Abstract
Many genes are of unknown functions in any sequenced genome. A combination of chemical and genetic perturbations has been used to investigate gene functions. Here we present a case that such “chemogenomics” information can be effectively used to identify missing genes in a defined biological pathway. In particular, we identified the previously unknown enzyme diphthamide synthetase for the last step of diphthamide biosynthesis. We found that yeast protein YLR143W is the diphthamide synthetase catalyzing the last amidation step using ammonium and ATP. Diphthamide synthetase is evolutionarily conserved in eukaryotes. The previously uncharacterized human gene ATPBD4 is the ortholog of yeast YLR143W and fully rescues the deletion of YLR143W in yeast.
Keywords: protein posttranslational modifications, DPH6, diphthine, yeast fitness, P-loop-like motif
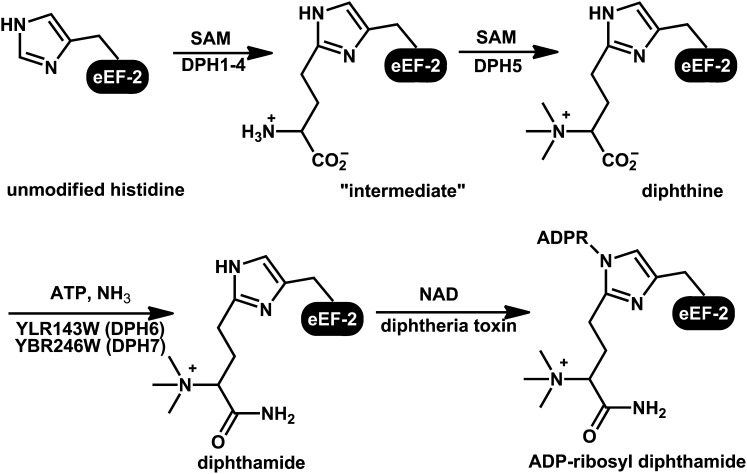
Diphthamide is a posttranslationally modified histidine residue found on archaeal and eukaryotic translation elongation factor 2 (eEF-2) (1–3). Diphtheria toxin (DT) and Pseudomonas exotoxin A recognize and ADP-ribosylate diphthamide, inhibiting eEF-2 and stopping ribosomal protein synthesis (4). Although diphthamide has been known for more than three decades, its biosynthesis and biological functions are still not completely understood. In eukaryotes, the diphthamide biosynthesis pathway consists of three steps (5–9) (Fig. 1). The first step is the transfer of a 3-amino-3-carboxypropyl group from S-adenosyl methionine to the histidine residue of eEF-2. Four proteins, DPH1–4 (DiPHthamide biosynthesis 1-4), are required for this step in eukaryotes. The second step is a trimethylation reaction to form diphthine, which is catalyzed by a single methyltransferase DPH5. Both the first and second steps have been reconstituted in vitro using purified proteins from a thermophilic archaea, Pyrococcus horikoshii (10–12). The third step is the amidation of diphthine to form diphthamide. For a long time, no proteins for the last step were identified. Recently, yeast YBR246W (WDR85 in human) was reported to be another protein required for diphthamide biosynthesis (13). We demonstrated that yeast cells lacking YBR246W accumulate diphthine, the intermediate that is one step away from diphthamide. Therefore, YBR246 is required for the last amidation step (14). However, we thought it was a scaffold or adaptor protein instead of the actual enzyme that catalyzes the amidation reaction because it lacks an ATP-dependent catalytic domain. Thus, the enzyme that catalyzes the amidation step remains unknown, even three decades after the discovery of diphthamide.
Fig. 1.
Biosynthetic pathway of diphthamide in eukaryotes.
In Saccharomyces cerevisiae, a systematic profiling of the growth fitness of ∼5000 homozygous gene deletion strains has been performed; the data are publically available at Yeast Fitness Database (http://fitdb.stanford.edu/) (15, 16). The growth was scored in response to ∼400 small molecules and environmental stresses. The similarity of growth fitness under various conditions between any two different deletion strains has been calculated as the cofitness (15, 16). Two genes that have similar biological functions are likely to have a high cofitness value. In this study, we explored the possibility of using the cofitness data to discover the missing diphthamide biosynthesis gene. We found that a previously uncharacterized yeast gene YLR143W is the diphthamide synthetase.
Results
Cofitness Analysis Revealed That YLR143W Is Closely Related to Diphthamide Biosynthesis.
