Abstract
Protein tyrosine nitration is a common biomarker of biological aging and diverse pathologies associated with the excessive formation of reactive oxygen and nitrogen species. Recently, we suggested a novel fluorogenic derivatization procedure for the detection of 3-nitrotyrosine (3-NT) using benzylamine derivatives to convert specifically protein or peptide bound 3-NT to a highly fluorescent benzoxazole product. In the current study, we applied this procedure to fluorogenic derivatization of protein 3-NT in sections from adult rat cerebellum in order to: (i) test this method in imaging nitrated proteins in fixed brain tissue sections, and (ii) compare the chemical approach to immunohistochemical labeling with anti-3-NT antibodies. Immunofluorescence analysis of cerebellar sections using anti-3-NT antibodies showed differential levels of immunostaining in the molecular, Purkinje, and granule cell layers of the cerebellar cortex; in agreement with previous reports, the Purkinje cells were most highly labeled. Importantly, fluorogenic derivatization reactions of cerebellar proteins with 4-(aminomethyl)benzenesulfonic acid (ABS) and K3Fe(CN)6 at pH 9, following sodium dithionite (SDT) reduction of 3-NT to 3-aminotyrosine (3-AT), showed a very similar pattern of relative intensity of cell labeling and improved resolution when compared with antibody labeling. Our data demonstrate that ABS-derivatization may be either a useful alternative or a complimentary approach to immunolabeling in imaging protein nitration in cells and tissues, including under conditions of dual labeling with antibodies to cell proteins, thus allowing for cellular co-localization of nitrated proteins and any protein of interest.
Keywords: Protein nitration, 3-nitrotyrosine, cerebellum, Purkinje cells, 4-(aminomethyl)benzenesulfonate (ABS), fluorogenic derivatization, fluorescence microscopy, immunohistochemistry, synaptophysin, SIN-1
Introduction
Tyrosine (Tyr) nitration to 3-nitrotyrosine (3-NT) represents a nitric oxide (•NO)-dependent protein modification that is recognized as a biomarker of numerous diseases and biological aging associated with oxidative stress [1–5]. Several proteomic studies have provided data about protein targets for nitration in vivo [6–12] demonstrating that the accumulation of 3-NT on proteins is not random but controlled by multiple parameters such as: (i) mechanisms of nitration, (ii) accessibility of Tyr residues, (iii) the nature of the sequence in which Tyr is located, (iv) the potential repair of 3-NT, and (v) protein turnover. While proteomic methods can locate and quantify 3-NT residues in target proteins, they usually do not provide details about the location of nitrated proteins in intact cells or tissues. However, such information is very important for studies of oxidative injury and protection against oxidative stress, as well as for redox signaling in vivo.
Thus far, data on the spatial distribution of 3-NT-containing proteins in cells and tissues have been obtained only by confocal fluorescence microscopy after immune labeling of permeabilized tissue sections using 3-NT-specific primary antibodies. Since the first report in 1994 by Beckman and co-workers [13,14], this approach has been continuously used to monitor •NO - and peroxynitrite-dependent protein nitration in plants [15] and animal models of pathological states associated with oxidative stress, such as cardiovascular disease [16–18], hypoxia and ischemia [19,20], spongiform encephalopathy [21], other neurodegenerative diseases [22–26], and aging-associated protein nitration [16,27–29]. In the brain, significant levels of protein 3-NT were detected in the cerebellum of adult animals, a brain region characterized by high expression levels and activities of different nitric oxide synthase (NOS) isoforms, by the release of •NO from nerve endings and the spatial regulation of synaptic excitability by •NO [30–32], and by the selective vulnerability of specific populations of neurons (such as Purkinje cells) towards oxidative stress [26,29,33,34].
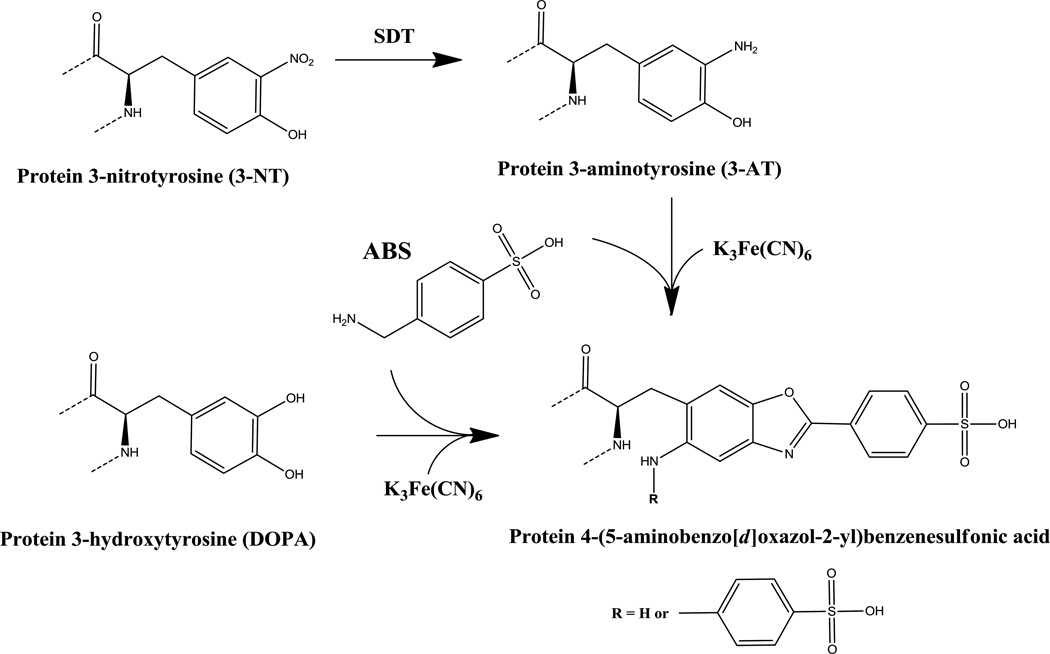
However, the potential for non-specific binding and the restricted accessibility of antibodies to target proteins in crowded and highly compartmentalized cellular structures are always a concern with regard to immunohistochemistry. Recently, we reported on the development of a fluorogenic chemical derivatization strategy specific for 3-amino and 3-hydroxy substituted Tyr residues, such as 3-aminotyrosine (3-AT) and 3,4-dioxyphenylalanine (DOPA), using 4-(aminomethyl) benzenesulfonic acid (ABS), [35,38,39]. The product of such a reaction is the highly fluorescent phenylbenzoxazole. A simplified scheme for the fluorogenic derivatization of 3-NT or DOPA with ABS is shown in Fig.1. The ABS reagent is not fluorescent and does not form fluorescent products with any isolated amino or hydroxyl groups of proteins and other biomolecules, except with 5-hydroxytryptophan [39]. It should be noted that 5-hydroxy-Trp may also be formed on proteins under conditions of oxidative stress. We previously demonstrated that ABS derivatization can be used for the fluorescent staining of either 3-NT or DOPA-containing proteins separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [39]. Specificity of the reaction of ABS with 3-NT is achieved through reduction of 3-NT to 3-AT by sodium dithionite (SDT) prior to derivatization with ABS. Conduct of the derivatization reaction without prior reduction with SDT may thus serve as a control for the specificity of the reaction with 3-NT.
