Introduction
One of the defining features of eukaryotic cells is the use of organelles to compartmentalize biochemical activities. This compartmentalization permits the separation of cellular functions that may not otherwise co-exist, such as proteolysis (which can occur in proteasomes and lysosomes) and protein synthesis (which occurs in ribosomes). There are numerous additional examples of organelles being utilized to separate biochemical activities, but one area where this idea has only recently gained attention is in the operation of the innate immune system (Barton and Kagan, 2009). The mammalian innate immune system is comprised of several families of microbial detection receptors (Akira et al., 2006), classically called Pattern Recognition Receptors (PRRs) (Janeway, 1989), which share little sequence similarity. Because different PRRs are located in different subcellular compartments (Barton and Kagan, 2009), they have the potential to provide fascinating insight into how signal transduction pathways operate within the general infrastructure of a eukaryotic cell. This Perspective will identify the organelles that permit innate immune signal transduction, and discuss the means by which these sites are “chosen”. I suggest that the innate immune system is designed to dissociate the sites of microbial detection from the sites of signal transduction. While the receptors of the innate immune system determine the sites of microbial detection, the sites of signaling are dictated by the localization of distinct membrane-bound proteins called sorting adaptors. A central conclusion of this essay is that PRRs are dependent on the cellular trafficking machinery to be delivered to both the sites of immune-surveillance and the sites of innate immune signal transduction. The regulators of PRR transport can therefore be categorized as either biosynthetic trafficking factors or microbe-inducible trafficking factors, the latter of which has only recently emerged as a critical control element for innate immune signal transduction.
Classifying microbe detection receptors based on their ability to induce inflammation
Some PRRs bind microbial products directly, such as the mannose receptor, mannose-binding lectin (MBL), C-reactive protein, serum amyloid protein and the complement system (Janeway and Medzhitov, 2002). A defining feature of these receptors is that they function primarily in the extracellular spaces of multicellular eukaryotes to promote the phagocytosis and/or killing of microbes they encounter. However, these proteins are not inflammation-inducing receptors in the sense that they have no intrinsic ability to induce the expression of inflammatory cytokines. This latter task is accomplished by a second class of PRRs that link microbial detection to the expression and/or secretion of chemokines, cytokines, interferons (IFNs) and other inflammatory mediators. This class includes the Toll-like Receptors (TLRs), NOD-like Receptors (NLRs), RIG-I like Receptors (RLRs), AIM2-like Receptors (ALRs) and the C-type Lectin Receptors of the Dectin family (Brown and Gordon, 2001; Inohara et al., 1999; Medzhitov et al., 1997; Unterholzner et al., 2010; Yoneyama et al., 2004). Since the purpose of this Perspective is to discuss the sites within mammalian cells that permit innate immune signal transduction, I will focus on this latter group of “signaling” PRRs, whose members can be found on multiple organelles.
The aforementioned families of signaling PRRs share little sequence homology, and as such, bioinformatic analyses alone would not permit the grouping of these receptors into a superfamily. However, detailed functional analysis of individual family members revealed themes that transcend the structural characteristics of each protein family. For example, all signaling PRRs exhibit the ability to recognize products of microbial metabolism (Brown et al., 2003; Girardin et al., 2003; Pichlmair et al., 2006; Poltorak et al., 1998; Unterholzner et al., 2010). These products, classically defined as Pathogen Associated Molecular Patterns (PAMPs) (Janeway, 1989), are largely unique to the microbial world and are often common to broad classes of microbes. Examples of PAMPs include bacterial lipopolysaccharide (LPS), lipoproteins, flagellin, double stranded RNA, unmethylated CpG-containing DNA, and RNA that contains a triphosphorylated 5’ end. For the most part, these molecular features are absent from mammalian cells, thus allowing their detection to indicate the presence of a microorganism (Janeway and Medzhitov, 2002). While many PRRs recognize PAMPs directly (Kang and Lee, 2011; Kowalinski et al., 2011; Latz et al., 2004), some require the assistance of accessory molecules that themselves are high affinity PAMP binding proteins (Gioannini et al., 2004). The second feature that is common to all signaling PRRs is their ability to induce the activation of proinflammatory transcription factors such as AP-1, NF-κB and various IFN regulatory factors (IRFs) (Kumar et al., 2011). The combinatorial actions of these transcription factors determine the precise gene expression program that is induced by a given PRR. The third common feature of these PRRs is that none of them are signaling enzymes, yet that all induce an enzyme-dependent signaling response that activates inflammatory transcription factors. Innate immune signaling pathways therefore enlist the help of adaptor proteins to link active receptors to the downstream kinases and ubiquitin ligases to promote inflammation.
Organelles that permit pathogen detection in the innate immune system
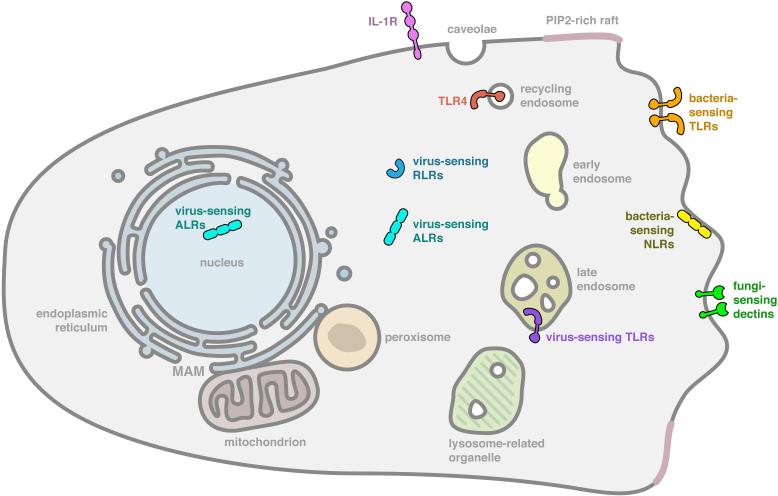
The fundamental purpose of the innate immune system is to detect the presence of microbes and their products. This rationale has been used to explain the diversity of PAMPs that are detected by PRRs (Medzhitov, 2009). While we have an increasingly detailed understanding of the molecular interactions that occur between PAMPs and their receptors (Jin and Lee, 2008), insight into where within mammalian cells these interactions occur has only begun to emerge. In fact, it is now clear that most membrane-bound organelles harbor one or more PRRs (Figure 1). For example, the plasma membrane is occupied by the transmembrane domain-containing receptors that either detect bacterial products (e.g. TLRs 1, 2, 4, 5 and 6) or fungal products (Dectins 1 and 2) (Brown and Gordon, 2001; Du et al., 1999; Gewirtz et al., 2001; Ozinsky et al., 2000; Underhill et al., 1999). Interestingly, while the NLR family is generally believed to encode cytosolic proteins, NOD1 and NOD2 have been localized to the inner leaflet of the plasma membrane (Barnich et al., 2005; Travassos et al., 2010). There are two features that link all plasma membrane localized PRRs. First, they detect microbial cell surface components, such as LPS (e.g. TLR4), lipoproteins (TLR2/1 and TLR2/6 heterodimers), flagellin (TLR5), peptidoglycans (NOD1 and NOD2) and fungal cell wall components (Dectins 1 and 2) (Akira et al., 2006). It seems reasonable to propose that the localization of these receptors to the cell surface is linked to the ligands that they detect, as the most rapid means of detecting microbial cell surface components would be to localize PRRs that detect said components to the surface of mammalian cells. The second feature that links all plasma membrane localized PRRs is that they have the ability to be recruited to sites of phagocytosis (Brown and Gordon, 2001; Ozinsky et al., 2000; Tissieres et al., 2008; Underhill et al., 1999). In some instances (e.g. Dectin family members), these receptors direct the phagocytic process (Goodridge et al., 2012). Interestingly, at least some of these receptors (e.g. TLR2) can be recruited to phagosomes containing any type of cargo (Underhill et al., 1999). Thus, it is likely that the function of phagosomal recruitment is to determine if microbial cargo is present within the phagosome-to-be (Underhill et al., 1999). Both of these features of plasma membrane localized PRRs ensure rapid detection of microbial encounters in the extracellular spaces.