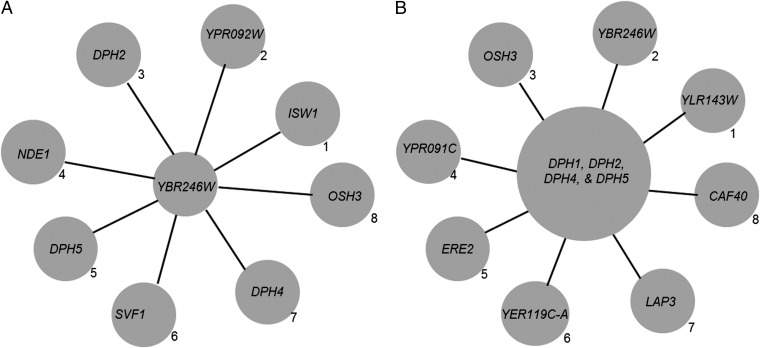
We reasoned that the strain lacking the unknown diphthamide synthetase gene should have high cofitness values with strains lacking other diphthamide biosynthesis genes. To validate this approach, we first analyzed the cofitness data of Δybr246w strain, which has high cofitness value with Δdph2 (0.59624, ranked #3), Δdph4 (0.55343, ranked #7), and Δdph5 (0.57422, ranked #5) strains (Fig. 2A). DPH3 is a fairly small ORF (249 bp) and was not included in the original deletion strain collections. Based on the cofitness data, it was clear that YBR246W has a close relation with the diphthamide biosynthetic pathway. However, it was less clear which gene on the cofitness list of Δybr246w could be the diphthamide synthetase.
Fig. 2.
The eight genes having highest cofitness values to (A) YBR246W deletion strain and (B) DPH1, 2, 4, and 5 deletion strains as a whole group. The rank of each correlation is labeled to the bottom right of each circle.
To provide more accurate prediction of the diphthamide synthetase gene, we reasoned that instead of examining the cofitness to any individual DPH gene deletion strain, looking at the cofitness to all of the DPH gene deletion strains may lead to more reliable predictions. This was done by adding up the cofitness values of each homozygous deletion strain to each of the four DPH gene (dph1, 2, 4, and 5) deletion strains (Dataset S1). The grouped cofitness values were ranked from the highest to the lowest. Excluding the four DPH gene deletion strains, Δybr246w had the second highest sum cofitness value, showing that the sum of the cofitness values was effective in picking up genes in diphthamide biosynthesis. More interestingly, Δylr143w showed a higher cofitness value than Δybr246w to other DPH deletion strains (Fig. 2B). Δylr143w also has high cofitness values with strains lacking known DPH genes (Fig. S1). BLAST search indicated that YLR143W is conserved in both eukaryotes and archaea, but not in bacteria, similar to other proteins involved in diphthamide biosynthesis. The human ortholog is ATP-binding domain-containing protein 4 (ATPBD4), which is consistent with the report that the amidation step of diphthamide biosynthesis is ATP-dependent (7). The analysis of the cofitness data thus pointed to the possibility that yeast YLR143W or human ATPBD4 is the diphthamide synthetase.
YLR143W Is Required for Diphthine Amidation in Vivo.
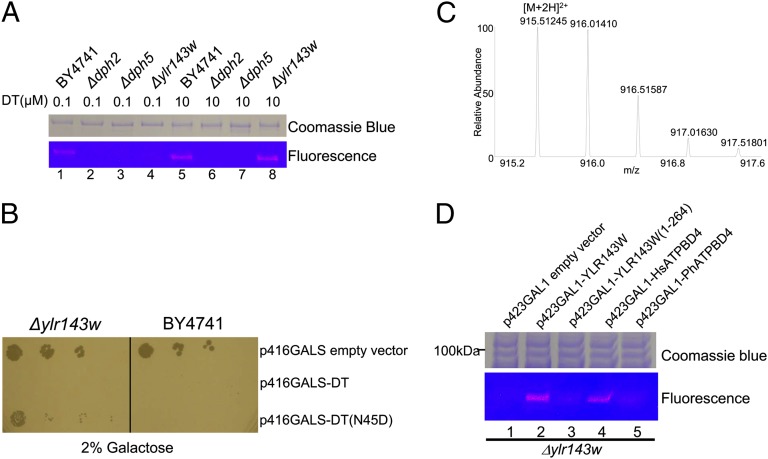
To test whether YLR143W is indeed required for the last step of diphthamide biosynthesis, we first showed that yeast strain lacking YLR143W did not make diphthamide. We obtained the Δylr143w strain and determined the modification on eEF-2 using DT and a fluorescent NAD analog, rhodamine-NAD (Rh-NAD). We have previously demonstrated that diphthamide can be labeled at both high (10 μM) and low (0.1 μM) DT concentrations, diphthine (from Δybr246w strain) can only be labeled at high (10 μM) DT concentration, whereas unmodified or 3-amino-3-carboxypropyl–modified eEF-2 (from Δdph1, Δdph2, Δdph3, Δdph4, or Δdph5 strains) cannot be labeled even at high DT concentration (14). When labeling purified eEF-2 from WT and mutant strains with DT and Rh-NAD, eEF-2 from WT can be labeled at both high and low DT concentrations. In contrast, eEF-2 from Δylr143w strain can only be labeled at high DT concentration, just like the eEF-2 from Δybr246w strain (Fig. 3A). Similar results were obtained when labeling endogenous eEF-2 in the crude cell extract (Fig. S2). The data suggested that eEF-2 from Δylr143w strain contains diphthine instead of diphthamide, similar to the eEF-2 from Δybr246w.