Figure 1.
Reaction scheme for fluorogenic derivatization of protein 3-NT or DOPA with ABS to form a protein-linked sulfophenylbenzoxazole product. The introduction of either an amino (R = H) or a second benzylamino (R = CH2-Ph-SO3H) substituent into the final benzoxazole depends on the reaction conditions [39].
The aim of the current study was to apply this chemical method of 3-NT labeling to the visualization of cellular protein 3-NT by confocal microscopy. Cryotome sections of rat brain cerebellar cortex were selected for an examination of in situ cellular labeling using the ABS reaction with protein 3-NT. The choice of the cerebellar region for these studies was based on reasons described previously [27,40], including high levels of expression of NOS in granule cell neurons of this brain region and the importance of •NO in shaping long-term changes in synaptic activity of key Purkinje neurons [30–32,36,37]. Fluorogenic derivatization of fixed and permeabilized cerebellar sections with ABS following SDT reduction of 3-NT to 3-AT, produced a pattern of fluorescence labeling that correlated with that of 3-NT immunofluorescence. The formation of benzoxazole derivatives was confirmed by the spectral properties of the fluorescent images obtained after ABS derivatization, and by the observed reduction of fluorescent signals when the SDT reduction step was omitted. The yield of benzoxazole was increased when unfixed cerebellar slices were exposed to SIN-1, a reagent that releases •NO and superoxide [34,41,42]. Importantly, the 3-NT derivatization strategy was shown to produce specific labeling when used in dual labeling, fluorescence studies with a protein-specific antibody, anti-synaptophysin antibody, thus allowing for the co-localization of 3-NT-modified proteins and other cellular constituents, such as the vesicular protein synaptophysin which marks nerve endings that are likely to be the sources of •NO released upon synaptic activation [30–32,37,43]. Such co-localization studies are, of course, essential for revealing not only protein targets of Tyr nitration in tissues but also of the type of protein modification that may drastically alter intracellular signaling, such as that by Tyr phosphorylation.
Materials and methods
Materials
ABS was synthesized as described previously [39]. The antibodies used in the present studies were: anti-3-NT mouse monoclonal from GenWay Biotech (San Diego, CA), anti-synaptophysin from Chemicon (Temecula, CA), Alexa 568-conjugated goat anti-rabbit from Life Technologies (Grand Island, NY), Alexa 488 goat anti-mouse from Molecular Probes (Carlsbad, CA), and HRP-conjugated goat anti-mouse from Thermo Fisher Scientific (Rockford, IL). Rabbit muscle phosphorylase b (Ph-b) was obtained from Prozyme (San Leandro, CA). Electrophoresis buffer systems used were from Invitrogen (Carlsbad, CA), 0.45 µm PVDF membranes from Millipore (Billerica, MA), and ECL or ECL-Plus detection kits from Amersham Biosciences (Piscataway, NJ). All other chemicals were from Sigma-Aldrich (St. Louis, MO).
Cerebellum immunohistochemistry
Twelve month-old Spague-Dawly Wistar rats were perfused and the tissues fixed following transcardial perfusion with 4% paraformaldehyde-phosphate-buffered saline (paraformaldehyde-PBS) solution [44]. The brains were removed, dissected and post-fixed in the same buffer at 4°C overnight, rinsed in PBS, cryopreserved in 30% sucrose-PBS solution, and stored at 4°C. Cerebellar sections (22 µm thick) were cut by cryotome sectioning, and immunostaining for 3-NT performed as follows. The sections were treated with fresh NaBH4 (1 mg/ml in PBS, three times for 10 min), rinsed in 2 mM EDTA in PBS (10 min at 23°C), incubated in 3% gelatin-PBS (1 hr at 37°C), rinsed with EDTA-PBS (three times for 10 min), permeabilized with 0.1% Triton X-100 in PBS (15 min at 23 °C), and rinsed again in EDTA-PBS (10 min at 23°C). The sections were subsequently incubated with anti-3-NT monoclonal antibody (1:500 dilutions) at 4°C overnight and for 1 hr at 23°C. After rinsing with EDTA-PBS, the sections were incubated (2 hrs at 37°C) with Alexa488 goat anti-mouse secondary antibody (1:1000 dilutions), rinsed in PBS buffer, and mounted on glass slides in 70% glycerol-PBS. In control experiments, the incubation of cerebellar sections with anti-3-NT antibodies was preceded by incubation with either 10 or 100 mM (10 min) SDT in order to reduce the 3-NT to 3-AT. This was followed by rinsing with EDTA-PBS (10 min at 23 °C) and processing as described above.
ABS derivatization reactions in cerebellar sections
Floating, permeabilized cerebellar sections were pre-treated with 10 mM SDT in 0.1 M sodium phosphate buffer (pH 9.0) for 30 min, rinsed with the same buffer (three times for 5 min), incubated with 2 mM ABS and 10 µM K3Fe(CN)6 (1 hr at 23°C), rinsed in PBS, and mounted on glass slides in 70% glycerol–PBS. ‘
Fluorescence microscopy
The fluorescence labeled tissue sections were viewed using a spinning disk in an Olympus 1X81 laser confocal microscope with various excitation/emission settings prior to the selection of the optimal wavelength combination for visualization. The filters used for excitation were 340±13, 377±25, and 387±13 nm, and for the emission: 447±25, 531±32, and 624±20 nm. The excitation/emission settings for immunostained sections were 488/510 nm for sections stained with Alexa488-conjugated antibodies, and 578/603 for Alexa568-stained sections. All images captured were processed using Slidebook 5.0 software. 3-D Images were reconstructed using 0.1 µm thick series of z-sections and were processed further using the Adobe Photoshop CS program.
In vitro protein tyrosine nitration in cerebellar slices
Non-fixed cerebellar slices (~300 µm thick) were incubated (1 hr at 37°C) in artificial cerebrospinal fluid (ACSF) containing 125 mM NaCl, 25 mM NaHCO3, 1.25 mM KCl, 1.25 mM KH2PO4, 1.5 mM MgCl2, 2 mM CaCl2, 16 mM glucose (pH 7.4) in the absence or presence of 3 or 10 mM SIN-1 [41,42]. Each cerebellar slice was divided into two parts, one was used for WB analyses and the other processed for fluorescence microscopy following derivatization with ABS or after labeling with anti-3-NT antibody as described above. The slices used for WB analyses were homogenized in ACSF in Eppendorf tubes using a Potter-Elvehjem homogenizer. Protein concentration was determined using the BioRad protein assay method (Bradford) as suggested by the manufacturer, and samples containing 100 µg of protein were mixed with equal volume of 2× reducing Sample Buffer, boiled for 2 min and subjected to electrophoretic separation on precast Bio-Rad 4–20% gradient gels using Tris-Glycine-SDS Running buffer (all from Invitrogen, Carlsbad, CA). The gels were then either stained for protein by Coomassie Blue or electro-blotted onto a 0.45 µm PVDF membrane prior to WB analysis. For WB analysis, anti-3-NT antibodies (1:4000 dilutions) and HRP-conjugated secondary goat anti-mouse antibody (1:10000 dilutions) were used. The bands were visualized with either the ECL or ECL-Plus detection kit according to the manufacturer’s procedure.