Figure 1. Organelles that serve as sites of microbial detection.
Illustrated is a typical mammalian macrophage that harbors innate immune receptors in specific subcellular locations. The subcellular sites indicated are the sites of receptor residence prior to any microbial encounter. As such, these are the most likely sites of microbial detection. At the plasma membrane are bacterial sensing TLRs such as TLRs 1, 2, 4, 5, and 6, and Dectins 1 and 2. Within the endosomal network is TLR4 and the virus sensing TLRs 3, 7, 8, 9 and 13. The RLRs RIG-I and MDA5, and the ALRs AIM2 are present in the cytosol, and the ALR IFI16 is present in the nucleus. Bacterial sensing NLRs NOD1 and NOD2 are present in the inner leaflet of the plasma membrane. Note, that the larger family of inflammasome activating NLRs are not indicated, but many are predicted to be cytosolic.
The endosomal network also represents a site of PRR residence (Figure 1). For example, in addition to its plasma membrane locale, TLR4 can be found in the early and recycling endosomes of macrophages (Husebye et al., 2010; Kagan et al., 2008). TLRs 3, 7, 8, 9 and 13 are also found in late endosomal compartments (Barton and Kagan, 2009). TLR9 is the best characterized in this regard, and it is found in late endosomes harboring phosphatidylinositol (PI)-3,5 bisphosphate (PI(3,5)P2) and in lysosomes-related organelles (LROs) (Sasai et al., 2010). With the exception of TLR4, the unifying feature of the endosomal TLRs is their ability to detect microbial nucleic acids such as double stranded RNA (e.g. TLR3), single stranded RNA (TLRs 7 and 8), unmethylated CpG DNA (TLR9) or ribosomal RNA (TLR13) (Alexopoulou et al., 2001; Diebold et al., 2004; Hemmi et al., 2000; Hidmark et al., 2012; Oldenburg et al., 2012). Since nucleic acids are hidden from host receptors until the microbe is degraded, the placement of these receptors in late endosomal vesicles likely ensures rapid detection once they are released by the hydrolytic late endosomal environment. Thus, just as was argued for the plasma membrane-localized PRRs, the localization of endosomal TLRs reflects a need for rapid responsiveness to a microbial presence. The second unifying feature of the endosomal TLRs (again with the exception of TLR4) is that they are synthesized as pro-proteins whose ectodomains must be cleaved by one or more endosomal proteases to generate a signaling-competent receptor (Ewald et al., 2011; Ewald et al., 2008; Park et al., 2008).
Within the secretory pathway, there is limited evidence of PRR residence, other than during the biosynthetic trafficking of the transmembrane receptors (Ewald et al., 2008). The mechanisms by which different PRRs are delivered to post-Golgi compartments such as the plasma membrane or endosomes are only now being revealed, with the best-characterized example being that provided by the protein Unc93B1. Unc93B1 was initially proposed to not exhibit any transport function, but to rather act as a signaling protein that regulates the functions of endosomal TLRs uniquely (Tabeta et al., 2006). Recent work firmly established its primary function as a regulator of TLR trafficking (Brinkmann et al., 2007; Kim et al., 2008). Unc93B1 binds to all nucleic acid sensing TLRs after they are synthesized on the endoplasmic reticulum (ER), and stays associated with these proteins as they transit the secretory pathway to be delivered to endosomes (Brinkmann et al., 2007; Kim et al., 2008). Evidence supporting a direct transport function of Unc93B1 for TLRs was provided by studies showing that forcing Unc93B1 localization to the cell surface also results in the delivery of nucleic acid sensing TLRs to this location (Kim et al., 2008). Thus, the biosynthetic transport of nucleic acid sensing TLRs is mediated by Unc93B1. Although there is some question over whether all nucleic acid sensing TLRs occupy the same endosomal vesicles, the fact that a single chaperone (Unc93B1) delivers all these receptors to endosomes, and remains associated with them, makes it likely that there is significant overlap in their subcellular distribution.
In addition to Unc93B1, gp96 and PRAT4A regulate the TLR trafficking (Takahashi et al., 2007; Yang et al., 2007). Unlike Unc93B1, gp96 and PRAT4A regulate the transport of all TLRs. Gp96 is an ER paralog of the Hsp90 family that acts as a chaperone for multiple protein substrates, including immunoglobulins, some integrins, and TLRs (Yang et al., 2007). Macrophages deficient for gp96 are unresponsive to all TLR ligands, indicating that gp96 is required for signaling via TLRs found at both the plasma membrane and endosomes (Yang et al., 2007). Similarly, PRAT4A deficient cells are unresponsive to ligands from all TLRs, except TLR3 (Takahashi et al., 2007).
Although TLRs can be found in the ER and Golgi compartments, there is little evidence that these proteins can signal from these locations. Thus, it is likely that their residence in secretory organelles is simply a reflection of their biosynthetic trafficking path that will lead them to an organelle poised for exposure to microbial contents.