Fig. 3.
Diphthine accumulates in Δylr143w yeast strain. (A) Labeling of eEF-2 purified from various strains using Rh-NAD and DT. The strains used are specified above each lane. In lanes 1–4, low DT concentration (0.1 μM) was used. In lanes 5–8, high DT concentration (10 μM) was used. (B) Diphtheria toxin sensitivity assay. The strains used are specified at the top. Plasmids used in the transformation are listed to the right. The cells were grown on 2% galactose. Each row represents a serial dilution from left to the right. (C) The mass spectrum of the eEF-2 peptide containing diphthine from the Δylr143w strain. (D) Ectopic expression of YLR143W or human ATPBD4 restores diphthamide biosynthesis in the Δylr143w strain. The plasmids used are shown above each lane. HsATPBD4 is the human ATPBD4. PhATPBD4 is the ortholog from P. horikoshii (PH1257). Low DT concentration (0.1 μM) was used in all lanes.
We also confirmed the deficiency of diphthamide biosynthesis in Δylr143w strain using an in vivo assay. A truncated version of DT has been used to select strains that are deficient in diphthamide biosynthesis and thus resistant to DT (17). This method allowed the identification of DPH1–5 (9). However, this assay was not suitable for the identification of YBR246W because diphthine can be ADP-ribosylation at high DT concentration (14). To detect yeast mutants defective in the last step of diphthamide biosynthesis, a better assay was needed. We developed such an assay by using a weaker GALS promoter (18) to decrease DT expression and by using a less active Asn45Asp (N45D) mutant of DT (see SI Materials and Methods and Figs. S3 and S4 for the development of this assay) (19, 20). In this assay, a Δybr246w strain containing diphthine can grow, but the WT strain cannot (Fig. S3B). Similarly, Δylr143w strain was able to grow in this assay when DT(N45D) was present, but not when WT DT was present (Figs. 3B and S4). These in vivo assay data are consistent with the in vitro fluorescent labeling data and suggested that the eEF-2 from Δylr143w strain contained the same modification (diphthine) as the eEF-2 from Δybr246w strain.
We further confirmed that the modification in eEF-2 from Δylr143w strain is diphthine using mass spectrometry (MS). The peptide (686-VNILDVTLHADAIH*R-700) containing the modified His residue (indicated by *) from Δylr143w strain has an observed m/z of 915.51245 (doubly charged, Fig. 3C). The calculated m/z is 915.51580, whereas the corresponding diphthamide containing peptide has an m/z of 915.02351 (doubly charged) (14). The observed m/z value is consistent with the modification in Δylr143w being diphthine. Tandem MS further confirmed the modification and sequence of the peptide (Fig. S5). Thus, all data suggested that eEF-2 from the Δylr143w strain contains diphthine.
YLR143W and Human Ortholog ATPBD4 Restores Diphthamide Biosynthesis in Δylr143w Strain.
To confirm that the lack of diphthamide in Δylr143w is indeed due to the lack of YLR143W, we tested whether diphthamide biosynthesis can be restored in Δylr143w strain by introducing YLR143W back to the cells. When Δylr143w was transformed with YLR143W, diphthamide can be detected by the labeling with low concentration of DT and Rh-NAD (Fig. 3D and Fig. S6). Transformation with an empty vector failed to restore diphthamide biosynthesis. These results demonstrated that the lack of YLR143W was the direct cause for the lack of diphthamide biosynthesis in Δylr143w and provided further support that YLR143W is required for the last step of diphthamide biosynthesis. Furthermore, human ATPBD4 can also fully restore diphthamide biosynthesis in Δylr143w, demonstrating that human ATPBD4 is also a diphthamide synthetase (Fig. 3D). It is noteworthy that YLR143W is almost three times larger than human ATPBD4. The two proteins are similar at N-termini, but YLR143W has a long C-terminal extension. The C-terminal extension appears to be important for the proper function of YLR143W, because the truncated version of YLR143W failed to restore diphthamide biosynthesis (Fig. 3D). This could be due to poor solubility or misfolding of truncated YLR143W. P. horikoshii ATPBD4 (PH1257) also failed to restore the diphthamide biosynthesis in Δylr143w. We think this is likely because of its inability to recognize yeast eEF-2.