Preparation of a nitrated protein standard for Western blot analysis
Peroxynitrite (ONOO−) was generated by the ozonolysis of sodium azide solutions as described earlier [45]; the ONOO− content in 0.1% NaOH (pH 12) stock solutions was quantified using ε302=1,670 M−1cm−1. Prior to protein tyrosine nitration, 1 mg/mL (~10 µM) rabbit muscle phosphorylase b (Ph-b) dissolved in a buffer containing 1% (w/v) SDS, 50 mM NH4HCO3, pH 7.8, was reductively alkylated through incubation with 2 mM DTT for 30 min at 50°C, followed by the addition of iodoacetic acid at a final concentration of 5 mM and incubation for an additional 30 min at room temperature. The alkylated protein was separated from unreacted compounds and side products by precipitation in ten volumes of cold ethanol for 4 h at −20°C followed by centrifugation at 14,000 g for 2 min. Reconstituted 100-µL (100-µg) Ph-b aliquots in 0.1 M phosphate buffer containing 1% SDS were then nitrated by the addition of 10 µL alkaline stock solution of ONOO− (final concentration 3 mM) while vortexing to yield ca. 100 µM protein 3-NT. The content of 3-NT in Ph-b was quantified by UV-spectroscopy (ε430=4,400 M−1cm−1 at pH>9) [2] using non-nitrated protein samples for background control.
Double-labeling of cerebellar sections using the ABS derivatization reaction and synaptophysin antibody
Cerebellar sections were first reacted with ABS as described above, and then labeled with anti-synaptophysin antibodies (1:1500 dilutions). After overnight incubation at 4°C, the sections were rinsed with PBS (15 min at 23°C), incubated with Alexa 568-conjugated goat anti-rabbit antibody (1:1000 dilution, 2 hr at 37°C), rinsed in PBS (three times for 5 min at 23°C), and mounted on glass slides as described above.
Results
3-NT reactivity in intact rat cerebellar slices
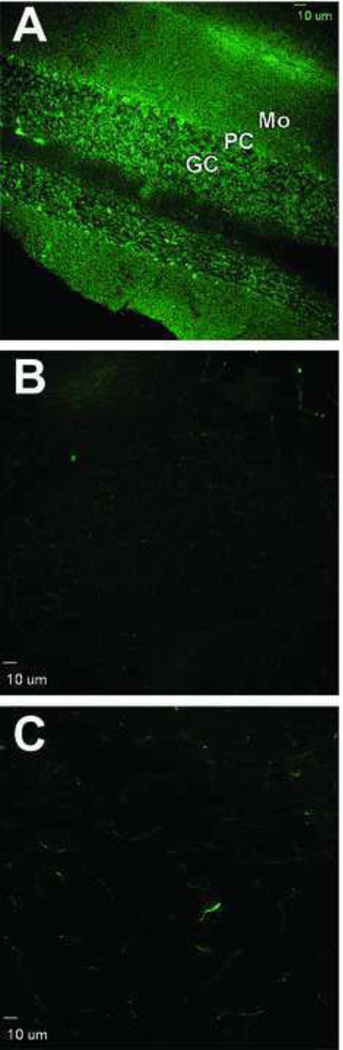
Labeling with anti-3-NT-antibodies produced differential immunostaining patterns in rat cerebellar cortex neurons. Immune reactivity with putative nitrated proteins was prominent in Purkinje neurons and less so in granule cells and in cells within the molecular layer of the cerebellum (Fig.2A). The more pronounced levels of immune staining of Purkinje neurons as compared with those of granule cell neurons that we observed were consonant with those previously reported by others [27,29,33,34]. The observed levels of immune staining with anti-3-NT antibodies also correlated well with the differential expression levels and activities of NOS in these two cell types, as well as the differential sensitivity of cerebellar neuronal populations to reactive oxygen and nitrogen species [26–28,46,47]. The specificity of 3-NT immunostaining was validated in our studies by the fact that labeling of cells in cerebellar sections was significantly decreased in sections that were pre-treated with SDT (Fig.2B and 2C), a chemical treatment that was expected to reduce protein 3-NT to 3-AT [14].
Figure 2.
Labeling of cerebellar cortex cells by anti-3-NT monoclonal antibody and characterization of the specificity of such labeling. A) Low magnification image of cerebellar cortex that was exposed to anti-3NT antibody and Alexa 488-conjugated anti-mouse secondary antibody. B and C) Same low magnification image as in (A) except that the tissue sections were pre-reacted with either 10 mM (B) or 100 mM (C) SDT in order to convert 3-NT to 3-AT. Pretreatment with SDT markedly decreased immune labeling of cells in cerebellar sections and was, therefore, used as a control for antibody specificity toward 3-NT. GC, granule cell layer; PC, Purkinje cell layer; Mo, Molecular layer. Bar: 10 µm.