At a superficial level, it would seem redundant for a single cell to express RLRs (which detect viral RNA), NLRs (bacterial products) and TLRs (bacterial products and viral nucleic acids). However, these receptor families survey different compartments of the cell. While the TLRs survey the extracellular and lumenal endosomal networks (Barton and Kagan, 2009), the RLRs and NLRs survey the cytosol for microbial products (Kanneganti et al., 2007; Nakhaei et al., 2009). For example, the proinflammatory RLR family members RIG-I and MDA5 are soluble cytoplasmic proteins that survey this compartment for the presence of viral (and in some instances bacterial) RNA (Nakhaei et al., 2009). Likewise, most NLRs are thought to be cytosolic proteins (Kanneganti et al., 2007), although this family is poorly characterized from a cell biological perspective.
When considered in the context of an actual infection, the differential localizations of TLRs and RLRs/NLRs have functional implications. The most common type of microbe encountered by our innate immune system is likely to be non-pathogenic. These microbes have the potential to compete for nutrients of the host, but encode no sophisticated weapons of colonization or immune evasion. As such, these microbes are often recognized by PRRs that survey the extracellular and endosomal environments during the process of their removal by phagocytosis. In contrast, pathogens, almost by definition, must interact with host cells intimately (Lamkanfi and Dixit, 2009; Vance et al., 2009). For bacteria, these intimate interactions are often mediated by secreted protein toxins or specialized secretion systems such as Type III or Type IV systems that deliver effector molecules into the host cytosol to manipulate various signaling pathways (Cambronne and Roy, 2006; Galan and Wolf-Watz, 2006). In the case of viruses, these pathogens must enter the cytosol to gain access to the translation machinery of the host. Thus, a cell biological distinction between non-pathogen and pathogen is the ability of the latter to deliver itself or its information (effectors) into the host cytosol. Since all microbes are extracellular at the earliest stage of infection, all microbes should be detected by PRRs surveying the extracellular environment. In contrast, RLRs and NLRs only survey the compartment that can be accessed by a pathogen (the cytosol). The activation of these receptor families is therefore indicative of an encounter with a virulent microbe. An example of this principle comes from studies of the bacterium Listeria monocytogenes, which replicates in the cytosol of mammalian cells (Cossart, 2011). Wild type Listeria activates PRRs that survey the extracellular environment and PRRs that survey the cytosol (McCaffrey et al., 2004). In contrast, Listeria mutants that cannot enter the cytosol (and therefore mimic a non-pathogenic encounter) cannot engage cytosolic PRRs (McCaffrey et al., 2004). Thus, TLRs and Dectin family members can be considered Microbe-Detection Receptors, whereas cytoplasmic RLRs and NLRs can be considered Pathogen-Detection Receptors.
The logical consistency between where PRRs are located and where their target PAMPs are revealed to the host raises a strong argument that the primary selective pressure for diversifying receptor locale was to ensure rapid detection of microbial products. Interestingly, as will be described below, recent evidence indicates that the site of receptor-ligand interaction is not necessarily the site of innate immune signal transduction. Rather, microbe-induced trafficking events must often occur before a ligand-bound receptor can induce its cognate transcriptional response.
Signaling organelles of the innate immune system
It has become somewhat dogmatic to use the localization of TLRs as an indication of where innate immune signal transduction occurs. In this section, several lines of evidence will be presented to suggest that the site of receptor-ligand interaction does not necessarily indicate the site of signaling. Rather, many PRRs must be transported to a second location for signal transduction to occur.
The first example of a microbe-inducible trafficking event came from studies of the LPS receptor TLR4 (Poltorak et al., 1998). TLR4 is found at the plasma membrane of many mammalian cell types, but has been best characterized on phagocytes such as macrophages and dendritic cells (DCs). The immunostimulatory structure of LPS is lipid A, a hydrophobic motif consisting of acyl chains of varying lengths and numbers (depending on the bacteria) (Gioannini and Weiss, 2007). Lipid A is extracted from the outer membrane of gram-negative bacteria by the LPS-binding protein (LBP) by a process facilitated by albumin (Gioannini et al., 2005). LPS is then transferred to a second LPS-binding protein called CD14 (Gioannini et al., 2004; Gioannini et al., 2005), which can exist as a soluble extracellular protein or a GPI-anchored protein attached to the outer leaflet of the plasma membrane (Wright et al., 1990). CD14 then transfers LPS to a high-affinity LPS-binding protein called MD-2, which forms a stable complex with TLR4 (Gioannini et al., 2004; Gioannini et al., 2005). Recognition of LPS by TLR4-MD-2 leads to the dimerization of the Toll-IL-1 Receptor (TIR) domains present in the cytosolic tail of the receptor, which activates inflammatory signal transduction (Park et al., 2009).
TLR4 signaling is initiated when its dimerized TIRs engage two pairs of TIR-domain containing adaptor proteins that activate distinct transcriptional responses. The adaptors MyD88 and TIRAP activate signaling events that lead to NF-κB and AP-1 dependent expression of inflammatory cytokines such as IL-1β, IL-6 and IL-12 (Horng et al., 2002; Kawai et al., 1999; Yamamoto et al., 2002). The adaptors TRAM and TRIF activate signaling events that lead to the IRF3-dependent expression of Type I IFNs (Yamamoto et al., 2003a; Yamamoto et al., 2003b). Studies using the endocytosis inhibitor dynasore (which inactivates dynamin GTPases) revealed that LPS-induced TLR4 endocytosis was required for activation of the TRIF pathway to Type I IFNs (Kagan et al., 2008). Subsequent work on the IFN-inducing E3 ubiquitin ligase TRAF3 provided a biochemical explanation for this phenotype. TLR4 endocytosis is necessary for the attachment of lysine-63 linked ubiquitin chains to TRAF3, a process that is necessary for its ability to propagate TRIF signaling (Tseng et al., 2010). In contrast, endocytosis was not required for TLR4 to induce MyD88-dependent signaling in response to LPS or gram-negative bacteria (Kagan et al., 2008; Zanoni et al., 2011). However, in order to induce MyD88-dependent responses, TLR4 must be first transported to subdomains of the cell surface called lipid rafts (Triantafilou et al., 2002). These results suggest that the initial site of LPS detection by TLR4 (the plasma membrane) is not the site of signaling. Rather TLR4 must first be mobilized into lipid rafts to induce MyD88-dependent signal transduction, and then internalized into endosomes to induce TRIF-dependent signaling. Interestingly, TLR2 must also be delivered to lipid rafts to promote MyD88-dependent signaling responses (Triantafilou et al., 2006). These data suggest that a common feature of PRRs at the cell surface is the spatial separation of sites of ligand binding and signal transduction.