YLR143W Amidates Diphthine in Vitro.
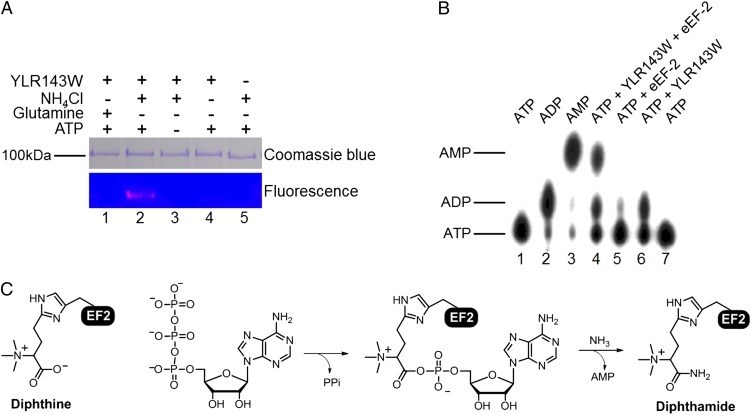
To provide direct evidence that YLR143W is the enzyme catalyzing the last step of diphthamide biosynthesis, we reconstituted the diphthine amidation in vitro using purified proteins. Hexahistidine (His6)-tagged YLR143W was expressed in Escherichia coli and purified using metal affinity purification. The reconstitution reaction was carried out by incubating diphthine-containing eEF-2 with YLR143W, ATP, and either glutamine or ammonium chloride as the nitrogen sources. The formation of diphthamide was detected with Rh-NAD and a low concentration of DT. As shown in Fig. 4A, diphthamide was formed when ammonium was used as the nitrogen source in the presence of both ATP and YLR143W. When glutamine was used as the nitrogen source, no diphthamide formation was detected.
Fig. 4.
YLR143W uses ATP and NH4+ for the amidation reaction. (A) YLR143W in vitro reconstitution detected using DT (0.1 μM) and Rh-NAD. All reactions contain eEF-2 purified from the Δylr143w strain. Purified YLR143W and small molecule substrates used are indicated above each lane. (B) AMP is formed in the diphthine amidation reaction. All lanes contained [α-32P]-ATP. ADP and AMP standards in lanes 2 and 3 were generated enzymatically from ATP. (C) Proposed reaction pathway for YLR143W-catalyzed diphthine amidation reaction.
Sequence analysis of ATP-binding domain of YLR143W revealed that it belongs to the adenine nucleotide alpha hydrolases superfamily. The members of this superfamily include GMP synthetases, argininosuccinate synthetases, and asparagine synthetases. All of these enzymes catalyze the cleavage of the bond between the α and β phosphates of ATP to form AMP as the end product. The P-loop-like motif SGGXD(S/T) is the signature of the ATP pyrophosphatase domains (21). The SGGKDS motif is strictly conserved among all of the YLR143W orthologs (Fig. S7). Therefore, we speculated that YLR143W should form AMP in the reaction. To gain further insights into the catalytic mechanism of YLR143W, we determined the reaction product generated from ATP. [α-32P]-ATP was used to allow the detection of reaction product by autoradiography. Standard spots of AMP and ADP were generated by acyl-CoA synthetase and glutamine synthetase, respectively. We detected the formation of AMP only when YLR143W and eEF-2 were both present, showing that AMP is the enzymatic product (Fig. 4B). eEF-2 by itself generated ADP spot, probably because of its GTPase/ATPase activity (22). It is less clear why ADP was generated by YLR143W in the absence of eEF-2. Nevertheless, AMP was found to be the major product of the diphthine amidation reaction. Based on the dependence on ATP and NH4+ and the formation of AMP, we propose the reaction pathway for YLR143W as shown in Fig. 4C.