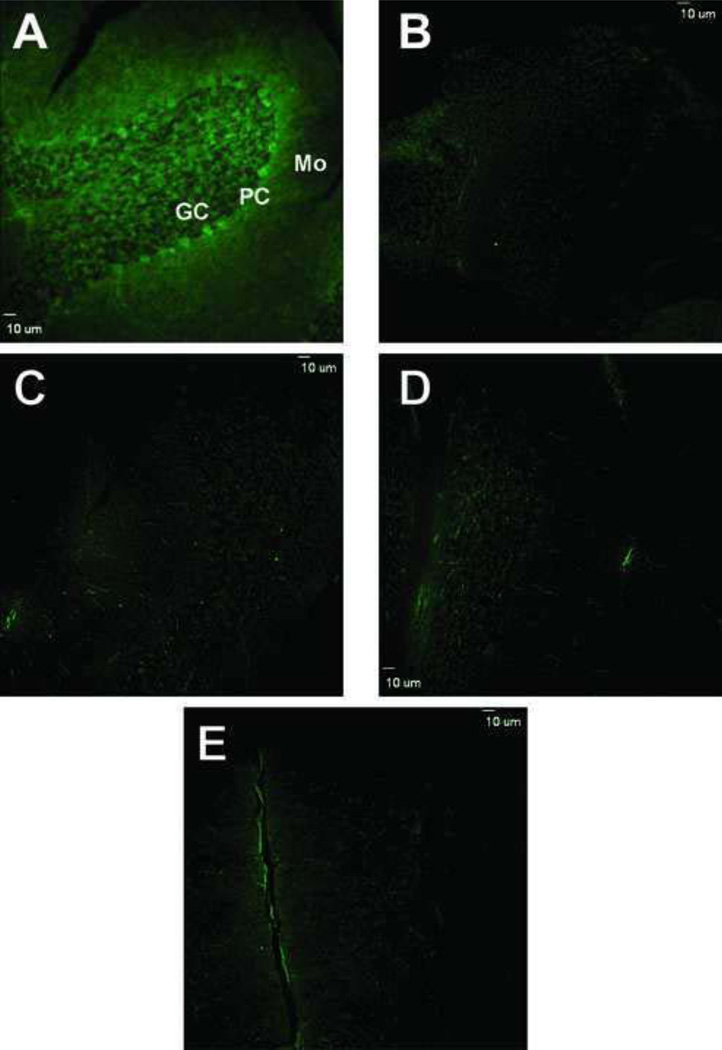
Fluorogenic derivatization of permeabilized cerebellar sections with 2 mM ABS and 10 µM K3Fe(CN)6, after SDT reduction of 3-NT to 3-AT, led to a very similar pattern of cell labeling as that observed in sections immunostained with anti-3-NT antibodies (Fig.3A). The maximal fluorescence intensity was detected at excitation and emission wavelengths of the microscope filters set at 387 ± 11 and 470 ± 30 nm. These excitation and emission wavelengths were in good agreement with the excitation and emission maxima of approximately 360 and 480–490 nm, respectively, of ABS-tagged protein 3-NT in solution [39]. Furthermore, the specificity of fluorogenic derivatization of 3-NT in situ was confirmed in control experiments where either all reagents or only one of the reagents necessary for 3-NT fluorogenic derivatization were excluded (Fig.3, B–E). In addition, it should be noted that without SDT reduction, ABS derivatization did not result in significant fluorescent product formation (Fig.3D). This is an indication that the in situ ABS reaction led to relatively low levels of other potential derivatization targets, such as protein DOPA or 5-hydroxy-Trp (no other amino acids generate fluorescent products upon reaction with ABS). The low levels of background fluorescence in these experiments also excluded the possibility of the formation in cerebellar sections of additional products of protein Trp oxidation, such as N-formylkynurenine and kynurenine. Such products, if formed, would have fluorescence spectral properties comparable to the ABS derivatization product, benzoxazole [39]. However, in the control samples, the levels of intrinsic fluorescence were negligible (Fig.3B). Overall, the fluorescence imaging obtained following the reaction of cerebellar proteins with ABS was strongly indicative of the fluorogenic derivatization of protein 3-NT by ABS and thus could be considered as being comparable in terms of sensitivity to that of immunofluorescence with anti-3-NT-specific antibodies.
Figure 3.
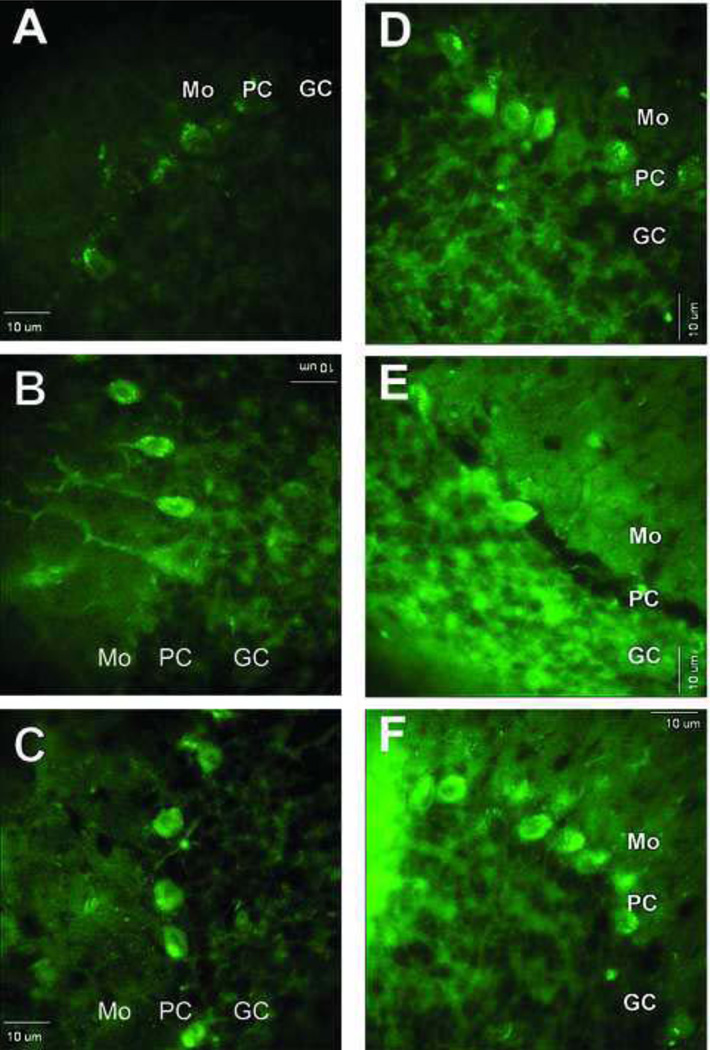
Confocal fluorescence microscopy images of protein 3-NT derivatized with ABS in sections of cerebellar cortex. A) Low magnification image of cerebellar cortex that was exposed to reduction with 10 mM Na2S2O4 followed by derivatization with 2 mM ABS and 10 µM K3Fe(CN)6 as described under Methods. Controls for the specificity of labeling are shown in panels B–E where derivatization either was not performed (B) or was performed excluding ABS (C), SDT (D) or K3Fe(CN)6 (E), respectively. Bar: 10 µm.