Studies of LPS-inducible TLR4 transport revealed that the aforementioned LPS-binding protein CD14 controls these events. CD14 is required for the delivery of TLR4 into lipid rafts after LPS binding (Triantafilou et al., 2002), and it is required to deliver TLR4 to endosomes (Zanoni et al., 2011). Interestingly, the mechanism by which TLR4 is delivered to endosomes does not require TLR4 signaling, nor does it require the TIR domain containing adaptor proteins (Zanoni et al., 2011). Thus, CD14 induces a novel LPS response pathway that leads to the rapid endocytosis of TLR4, which is necessary for TRIF-dependent signaling. This third LPS response pathway is a calcium-dependent process that involves several general regulators of cellular trafficking, such as the tyrosine kinase Syk, the ITAM-containing adaptors DAP12 and FcεRγ, and PLCγ2 (Chiang et al., 2012; Zanoni et al., 2011). These data indicate that TLR4 does not control all cellular responses to LPS; rather it controls all transcriptional responses to LPS. Since the transcriptional responses induced by LPS require CD14-dependent transport of TLR4 into lipid rafts and endosomes, CD14 can be considered a master regulator of TLR4-mediated signal transduction. Thus, for MyD88-dependent signaling from the plasma membrane, lipid rafts represent the subcellular site where TLR4- and TLR2-dependent signaling platforms can be assembled (Figure 2).
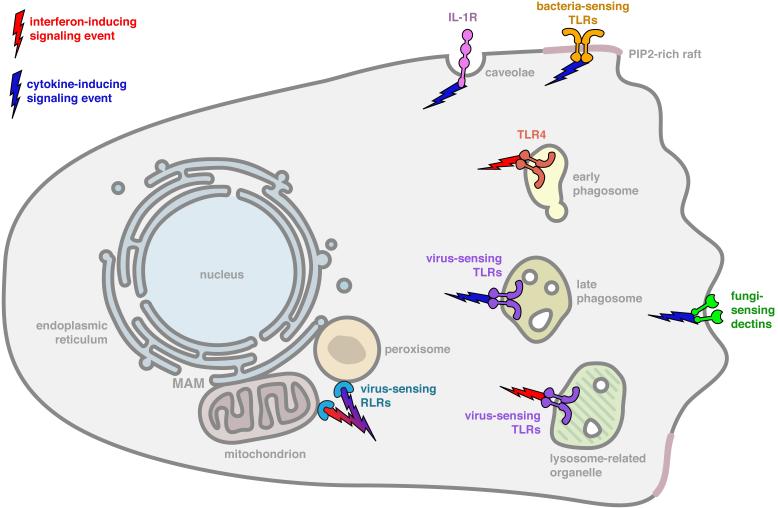
Figure 2. Signaling organelles of the innate immune system.
Illustrated is a typical mammalian macrophage that harbors innate immune receptors in specific subcellular locations. The subcellular sites indicated are the sites of receptor residence after microbe-induced receptor transport. The legend indicates the type of signaling pathway activated from each location. Note in most cases, the sites of receptor signaling differ from the sites of microbial detection, as illustrated in Figure 1. At the plasma membrane rafts are bacterial sensing TLRs such as TLRs 1, 2, 4, 5, and 6. Within early endosomes is TLR4. The virus sensing TLRs 3, 7, 8, 9 and 13 may signal from late endosomes and lysosomes-related organelles, although this has been only formally demonstrated for TLR9. The RLRs RIG-I and MDA5 signal from mitochondria and peroxisomes that are docked at the MAM. Note, that some of the receptors indicated in Figure 1 are not illustrated here, because not enough information is available to define their sites of signaling.
Recent work on the adaptor protein TRAM, which acts with TRIF to induce TLR4-dependent IFN expression, suggests that signaling probably occurs in early endosomes (or early phagosomes) (Doyle et al., 2012; Husebye et al., 2010; Palsson-McDermott et al., 2009). Evidence supporting this statement comes from work on a splice variant of TRAM called TAG (Doyle et al., 2012; Palsson-McDermott et al., 2009). Whereas TRAM functions to promote TRIF signaling, TAG functions to inactivate these signaling events (Doyle et al., 2012; Palsson-McDermott et al., 2009). In contrast to the proinflammatory TRAM protein, which is located on early endosomes and the plasma membrane, the TAG splice variant is located on late endosomes. Since the inhibitor of this pathway is located on late endosomes, it is likely the early endosomal compartments are the primary site of TRAM-TRIF dependent signal transduction. Thus, for TRIF-dependent signaling, early endosomes (or phagosomes) are the subcellular site where TLR4-dependent signaling platforms can be assembled (Figure 2).
A second example of PRRs that must be transported after ligand recognition to induce signal transduction is provided the CpG DNA sensor TLR9 (Hemmi et al., 2000). In most phagocytic cells, TLR9 signaling activates the expression of inflammatory cytokines but does not induce Type I IFN expression (Honda et al., 2005). A notable exception to this principle comes from studies of plasmacytoid dendritic cells (pDCs), within which TLR9 induces the expression of both cytokines and IFNs (Honda et al., 2005). Thus, in pDCs, TLR9-dependent transcriptional responses resemble those activated by TLR4. Also like TLR4, the site of ligand binding by TLR9 can be dissociated from the site of signal transduction, at least in the case of IFN expression (Sasai et al., 2010). When TLR9 is present within late endosomes that contain PI(3,5)P2, it is capable of inducing the expression of NF-κB-dependent cytokines such as IL-6 and IL-12 (Sasai et al., 2010) (Figure 2). However, the ability to induce IFN expression requires the transport of TLR9 to lysosomes-related organelles (LROs) (Figure 2). Transport of TLR9 to LROs is mediated by a protein complex called adaptor protein-3 (AP-3) (Sasai et al., 2010). Cells lacking AP-3 are unable to deliver TLR9 to LROs and consequently, TLR9 is only capable of inducing the expression of cytokines; no IFNs are expressed (Sasai et al., 2010). It remains unclear if PI(3,5)P2-rich endosomes are the site where TLR9 first detects its DNA ligand, and as such, it is not known if microbe-induced trafficking is required for all TLR9-dependent signaling events, or just signaling events that lead to IFN expression. Nevertheless, it is clear that in the case of TLRs 4 and 9, the ability to induce IFN expression requires the movement of the receptor to subcellular location distinct from the initial site of ligand binding.
