. Radiol. Surg. 4, 147–155 (2009). 10.1007/s11548-009-0285-2 [DOI] [PubMed] [Google Scholar]
- Wu Z., Paulsen K. D., and J. M.SullivanJr., “Model initialization and deformation for automatic segmentation of T1-weighted brain MRI data,” IEEE Trans. Biomed. Eng. 52, 1128–1131 (2005) 10.1109/TBME.2005.846709 [DOI] [PubMed] [Google Scholar]
- Ji S., Roberts D. W., Hartov A., and Paulsen K. D., “Real-time interpolation for true 3-dimensional ultrasound image volumes,” J. Ultrasound Med. 30, 241–250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartov A., Paulsen K. D., Ji S., Fontaine K., Furon M., Borsic A., and Roberts D. W., “Adaptive spatial calibration of a 3D ultrasound system,” Med. Phys. 37(5), 2121–2130 (2010). 10.1118/1.3373520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartov A., Roberts D. W., and Paulsen K. D., “A comparative analysis of coregistered ultrasound and magnetic resonance imaging in neurosurgery,” Neurosurgery 62(3 Suppl 1), 91–99 (2008). 10.1227/01.neu.0000317377.15196.45 [DOI] [PubMed] [Google Scholar]
- Roche A., Malandain G., Pennec X., and Ayache N., “The correlation ratio as a new similarity measure for multimodal image registration,” in Proceedings of Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science Vol. 1496 (Springer-Verlag, Berlin Heidelberg, 1998), pp. 1115–1124.
- Insight Segmentation and Registration Toolkit. (URL: http://www.itk.org/) [DOI] [PubMed]
- Ji S., Roberts D. W., Hartov A., and Paulsen K. D., “Combining multiple volumetric true 3D ultrasound volumes through re-registration and rasterization,” in Proceedings of Medical Image Computing and Computer-Assisted Intervention, Part I, edited by Yang G.-Z.et al. , Lecture Notes in Computer Science Vol. 5761 (Springer-Verlag, Berlin Heidelberg, 2009), pp. 795–802. [DOI] [PMC free article] [PubMed]
- Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Mercier L., Del Maestro R. F., Petrecca K., Kochanowska A., Drouin S., Yan C. X. B., Janke A. L., Chen S. J., and Collins D. L., “New prototype neuronavigation system based on preoperative imaging and intraoperative freehand ultrasound: System description and validation,” Int. J. Comput. Assist. Radiol. Surg. 6(4), 507–522 (2011). 10.1007/s11548-010-0535-3 [DOI] [PubMed] [Google Scholar]
- Reinertsen I., Lindseth F., Unsgaard G., and Collins D. L., “Clinical validation of vessel-based registration for correction of brain-shift,” Med. Image Anal. 11(6), 673–684 (2007). 10.1016/j.media.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Medical Image Registration, 1st ed., edited by Hajnal J., Hill D., and Hawkes D. (CRC, CRC Press, 2001). [Google Scholar]
- Danilchenko A. and Fitzpatrick J. M., “General approach to first-order error prediction in rigid point registration,” IEEE Trans. Med. Imaging 30, 679–693 (2011). 10.1109/TMI.2010.2091513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S., Fan X., Hartov A., Roberts D. W., and Paulsen K. D., “Estimation of intraoperative brain deformation,” in Studies in Mechanobiology, Tissue Engineering and Biomaterials, edited by Payan Y. (Springer-Verlag, Berlin, 2012), Vol. 11, pp. 97–133. [Google Scholar]
- Shekhar R. and Zagrodsky V., “Mutual information-based rigid and nonrigid registration of ultrasound volumes,” IEEE Trans. Med. Imaging 21(1), 9–22 (2002). 10.1109/42.981230 [DOI] [PubMed] [Google Scholar]
- Slomka P. J., Mandel J., Downey D., and Fenster A., “Evaluation of voxel-based registration of 3-D power Doppler ultrasound and 3-D magnetic resonance angiographic images of carotid arteries,” Ultrasound Med. Biol. 27(7), 945–955 (2001). 10.1016/S0301-5629(01)00387-8 [DOI] [PubMed] [Google Scholar]