Protein 3-NT derivatization with ABS in cerebellar slices after exposure to SIN-1
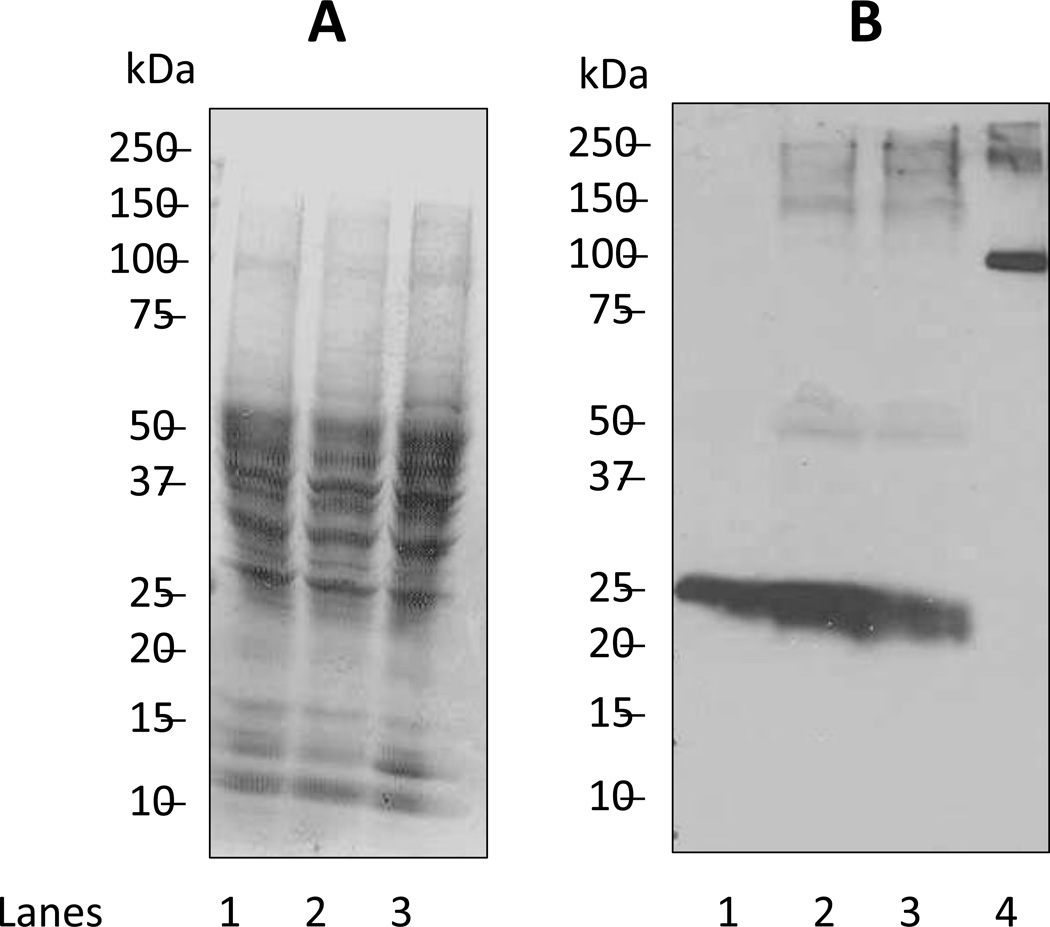
To validate the use of the ABS-derivatization method in fluorescence microscopy of protein nitration in tissue sections, we incubated living cerebellar cortex slices with SIN-1, a compound that releases •NO and superoxide (O2•−) [41,42,48]. As shown in Fig. 4, SIN-1 caused a concentration-dependent increase in protein nitration levels as assessed by WB analyses. The overall protein 3-NT content was below 5 pmol per 100 µg protein based on a comparison with a nitrated standard protein, rabbit Ph-b. We note that a band of ca. 25kDa shows significant affinity to the anti-3-NT antibody in tissue not exposed to SIN-1 (Fig. 4B, lane 1). The intensity of anti 3-NT-antibody staining of this 25 kDa band increases after the exposure to SIN-1, suggesting that this band represents a nitration-sensitive target in our cerebellar cryosections. The fact that SDT treatment of cerebellar cryosections abolishes the reactivity of cerebellar cryosections towards the anti-3-NT antibody (Fig. 2) suggests that the anti-3-NT antibody shows little non-specific reactivity towards non-nitrated proteins in cerebellar cryosections.
Figure 4.
Biochemical analysis of protein nitration in cerebellar slices pre-exposed to SIN-1. Migration of molecular size standards are shown on the left of each electropherogram/Western blot. A) Separation of proteins by SDS-PAGE and visualization by Coomassie Blue staining. To each lane of the gel we added 100 µg protein extracted from cerebellar slices that were either not incubated or incubated with SIN-1 as indicated: Lane 1, no exposure to SIN-1; Lane 2, incubated with 3 mM SIN-1; Lane 3, incubated with 10 mM SIN-1. B) Immunoblot detection of nitrated proteins using anti-3-NT monoclonal antibody as described under Methods. Lanes 1–3 as in Figure 4A; lane 4 represents 0.1 µg of nitrated rabbit myophosphorylase b standard (5 pmol of 3-NT).
Images of ABS labeling of cells in cerebellar sections obtained from control, i.e. non-SIN-1 treated slices, were compared with those of sections derived from slices pre-incubated with SIN-1. The sections from SIN-1-pre-incubated slices exhibited more extensive fluorescent staining, particularly of the Purkinje cell layer (Fig.5A–C). In the Purkinje cells of the SIN-1-treated sections, both the cell body and the processes (dendrites) were strongly fluorescent when compared with the surrounding tissue (cf. especially Fig. 5B, F), while tissues not treated with SIN-1 showed predominantly labeling within the cell body (Fig. 5A, D). It should be noted though, that both ABS and anti-NT labeling in sections that were not pre-exposed to SIN-1, labeled primarily Purkinje cell bodies (Fig. 5A, D), whereas after SIN-1 treatment, they labeled both cell bodies and dendrites (Fig. 5B, C, E, F). An increase in 3-NT staining following exposure of the cerebellar slices to SIN-1 was also observed using anti-3-NT antibodies (Fig.5D–F). A direct comparison between these two approaches could not be made because different tissue sections were used for ABS labeling and immunolabeling. In general, protein 3-NT images obtained by ABS derivatization displayed a higher contrast than those following immunolabeling. The fact that nearly only Purkinje cells are labeled with ABS in Fig. 5A while the anti-3-NT antibody labels both Purkinje and granule cells in Fig. 5D is likely due to some variation in the specific tissue sections that were used to monitor fluorescence labeling in these experiments. As shown above under low magnification in Fig. 3A, ABS clearly labeled both Purkinje and granule cells, though granule cells were not homogeneously labeled throughout the tissue section (Fig. 3A).
Figure 5.
Images of fluorescent labeling following either derivatization of 3-NT-containing proteins with ABS (A–C) or immunolabeling of same proteins with anti-3-NT antibody (D–F) in cerebellar cortex from slices that were pre-incubated without or with SIN-1 at varying concentrations. Sections shown in images A and D were from slices not exposed to SIN-1, whereas sections in B, C, E, and F were from slices pre-treated with either 3 mM SIN-1 (B and E) or 10 mM SIN-1 (C and F). Bar: 10 µm.
Conjoint use of protein 3-NT ABS tagging and protein immunolabeling in cerebellar sections
In order to probe for the specific cellular compartmentation of proteins that are labeled following ABS derivatization of 3-NT, it would be necessary to perform studies of combined labeling with ABS and an antibody that is targeted to a specific cellular domain of interest. Such co-localization studies are routinely performed using confocal microscopy following tissue labeling with two different primary and secondary antibodies. In previous studies, a double immunofluorescence approach using polyclonal antibodies against α-synuclein and monoclonal antibodies against protein 3-NT was used to study Tyr nitration of α-synuclein in the rat brain under hyper- and hypoglycemic conditions [49]. A similar approach was used to probe for the co-localization of neuronal protein markers involved in neurotransmitter metabolism (e.g., dopamine transporters or tyrosine hydroxylase) with protein 3-NT for the assessment of the role of nitrative damage in the selective vulnerability of neurons to degeneration during the aging process [28]. These types of studies require multiple species-specific primary antibodies, the use of which may be limited by different sensitivities and/or specificities of mono- and polyclonal antibodies.