The need for receptor movement to a second subcellular location to activate signal transduction extends beyond the endosomal network. The RLRs RIG-I and MDA5 survey the cytosolic compartment for the presence of viral RNA (Nakhaei et al., 2009). RIG-I detects structures that are usually absent from host RNAs but common to viral RNAs, such as those containing 5’ triphosphate groups, short double stranded regions and/or uridine-rich 3’ regions (Pichlmair et al., 2006; Saito et al., 2008; Uzri and Gehrke, 2009). The precise ligand detected by MDA5 remains unclear, but it is generally believed that this PRR recognizes long sequences of double stranded RNA (Kato et al., 2008; Pichlmair et al., 2009). Despite the differences in RNA structures detected by these RLRs, a common mechanism of signaling pathway activation exists. RLR activation occurs via the sequential binding of RNA and unanchored lysine-63 linked polyubiquitin chains (Jiang et al., 2012). Polyubiquitin binding by RLRs promotes their oligomerization, which somehow leads to the activation of inflammatory cytokines and IFNs.
There are no clear “rules” to explain when and where during the lifecycle of viruses their RNA is exposed to the cytosol. As such, it is unknown exactly where in the cell RLRs first detect viral RNA. Since the lifecycles of RNA viruses are incredibly diverse, it is likely that RLRs detect viral RNA present in many locations. Once viral RNA is recognized, RLRs must engage an adaptor protein called MAVS, which is a tail-anchored protein that is located on the limiting membranes of mitochondria, peroxisomes and mitochondria-associated ER membranes (MAM) (Dixit et al., 2010; Horner et al., 2011; Seth et al., 2005). Thus, while RLRs probably have the ability to detect RNA in many locations of the cell, these PRRs must be transported to the aforementioned organelles to promote MAVS-dependent signaling (Figure 2). Recent work indicates that upon binding to viral RNA, RIG-I forms a complex with the E3 ubiquitin ligase TRIM25 and the cytosolic chaperone 14-3-3ε (Gack et al., 2007; Liu et al., 2012). Whereas in wild type cells, RNA-binding likely causes the transport of RIG-I to membrane fractions that likely contain MAM/mitochondria and peroxisomes (Dixit et al., 2010; Horner et al., 2011), this transport event is defective in cells lacking 14-3-3ε (Liu et al., 2012). Thus, like the TLRs, the subcellular sites of RLR-ligand interaction can be dissociated from the sites of signal transduction, and the movement between these two locations is a regulated event.
A final example of microbe-induced receptor transport comes from recent work on the innate immune responses to intracellular (i.e. cytosolic or nuclear) DNA. While the identity of the sensor(s) of intracellular DNA remains unclear, ample evidence exists to support a role for the endoplasmic reticulum (ER) localized protein STING (Ishikawa and Barber, 2008; Ishikawa et al., 2009). STING has the ability to bind both the IFN-inducing transcription factor IRF3 and its upstream kinase TBK1 (Tanaka and Chen, 2012). The ability of STING to bind both of these factors probably facilitates IRF3 phosphorylation, which leads to its translocation into the nucleus where Type I IFN expression can be induced. The importance of STING in antiviral innate immunity is underscored by findings that STING-deficient cells are unable to induce Type I IFN or IFN Stimulated Gene (ISG) expression in response to numerous DNA viruses and some bacteria (Ishikawa and Barber, 2008; Ishikawa et al., 2009). STING has been proposed to serve as either an adaptor protein that is engaged by upstream DNA-binding receptors, or a direct sensor of bacterial cyclic dinucleotides (Burdette et al., 2011; Ishikawa and Barber, 2008; Ishikawa et al., 2009). Thus, for the purpose of this discussion, STING can be considered a “receptor”. Upon DNA transfection or viral infection, STING will move from the ER to poorly characterized vesicles that contain the IRF3 kinase TBK1 (Ishikawa et al., 2009; Saitoh et al., 2009). Blocking the transport of STING either genetically or pharmacologically prevents the expression of Type I IFNs and ISGs in response to transfected DNA (Ishikawa et al., 2009; Saitoh et al., 2009). While the mechanism of DNA-induced STING transport remains unclear, these data suggest that like the cytosolic sensors of viral RNA (RLRs), the cellular machinery involved in DNA detection must also be transported to a second subcellular site to induce signal transduction.
Recent work on Herpes Simplex Virus (HSV) infections suggest that STING is not the only protein that must be inducibly transported for innate immune signaling to occur. HSV is an enveloped virus that injects genomic DNA into the host nucleoplasm. This nuclear viral DNA then activates innate immune signaling pathways that depend on STING (Orzalli et al., 2012). The protein IFI16 is a member of the newest family of PRRs, called AIM2-like Receptors (ALRs) (Unterholzner et al., 2010). ALRs detect viral DNA, with the founding family member AIM2 functioning in the cytosol to induce inflammasome activation after DNA sensing (Rathinam et al., 2010). IFI16 recognizes HSV DNA in the host cell nucleus and is required for the STING-dependent expression of IFNs and ISGs after infection (Li et al., 2012; Orzalli et al., 2012). This suggests that the site of HSV DNA detection by IFI16 (nucleus) is distinct from the site of signaling (cytosol). It is therefore possible that IFI16 (or some other nuclear factor) is transported to the cytosol by a specific chaperone to induce STING/TBK1-mediated gene expression.
To conclude this section, it is worth noting that in most/all of the examples listed above, it is likely that both the receptor and its microbial ligand are transported to a signaling-competent organelle. How can one distinguish the importance of receptor transport from ligand transport for the induction of cytokine and antiviral gene expression? In this regard, studies of TLR4 in CD14-deficient macrophages have proved informative. In wild type macrophages, both LPS and TLR4 will be internalized into endosomes that are competent for TRIF-dependent signal transduction. CD14-deficient macrophages, in contrast, internalize neither LPS nor TLR4. Interestingly, CD14-deficient macrophages are able to internalize LPS-coated beads (or gram-negative bacteria), but they will not permit the endocytosis of TLR4 (Zanoni et al., 2011). Under these conditions, TRIF signaling does not occur. These data therefore provide definitive proof that the receptor itself (TLR4) must be delivered to endosomes in order to initiate TRIF signaling.
Molecules that identify signaling organelles of the innate immune system
The above examples illustrated that several distinct families of PRRs must be delivered to an organelle distinct from the site of ligand binding to promote signal transduction. In this section, several lines of evidence will be presented to indicate that the site of signal transduction is defined by a structurally diverse set of membrane proteins called sorting adaptors.
Generally, sorting adaptors are defined as signaling proteins that are located at the site of signal transduction prior to any microbial encounter. In contrast, upstream receptors and downstream signaling proteins are not often found at the sites of signaling prior to microbial detection. Sorting adaptors are therefore unique in that they serve as a cell biological “landmark” that identifies the subcellular site of signal transduction. Microbe-inducible delivery of a PRR to an organelle harboring the correct sorting adaptor leads to the recruitment of downstream regulatory proteins to activate signal transduction. The prepositioning of sorting adaptors at the sites of signaling is important, since mislocalization of these adaptors to different subcellular compartments renders the innate immune signaling pathway defective.
The proteins TIRAP and TRAM represent the known sorting adaptors in the TLR system (Kagan and Medzhitov, 2006; Kagan et al., 2008). As described above, these adaptors function together with other TIR-domain containing proteins to promote TLR4 signaling. TIRAP acts together with MyD88 to induce NF-κB-dependent cytokine expression downstream of plasma membrane-localized TLR4 and TLR2 (Horng et al., 2002; Yamamoto et al., 2002). TRAM acts with TRIF to induce IRF3-dependent Type I IFN expression downstream from endosomal TLR4 (Yamamoto et al., 2003b). Whereas the upstream receptors must rely on microbe-induced trafficking factors for their delivery to lipid rafts and endosomes to induce signal transduction (Triantafilou et al., 2006; Triantafilou et al., 2002; Zanoni et al., 2011), the sorting adaptors TIRAP and TRAM are positioned at these locations at steady state (Kagan and Medzhitov, 2006; Kagan et al., 2008). TIRAP localizes to PI(4,5)P2-rich regions of the plasma membrane, such as membrane ruffles and lipid rafts by means of an N-terminal phosphoinositide-binding motif that selectively binds to PI(4,5)P2 (Kagan and Medzhitov, 2006). TRAM is localized to the plasma membrane and early endosomes through the actions of a bipartite localization domain consisting of an N-terminal myristoylation sequence and an adjacent phosphoinositide-binding domain (Kagan et al., 2008; Rowe et al., 2006). Mutations that abolish the integrity of the localization domains in TIRAP or TRAM render these proteins cytosolic (Kagan and Medzhitov, 2006; Kagan et al., 2008; Rowe et al., 2006). Under these conditions, TLR4 signaling is defective (Kagan and Medzhitov, 2006; Kagan et al., 2008; Rowe et al., 2006), even though these conditions do not prevent CD14-dependent transport of TLR4 (Zanoni et al., 2011). Thus, microbe-induced transport of TLR4 is not sufficient for signaling to occur. The sorting adaptors must be localized in the appropriate organelle in order to detect the receptor and assemble a platform for signal transduction.
Recent work indicates that although TRAM functions from endosomes to assemble a signaling platform for TLR4-dependent TRIF signaling, it has a distinct function at the plasma membrane. From this latter location, TRAM facilitates the signaling events activated by the Interleukin-18 Receptor (IL-18R), a TLR-superfamily member (Ohnishi et al., 2012). These results provide a potential explanation for why TRAM is located in two subcellular locations—it has distinct sorting adaptor functions from each. Additional work remains to be done to determine if a cytokine-induced trafficking event is needed to deliver IL-18R to the as-yet undefined subdomain of the plasma membrane that harbors TRAM. In addition to the IL-18R, the better understood IL-1R (another TLR superfamily member) may use a sorting adaptor to signal from the plasma membrane. Binding of the IL-1R to its cytokine ligand IL-1β induces the transport of this receptor to a subdomain of the plasma membrane called caveolae (Blanco et al., 2008; Liu and Anderson, 1995). It is from caveolae that MyD88-dependent cytokine expression is induced. Relative to lipid rafts and sites of phagocytosis, caveolae contain low (but detectable) levels of PI(4,5)P2 (Watt et al., 2002). Thus, the sorting adaptor TIRAP should not be concentrated in caveolae, which may explain why TIRAP is not required for IL-1R signaling (Horng et al., 2002; Yamamoto et al., 2002). Rather, a distinct sorting adaptor may assemble platforms for IL-1R signaling from caveolae. Detailed biochemical analysis suggests that the IL-1R accessory protein (IL-1RacP) is a core component of the IL-1R signaling complex (Brikos et al., 2007). Although this protein has some role in ligand binding, it most impressive function is to act together with MyD88 to induce signal transduction (Brikos et al., 2007; Korherr et al., 1997; Radons et al., 2002). In this regard, IL-1RacP is similar to TIRAP and TRAM, in that it acts with a downstream adaptor to assemble a signaling platform in a specific subcellular location. For this reason, I speculate that the plasma membrane contains at least three sites of innate immune/cytokine signaling and each site contains a unique sorting adaptor. The three sites are 1) PI(4,5)P2-rich lipid rafts occupied by TIRAP, 2) PI(4,5)P2-poor caveolae occupied by IL-1RacP and 3) an unknown compartment occupied by TRAM.
In addition to the TLR and cytokine receptors, the RLR family also appears to utilize sorting adaptors to define the site of signaling and activate a platform for signal transduction. As discussed earlier, RLR signaling depends on the MAVS adaptor (Kumar et al., 2006; Sun et al., 2006), which is located on the limiting membranes of mitochondria, peroxisomes and the MAM (Dixit et al., 2010; Horner et al., 2011; Seth et al., 2005). MAVS localization to each of these compartments is important for signaling, and it is thought that the MAM provides a scaffold where peroxisomes and mitochondria dock to create a signaling platform for antiviral responses. The MAVS-dependent signaling platform depends on its ability to oligomerize into prion-like structures that are potent inducers of antiviral gene expression (Hou et al., 2011). Mislocalizing MAVS to the cytosol renders cells incapable of inducing RLR signaling (Dixit et al., 2010; Seth et al., 2005), even though these conditions do not prevent 14-3-3ε-dependent delivery of RLR to these signaling organelles (Liu et al., 2012). Thus, as was observed for TLR4, microbe-induced transport of RLRs is not sufficient for signal transduction. The sorting adaptor MAVS must be localized in the appropriate organelle(s) in order to detect the receptor and assemble a platform for signal transduction.