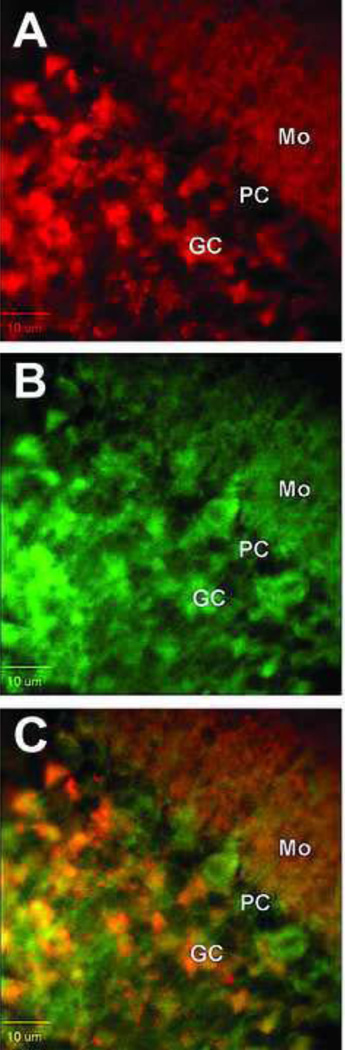
In the present study, we tested whether ABS tagging of protein 3-NT can be used for co-localization studies, based on the observations that the derivatization approach was as sensitive as that of labeling tissue proteins with monoclonal anti- 3-NT antibodies. Shown in Fig. 6, are the images obtained from double labeling using ABS derivatization and anti-synaptophysin antibodies. Synaptophysin was selected for this double-labeling study in order to map the distribution of nerve terminals in the granule cell, Purkinje cell, and molecular layers. Some of these nerve terminals, such as those of the climbing and of parallel fibers making synapses on Purkinje neurons, are considered to be the primary generators of the NO released in the cerebellar cortex. Thus, the question considered in this study was whether protein nitration in Purkinje neurons detected by ABS derivatization of 3-NT, was more prominent at post-synaptic sites to nerve terminals labeled by synaptophysin. Microscopic examination of sections double labeled as described above revealed an extensive pattern of synaptophysin labeling in the cerebellar cortex (Fig. 6). Purkinje cell dendrites were surrounded by nerve endings labeled with anti-synaptophysin antibodies. However, there were comparatively few synaptophysin-labeled nerve fibers or terminals on Purkinje cell bodies, yet the cell bodies were strongly labeled, once again, by the reaction products of ABS derivatization. Thus, ABS tagging of protein 3-NT can be applied together with any protein–specific antibodies in order to characterize the cellular compartmentation of proteins that may be nitrated in vivo. With respect to the cerebellar sections examined in this study, protein nitration appeared to be occurring within a larger volume or spatial distribution within neuronal cell bodies and was not confined to sites post-synaptic to such terminals.
Figure 6.
Double labeling of cerebellar sections by derivatization with ABS (green) and immunostaining with anti-synaptophysin antibody (red). A) Image of labeling by the ABS reaction; B) Image of immune labeling by anti-synaptophysin antibody; and C) superimposition of images from A and B. Mo, PC, and GC. All designations are the same as in Figure 1. Bar: 10 µm.
Discussion
Cerebellum represents a brain structure exposed to relatively high levels of •NO due to the expression of both nNOS and iNOS in the cells of this region [27,40]. The primary neuronal populations in the cerebellum that express NOS are the granule cell neurons [37]. These neurons send their axonal processes, the parallel fibers, to the molecular layer where they form excitatory, glutamatergic synapses on Purkinje cell dendrites. The second population of neurons that express high levels of NOS are the stellate and basket cells within the molecular layer [37]. These neurons form inhibitory, gabaergic synapses that surround the cell body of Purkinje neurons. Purkinje cells do not appear to express NOS that is detectable by immunostaining methods [37]. Yet, despite the low expression levels of NOS in Purkinje neurons, these cells display high levels of protein 3-NT. We assume that the nitrated proteins detected in Purkinje cells represent a target of the •NO signaling initiated by the release of •NO from the nerve terminals of granule cell and basket cell neurons, or possibly other neuronal types that form synapses on Purkinje neurons.
It has been estimated that •NO levels following parallel fiber stimulation in cerebellum may reach nanomolar (1–50 nM) and up to micromolar concentrations [31,51,52]. The volume of distribution of •NO that is released from the nerve terminals of parallel fibers has been estimated to be 1–5 µm in all directions from the point of its release [30,31,52]. The •NO that permeates post-synaptic neurons may form secondary reactive nitrogen species which would be expected to cause protein nitration, possibly at sites distant to the nerve terminal synapses. The distance from the sites of •NO release that one might find nitrated proteins would depend not only on the volume within which •NO might diffuse, but also on the intracellular compartmentation of proteins susceptible to Tyr nitration and the transport and degradation processes that are involved in the trafficking of these proteins. In addition, the activation of endothelial and inducible isoforms of NOS from microglia (mostly localized in the molecular layer of the cerebellum) may contribute to the total •NO flow in the cerebellar cortex and to protein nitration associated with neuronal damage [26].
Our observations during the exposure of cultured tissue slices to external sources of reactive oxygen and nitrogen species, i.e., to SIN-1, confirms the expected outcome that the simultaneous formation of •NO and O2•− results in enhanced 3-NT formation in cerebellar cortex, particularly in neurons selectively vulnerable to oxidative stress, such as the Purkinje cells. These data suggest that the sensitivity of cells to protein nitration in intact tissue may be determined, in part, by higher rates of O2•− generation by mitochondria, and the subsequent reaction of superoxide with •NO˙ to form peroxynitrite [2,3]. Overall, our data demonstrate that the accumulation of 3-NT on proteins in the cerebellar cortex is not random, and represents an outcome of multiple cell-specific processes, which may ultimately determine long-term changes in cell signaling or the susceptibility to oxidative injury.