It should be noted that most biochemical studies on the localization and signaling functions of MAVS cannot distinguish its actions on the aforementioned organelles, as “mitochondria-enriched membrane fractions” are highly contaminated with peroxisomes and MAM (Horner et al., 2011). Additional work is therefore needed to determine if common or distinct mechanisms of signaling platform assembly occur on each MAVS-containing compartment. This issue of compartment-specific signaling is important, since MAVS induces the expression of different antiviral factors when localized to either mitochondria or peroxisomes. For example, mitochondrial MAVS induces the expression of Type I IFNs and ISGs, whereas peroxisomal MAVS induce ISG expression independently of Type I IFNs (Dixit et al., 2010). How (and why) RLRs induce different signaling pathways from each compartment remains unknown, but it is intriguing to consider this question in the context of the TLR pathways, which also induce location-specific transcriptional responses. For example, at the cell surface, TLR activation induces the expression of NF-κB dependent cytokines whereas endosomal TLRs induce both cytokine and Type I IFN expression (Barton and Kagan, 2009). Thus, for both the TLRs and RLRs, the site of signaling determines the type of transcriptional response activated during a host-microbe encounter.
The role of sorting adaptors in the control of innate immunity may extend beyond mammals. In the fruit fly Drosophila melanogaster, antibacterial defenses are governed by a Toll signaling pathway that is similar to the mammalian network (Hoffmann, 2003). Like in mammals, the receptor proximal regulators are adaptor proteins, one of which is called Tube and the other dMyD88. These adaptors mediate the assembly of a signaling platform consisting of serine/threonine kinases that activate NF-κB (Sun et al., 2002). Although dMyD88 was named for its structural similarities to the mammalian MyD88 protein (Horng and Medzhitov, 2001; Tauszig-Delamasure et al., 2002), dMyD88 exhibits all the cell biological properties of the sorting adaptor TIRAP (Marek and Kagan, 2012). dMyD88 is localized to PI(4,5)P2-rich regions of the plasma membrane prior to any microbial encounter and it acts to recruit its downstream partner Tube to this location. Also, like all other sorting adaptors, mislocalizing dMyD88 by ablating its lipid binding motif results in flies that are defective for Toll signaling and antibacterial immunity (Marek and Kagan, 2012). Thus, it appears that the Toll signaling pathway follows similar cell biological principles as the mammalian network. It remains to be determined if Toll must be inducibly recruited to PI(4,5)P2-rich regions of the cell surface to engage the sorting adaptor dMyD88.
There are some innate immune signaling pathways that are not yet understood enough to reliably predict where signal transduction occurs and if any regulator of signaling has sorting adaptor functions. For example, based on the ability of Dectins to regulate phagocytosis (Goodridge et al., 2012), signaling from the plasma membrane is certain to occur, but whether transport into subdomains of the cell surface are necessary for signaling is unclear. As such, it is not known whether any microbe-inducible trafficking factors or sorting adaptors control Dectin signaling. Similarly, while the IFI16-dependent signaling pathway activated by DNA viruses requires microbe-induced transport from the nucleus to the cytosol, which protein is actually transported is unknown.
Benefits of separating the site of ligand binding from the site of signal transduction
What would be the benefit to the cell of separating the sites of ligand binding from the sites of signal transduction? One of the central tenets of signal transduction is that ligand recognition often leads to the inducible oligomerization of receptors by a process referred to as induced proximity. The simplicity of induced proximity as a means to activate immune signal transduction likely explains is widespread use among diverse families of PRRs. However, transmembrane receptors can oligomerize at several stages of their “lifecycle”. For example, simultaneous binding of multiple TLRs by a single folding chaperone or trafficking factor (e.g. Unc93B1) would create a situation whereby the cytosolic TIR domains may also oligomerize. Since numerous experimental means of oligomerizing TLRs are sufficient to induce signal transduction, such as using chimeric receptors or crosslinking antibodies (Latz et al., 2002; Medzhitov et al., 1997), it does not appear that TLRs can distinguish mistaken from microbe-directed receptor oligomerization. Thus, when considered in the context of an intact cell, the induced proximity mechanism of signaling activation comes with significant regulatory challenges. How is signaling by innate immune receptors permitted only when the receptor is in the proper subcellular compartment?
I propose that within cells, specific checkpoints exist that distinguish biosynthetic from microbe-induced receptor oligomerization. One such checkpoint may be the obligatory separation of the subcellular sites of microbe detection and the sites of signal transduction. By making sorting adaptor localization the determinant of signaling, rather than PRR localization, the cell avoids the problem of distinguishing oligomerization as a result of microbe detection from some biosynthetic trafficking event.
The benefits described above may also explain the behavior of non-immune signaling receptors, such as epidermal growth factor (EGF) receptor family members. EGF engages its receptor(s) at the plasma membrane, an event that leads to the recruitment of an adaptor protein called KSR1. KSR1 functions to induce the Ras-dependent activation of ERK. EGFR is then internalized into the endosomal network where it encounters a second adaptor protein called MP1 (Teis et al., 2002). MP1 is recruited to late endosomes by the adaptor protein p14, and both function to induce a second wave of ERK activation from endosomes (Teis et al., 2002). Interestingly, ERK signaling from endosomes is essential for cell proliferation and early embryonic development (Teis et al., 2006). Thus, like the PRR families described above, some EGFR-induced cellular responses require the transport of the receptor to a subcellular site that differs from the site of ligand binding. It is possible that this signaling network also evolved to separate the sites of ligand binding and signal transduction to ensure the fidelity of signaling pathway activation, an event that is most critical when considering the role of ERK in cellular homeostasis and development. These data are similar to those generated from studies demonstrating G-protein coupled receptors (GPCRs) often must be endocytosed to promote signal transduction (Kovacs et al., 2009).
Overall, the separation of the sites of ligand binding from signal transduction appears to be a fundamental property of cellular signal transduction pathways, and not just a peculiarity of immune signaling events. It should be stated however, that this property does not apply to all signaling receptors. In fact, exceptions to this idea come from the study of signaling receptors that detect transmembrane proteins found on other cells, such as B-cell receptors and T-cell receptors. In these instances, the physical anchoring of a ligand on another cell may preclude the signaling receptor from undergoing the subsequent trafficking events. Thus, alternative mechanisms of ensuring the fidelity of signal transduction likely exist, such as the ligand-dependent exclusion of negative regulatory factors in the case of T-cell receptors (Rhee and Veillette, 2012).