In this study, we suggest a novel approach to study protein Tyr nitration in cells and tissues based on the selective fluorogenic tagging of protein 3-NT in situ. Our data show that ABS derivatization, proposed earlier for the quantitative proteomic analysis of protein and peptide 3-NT in cell lysates and tissue homogenates [35,38,39], can be applied to fluorescence microscopy imaging of nitrated proteins in fixed tissue samples. Fluorescence microscopy data for ABS-tagged protein 3-NT in rat cerebellum demonstrates a sensitivity comparable to that of anti-3-NT antibodies, and promises even higher specificity based on the selective derivatization procedure, as the tagging reagents are not fluorescent until reacted with 3-NT. This feature may be valuable for the development of high-resolution fluorescence microscopy directed toward the molecular localization of proteins that are fluorescently tagged [52]. Another advantage of this approach is the use of a low molecular weight tagging reagent which can easily diffuse through tissue sections. The demonstration in the present study that fluorogenic ABS tagging of protein 3-NT can be used in combination with protein immunofluorescence offers an additional benefit for protein 3-NT localization studies. Furthermore, the application of a fluorogenic 3-NT tagging reagent that contains an additional affinity tag [53] may provide an opportunity to pull down nitrated proteins from tissue sections after the completion of microscopic analyses. This would allow for proteomic identification of the nitrated protein sequences. Overall, we have shown that ABS-tagging of 3-NT in proteins may be a useful alternative to or a complementary approach for in situ imaging of protein nitration in cells and animal tissues.
Highlights.
3-Nitrotyrosine in brain tissue is converted to fluorescent benzoxazole.
Benzoxazole can be monitored by laser confocal microscopy.
Fluorogenic labeling is an alternative strategy to monitor 3-nitrotyrosine.
Acknowledgements
This research was supported by the NIA grants AG23551, AG25350, AG035982, 5P01AG012993, and NICHD grant HD02528.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2002;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 2.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turko IV, Murad F. Protein nitration in cardiovascular diseases. Pharmacol. Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds MR, Berry RW, Binder L. Nitration in neurodegeneration: deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 6.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. USA. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J. Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 8.Kanski J, Hong S-J, Schöneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J. Biol. Chem. 2005;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 9.Kanski J, Behring A, Pelling J, Schöneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: effects of biological aging. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 10.Gokulrangan G, Zaidi M, Michaelis ML, Schöneich C. Proteomic analysis of protein nitration in rat cerebellum: effect of biological aging. J. Neurochem. 2007;100:1494–1504. doi: 10.1111/j.1471-4159.2006.04334.x. [DOI] [PubMed] [Google Scholar]
- 11.Sacksteder CA, Qian W-J, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, Smith RD, Smith DJ, Squier TC, Bigelow DJ. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 12.Ghesquiere B, Colaert N, Hensens K, Dejager L, Vanhaute C, Verleysen K, Kas K, Timmerman E, Goethals M, Libert C, Vandekerckhove J, Gevaert K. In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol. Cell Proteom. 2009;8:2642–2652. doi: 10.1074/mcp.M900259-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckman JS, Ye YZ, Anderson P, Chen J, Accavetti MA, Tarpey MM, White CR. Biol. Chem. Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 14.Ye YZ, Strong M, Huang ZQ, Beckman JS. Antibodies that recognize nitrotyrosine. Methods Enzymol. 1996;269:201–209. doi: 10.1016/s0076-6879(96)69022-3. [DOI] [PubMed] [Google Scholar]
- 15.Valderrama R, Corpas F, Carreras A, Fernandez-Ocana A, Chaki M, Luque F. [Google Scholar]
- 16.Xu S, Ying J, Jiang B, Guo W, Sharov V, Lazar H, Menzoian J, Bigelow D, Schöneich C, Cohen R. Detection of sequence-specific tyrosine nitration of manganese SOD and SERCA in cardiovascular disease and aging. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2220–H2227. doi: 10.1152/ajpheart.01293.2005. [DOI] [PubMed] [Google Scholar]
- 17.Guo W, Adachi T, Matsui R, Xu S, Ying J, Jiang B, Zou M, Kirber M, Lieberthal W, Cohen R. Quantitative assessment of tyrosine nitration of manganese superoxide dismutase in angiotensin II-infused rat kidney. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H1396–H1403. doi: 10.1152/ajpheart.00096.2003. [DOI] [PubMed] [Google Scholar]
- 18.Parastatidis I, Thomson L, Fries DM, Moore RE, Tohyama J, Fu X, Hazen SL, Heijnen HF, Dennehy MK, Liebler DC, Rader DJ, Ischiropoulos H. Increased protein nitration burden in the atherosclerotic lesions and plasma of apolipoprotein A-I deficient mice. Circ Res. 2007;101:368–376. doi: 10.1161/CIRCRESAHA.107.157537. [DOI] [PubMed] [Google Scholar]
- 19.Aslan M, Ryan TM, Townes TM, Coward L, Kirk MC, Barnes S, Alexander CB, Rosenfeld SS, Freeman BA. Nitric oxide-dependent generation of reactive species in sickle cell disease. J. Biol. Chem. 2003;278:4194–4204. doi: 10.1074/jbc.M208916200. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigo J, Fernández AP, Serrano J, Peinado MA, Martínez A. The role of free radicals in cerebral hypoxia and ischemia. Free Radic. Biol. Med. 2005;39:26–50. doi: 10.1016/j.freeradbiomed.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Fernández AP, Serrano J, Rodrigo J, Monleó E, Monzón M, Vargas A, Badiola JJ, Martínez-Murillo R, Martínez A. Changes in the expression pattern of the nitrergic system of ovine cerebellum affected by scrapie. J. Neuropathol. Exp. Neurol. 2007;66:196–207. doi: 10.1097/01.jnen.0000248557.37832.b4. [DOI] [PubMed] [Google Scholar]