Genetic and cell biological rules of innate immune signal transduction
In this perspective, I proposed the existence of what appears to be cell biological “rules” that determine the subcellular sites of innate immune signal transduction. Rule one dictates that in order to execute their proinflammatory functions, PRRs must first be delivered to the most likely site of PAMP detection. Upon microbial detection, PRRs must then be delivered to an organelle permissive for signaling platform assembly (as defined by the presence of a sorting adaptor). These cell biological rules are useful because they help categorize proteins such as CD14, AP-3 and 14-3-3ε into a group called microbe-inducible trafficking factors. These proteins bear no similarity at the domain level, but they all function to regulate the microbe-induced trafficking of PRRs to a signaling organelle. Likewise, the sorting adaptors TIRAP, TRAM, MAVS, dMyD88, IL-1RacP share little sequence homology, yet they all function to define a given organelle as a site of innate immune signal transduction.
I propose that for every (most) innate immune signaling pathway(s), a microbe-induced trafficking factor exists to deliver a PRR to a sorting adaptor. The sorting adaptor then functions to recruit and assemble an innate immune signaling platform that permits signal transduction to occur. This model mandates that the rate of membrane (or protein) trafficking into and out of a signaling organelle would dictate the intensity or speed of signaling pathway activation. In this regard, a greater use of cell biological tools will likely reveal additional principles that help classify structurally diverse but functionally related signal transduction pathways.
Acknowledgments
This Perspective was not designed comprehensively cover all aspects of innate immunity. As such, several important areas of host defense are not discussed or cited. The views expressed reflect my personal opinion and should not be considered a consensus in the field. I would like to thank members of my lab for helpful discussions during preparation of this manuscript. The National Institutes of Health grants AI093589 and AI072955 support the work performed in the laboratory of J. Kagan. Dr. Kagan holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-{kappa}B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170:21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco AM, Perez-Arago A, Fernandez-Lizarbe S, Guerri C. Ethanol mimics ligand-mediated activation and endocytosis of IL-1RI/TLR4 receptors via lipid rafts caveolae in astroglial cells. J Neurochem. 2008;106:625–639. doi: 10.1111/j.1471-4159.2008.05425.x. [DOI] [PubMed] [Google Scholar]
- Brikos C, Wait R, Begum S, O'Neill LA, Saklatvala J. Mass spectrometric analysis of the endogenous type I interleukin-1 (IL-1) receptor signaling complex formed after IL-1 binding identifies IL-1RAcP, MyD88, and IRAK-4 as the stable components. Mol Cell Proteomics. 2007;6:1551–1559. doi: 10.1074/mcp.M600455-MCP200. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne ED, Roy CR. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic. 2006;7:929–939. doi: 10.1111/j.1600-0854.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- Chiang CY, Veckman V, Limmer K, David M. Phospholipase Cgamma-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem. 2012;287:3704–3709. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SL, Husebye H, Connolly DJ, Espevik T, O'Neill LA, McGettrick AF. The GOLD domain-containing protein TMED7 inhibits TLR4 signalling from the endosome upon LPS stimulation. Nat Commun. 2012;3:707. doi: 10.1038/ncomms1706. [DOI] [PubMed] [Google Scholar]
- Du X, Poltorak A, Silva M, Beutler B. Analysis of Tlr4-mediated LPS signal transduction in macrophages by mutational modification of the receptor. Blood Cells Mol Dis. 1999;25:328–338. doi: 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioannini TL, Teghanemt A, Zhang D, Levis EN, Weiss JP. Monomeric endotoxin:protein complexes are essential for TLR4-dependent cell activation. J Endotoxin Res. 2005;11:117–123. doi: 10.1179/096805105X35198. [DOI] [PubMed] [Google Scholar]
- Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- Goodridge HS, Underhill DM, Touret N. Mechanisms of fc receptor and dectin-1 activation for phagocytosis. Traffic. 2012;13:1062–1071. doi: 10.1111/j.1600-0854.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hidmark A, von Saint Paul A, Dalpke AH. Cutting Edge: TLR13 Is a Receptor for Bacterial RNA. J Immunol. 2012 doi: 10.4049/jimmunol.1200898. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc Natl Acad Sci U S A. 2001;98:12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E, et al. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Lee JO. Structural biology of the Toll-like receptor family. Annu Rev Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Korherr C, Hofmeister R, Wesche H, Falk W. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur J Immunol. 1997;27:262–267. doi: 10.1002/eji.1830270139. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes: guardians of cytosolic sanctity. Immunol Rev. 2009;227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M., Jr. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Anderson RG. Compartmentalized production of ceramide at the cell surface. J Biol Chem. 1995;270:27179–27185. doi: 10.1074/jbc.270.45.27179. [DOI] [PubMed] [Google Scholar]
- Marek LR, Kagan JC. Phosphoinositide binding by the Toll adaptor dMyD88 controls antibacterial responses in Drosophila. Immunity. 2012;36:612–622. doi: 10.1016/j.immuni.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Fawcett P, O'Riordan M, Lee KD, Havell EA, Brown PO, Portnoy DA. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci U S A. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Tochio H, Kato Z, Kawamoto N, Kimura T, Kubota K, Yamamoto T, Funasaka T, Nakano H, Wong RW, et al. TRAM is involved in IL-18 signaling and functions as a sorting adaptor for MyD88. PLoS One. 2012;7:e38423. doi: 10.1371/journal.pone.0038423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, et al. TLR13 Recognizes Bacterial 23S rRNA Devoid of Erythromycin Resistance-Forming Modification. Science. 2012 doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- Orzalli MH, Deluca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, Gong M, Golenbock D, Espevik T, O'Neill LA. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Radons J, Gabler S, Wesche H, Korherr C, Hofmeister R, Falk W. Identification of essential regions in the cytoplasmic tail of interleukin-1 receptor accessory protein critical for interleukin-1 signaling. J Biol Chem. 2002;277:16456–16463. doi: 10.1074/jbc.M201000200. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Veillette A. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol. 2012;13:439–447. doi: 10.1038/ni.2246. [DOI] [PubMed] [Google Scholar]
- Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O'Neill LA, Fitzgerald KA, Golenbock DT. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci U S A. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329:1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, Kobayashi T, Matsumoto F, Fukui R, Kouro T, et al. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med. 2007;204:2963–2976. doi: 10.1084/jem.20071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, et al. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol. 2006;175:861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- Tissieres P, Dunn-Siegrist I, Schappi M, Elson G, Comte R, Nobre V, Pugin J. Soluble MD-2 is an acute-phase protein and an opsonin for Gram-negative bacteria. Blood. 2008;111:2122–2131. doi: 10.1182/blood-2007-06-097782. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281:31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIGI/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Kular G, Fleming IN, Downes CP, Lucocq JM. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003a;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003b;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]