- 22.Liu D, Bao F, Wen J, Liu J. Mutation of superoxide dismutase elevates reactive species: comparison of nitration and oxidation of proteins in different brain regions of transgenic mice with amyotrophic lateral sclerosis. Neuroscience. 2007;146:255–264. doi: 10.1016/j.neuroscience.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 23.Barbeito LH, Pehar M, Cassina P, Vargas MR, Peluffo H, Viera L, Estévez AG, Beckman JS. A role for astrocytes in motor neuron loss in amyotrophic lateral sclerosis. Brain Res Brain Res Rev. 2004;47:263–274. doi: 10.1016/j.brainresrev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Butterfield DA, Reed TT, Perluigi M, De Marco C, Coccia R, Keller JN, Markesbery WR, Sultana R. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 26.Kyuhou S, Kato N, Gemba H. Emergence of endoplasmic reticulum stress and activated microglia in Purkinje cell degeneration mice. Neurosci. Lett. 2006;396:91–96. doi: 10.1016/j.neulet.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 27.Blanco S, Molina FJ, Castro L, Del Moral ML, Hernandez R, Jimenez A, Rus A, Martinez-Lara E, Siles E, Peinado MA. Study of the nitric oxide system in the rat cerebellum during aging. BMC Neurosci. 2010;11:78. doi: 10.1186/1471-2202-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanaan NM, Kordower JH, Collier TJ. Age-related changes in dopamine transporters and accumulation of 3-nitrotyrosine in rhesus monkey midbrain dopamine neurons: relevance in selective neuronal vulnerability to degeneration. Eur. J. Neurosci. 2008;27:3205–3215. doi: 10.1111/j.1460-9568.2008.06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung YH, Shin CM, Joo KM, Kim MJ, Cha CI. Immunohystochemical study on the distribution of nitrotyrosine and neuronal nitric oxide synthase in aged rat cerebellum. Brain Res. 2002;95:316–321. doi: 10.1016/s0006-8993(02)03261-4. [DOI] [PubMed] [Google Scholar]
- 30.Iino MJ. Ca2+dependent inositol 1,4,5-trisphosphate and nitric oxide signaling in cerebellar neurons. Pharmacol. Sci. 2006;100:538–544. doi: 10.1254/jphs.cpj06006x. [DOI] [PubMed] [Google Scholar]
- 31.Ogasawara H, Doi T, Doya K, Kawato M. Nitric oxide regulates input specificity of long-term depression and context dependence of cerebellar learning. PLOS Computational Biology. 2007;3:e179. doi: 10.1371/journal.pcbi.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelman GM, Gally JA. Nitric oxide: linking space and time in the brain. Proc. Nat. Acad. Sci. 1992;89:11651–11652. doi: 10.1073/pnas.89.24.11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigo J, Fernández AP, Serrano J, Monzon M, Monleon E, Badiola JJ, Climent S, Martinez-Murillo R, Martínez A. Distribution and expression pattern of the nitrergic system in the cerebellum of the sheep. Neurosci. 2006;139:889–898. doi: 10.1016/j.neuroscience.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 34.McFarland R, Blokhin A, Sydnor J, Mariani J, Vogel MW. Oxidative stress, nitric oxide, and the mechanisms of cell death in Lurcher Purkinje cells. Dev. Neurobiol. 2007;67:1032–1046. doi: 10.1002/dneu.20391. [DOI] [PubMed] [Google Scholar]
- 35.Pennington J, Schöneich C, Stobaugh JF. Selective fluorogenic derivatization with isotope coding of catechols and 2-aminophenol with benzylamine: a chemical basis for the relative determination of 3-hydroxytyrosine and 3- nitrotyrosine peptides. Chromatographia. 2007;66:649–659. [Google Scholar]
- 36.Garthwaite J, Charles SJ, Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- 37.Bredt DS, Hwang PM, Snyder SH. Nature. 1990;347:768–770. doi: 10.1038/347768a0. [DOI] [PubMed] [Google Scholar]
- 38.Sharov VS, Dremina ES, Pennington J, Killmer J, Asmus C, Thorson M, Hong SJ, Li XB, Stobaugh JF, Schöneich C. Selective fluorogenic derivatization of 3-nitrotyrosine and 3,4-dihydroxyphenylalanine (DOPA) in peptides: a method designed for quantitative proteomic analysis. Methods Enzymol. 2008;441:19–32. doi: 10.1016/S0076-6879(08)01202-0. [DOI] [PubMed] [Google Scholar]
- 39.Sharov VS, Dremina ES, Li X, Gerstenecker G, Galeva NA, Dobrowsky RD, Stobaugh JF, Schöneich C. Fluorogenic tagging of protein 3-aminotyrosine with 4-(aminomethyl)benzenesulfonic acid for quantitative analysis of protein tyrosine nitration. Chromatographia. 2010;71:37–53. doi: 10.1365/s10337-009-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siles E, Martinez-Lara E, Canuelo A, Sanchez M, Hernandez R, Lopez-Ramos JC, Del Moral ML, Esteban FJ, Blanco S, Pedrosa JA, Rodrigo J, Peinado MA. Age-related changes of the nitric oxide system in the rat brain. Brain Res. 2002;956:385–392. doi: 10.1016/s0006-8993(02)03575-8. [DOI] [PubMed] [Google Scholar]
- 41.Liu ZW, Lei T, Zhang T, Yang Z. Peroxynitrite donor impairs excitability of hyppocampal CA1 neurons by inhibiting voltage-gated potassium currents. Toxicol. Lett. 2007;175:8–15. doi: 10.1016/j.toxlet.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Larsen TR, Söderling A, Caidahl K, Roepstorff P, Gramsbergen JB. Nitration of soluble proteins in organotypic culture models of Parkinson’s disease. Neurochem. Int. 2008;52:487–494. doi: 10.1016/j.neuint.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Shibuki K, Kimura S. Dynamic properties of nitric oxide release from parallel fibres in rat cerebellar slices. J. Physiol. 1997;498:443–452. doi: 10.1113/jphysiol.1997.sp021870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal R, Agbas A, Bao X, Hui D, Leary C, Hunt J, Naniwadekar A, Michaelis ML, Kumar KN, Michaelis EK. Selective dendrite-targeting of mRNAs of NR1 splice variants without exon 5: identification of a cis-acting sequence and isolation of sequence-binding proteins. Brain Res. 2003;994:1–18. doi: 10.1016/j.brainres.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 45.Pryor WA, Cueto R, Jin X, Koppenol WH, Ngu-Schwemlein M, Squadrito GL, Uppu PL, Uppu RM. A practical method for preparing peroxynitrite solutions of low ionic strength and free of hydrogen peroxide. Free Radic. Biol. Med. 1995;18:75–83. doi: 10.1016/0891-5849(94)00105-s. [DOI] [PubMed] [Google Scholar]
- 46.Lopez IA, Acuna D, Beltran-Parrazal L, Lopez IE, Amarnani A, Cortes M, Edmond J. Evidence for oxidative stress in the developing cerebellum of the rat after chronic mild carbon dioxide exposure (0.0025% in the air) BMC Neurosci. 2009;10:53. doi: 10.1186/1471-2202-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbott LC, Nahm S. Neuronal nitric oxide synthase expression in cerebellum mutant mice. The Cerebellum. 2004;3:141–151. doi: 10.1080/14734220410031927. [DOI] [PubMed] [Google Scholar]
- 48.Luchovski P, Urbanska EM. SNAP and SIN-1 increase brain production of kynurenic acid. Eur. J. Pharmacol. 2007;563:130–133. doi: 10.1016/j.ejphar.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, He R. Fasting induces a high level of 3-nitrotyrosine in the brain of rats. Neurosci. Lett. 2010;472:204–209. doi: 10.1016/j.neulet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Hall CN, Garthwaite J. J. Physiol. 2006;577:549–567. doi: 10.1113/jphysiol.2006.118380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Namiki S, Kakizawa S, Hirose K, Iino M. J. Physiol. 2005;566:849–863. doi: 10.1113/jphysiol.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Mills JD, Periasamy A. Protein localization in living cells and tissues using FRET and FLIM. Differentiation. 2003;71:528–541. doi: 10.1111/j.1432-0436.2003.07109007.x. [DOI] [PubMed] [Google Scholar]
- 53.Dremina ES, Li X, Sharov VS, Stobaugh JF, Schöneich C. A methodology for simultaneous fluorogenic derivatization and boronate affinity enrichment of 3-nitrotyrosine containing peptides. Anal. Biochem. 2011;418:184–196. doi: 10.1016/j.ab.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]