Discussion
We have shown that the cofitness data are powerful in discovering missing members in a biosynthetic pathway. Moreover, our method takes advantages of existing knowledge of the diphthamide biosynthetic pathway by combining the cofitness to known DPH genes. This is an easy-to-perform yet reliable method because it eliminates correlations not resulting from functional relations. For example, for YBR246W, the highest correlation is to ISW1 (Imitation SWitch subfamily 1), which is the ATPase subunit of imitation switch class chromatin remodelers. ISW1 has no function in diphthamide biosynthesis. The reason ISW1 is showing a close relation to YBR246W in the cofitness data is that ISW1, which is the ORF YBR245C, is the neighboring ORF of YBR246W. Therefore, the disruption of ISW1 may affect the integrity of YBR246W or vice versa. Such nonfunctional relation may give high pairwise cofitness and may lower the rank of true functional partner. In fact, YLR143W ranks only number 13 in the correlation list of YBR246W. Fortunately, it seems that by combining the correlations to all of the known pathway members, the missing member will stand out. We think this is a useful method to guide the discovery of missing pathway members.
The physiological function of diphthamide is still not well understood. Yeast strains that lack diphthamide modification show no phenotype other than insensitivity to diphtheria toxin. However, DPH1, DPH3, and DPH4 do show important roles in mammal development (23–25). It has been suggested that diphthamide modification ensures translation fidelity (26, 27). Our correlation analysis shows that there may be a connection between diphthamide modification and phosphoinositide metabolism. OSH3 (OxySterol binding protein Homolog 3), SCS2 (Suppressor of Choline Sensitivity 2), and SLM1 (Synthetic Lethal with Mss4 1) are all on top of the correlation list. It would be interesting to find out whether diphthamide modification is connected to phosphoinositide metabolism, which may provide further insights into the function of diphthamide.
The discovery of the missing diphthamide synthetase may have practical implications. Exotoxin A, which is an ADP ribosylating toxin like DT, has been used in cancer chemotherapy. Recent research revealed a survival strategy of cancer cells treated with exotoxin A (28). In some patient samples, it was found that the DPH4 promoter region can be methylated and leads to the missing of diphthamide and renders the chemotherapy inefficient (28). This was an important research report that helps to make better evaluation of the efficacy of the exotoxin A–based chemotherapy. This report showed that DPH1, 2, 3, and 5 are not silenced by DNA methylation. However, because the knowledge of diphthamide biosynthesis pathway was not complete, we do not know if the epigenetic silencing happens to other diphthamide biosynthesis genes. The discovery of YBR246W and YLR143W as proteins required for the last step of the biosynthesis will be helpful to complete such studies.
YLR143W is conserved in eukaryotes. The ortholog in humans is ATPBD4. We have shown that ATPBD4 can complement the loss of YLR143W in yeast, demonstrating that ATPBD4 has diphthamide synthetase activity. Interestingly, in a human cancer genetic study, ATPBD4 is found to be significantly focally deleted in all epithelial cancers. ATPBD4 is deleted in 38% of all lung cancer samples BD4 (29). Another interesting observation was that ATPBD4 was found to be ubiquitylated in two independent studies (30, 31). These facts may provide crucial information to further understand the physiological role of ATPBD4 and diphthamide.
In summary, we discovered diphthamide synthetase, the enzyme that has remained unknown for more than 30 years after the diphthamide structure was determined. This was achieved by grouping the cofitness values to all known diphthamide biosynthetic genes. Compared with the cofitness analysis to an individual gene, this method provides more reliable results and is a powerful method to discover missing genes in defined pathways. During the preparation of the manuscript, we found that the alias DPH6 has been reserved on Saccharomyces Genome Database, but no detail regarding the biochemical function of YLR143W was revealed. We agree with this naming suggestion and propose the alias DPH7 to YBR246W.
Materials and Methods
Yeast Strains.
The yeast strains used in this study are listed in Table S1.
Cofitness Analysis.
The homozygous cofitness data of DPH1, 2, 4, and 5; YBR246W; and YLR143W deletion strains were acquired from Yeast Fitness Database (http://fitdb.stanford.edu/fitdb.cgi). The cofitness values were added and sorted using Microsoft Excel. The full calculation spreadsheet is in Dataset S1. The top 40 lines of the added cofitness are also listed in Dataset S1 with the gene name and function annotations from the Saccharomyces genome database (www.yeastgenome.org).
In Vitro ADP Ribosylation Using Rh-NAD.
Rh-NAD was prepared as described previously (32). Purified yeast eEF-2 and Rh-NAD (25 µM) were incubated with DT at 30 °C in 25 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 30 mM DTT, and 2 mM EDTA. The DT concentration was 100 nM and the reaction time was 15 min if not specified otherwise. The reaction mixture was resolved by SDS/PAGE. The rhodamine fluorescence signal from the protein gel was visualized on a Fisher Scientific UV transilluminator.
In Vitro Diphthine Amidation.
Purified eEF-2 from YLR143W deletion strain was used for the amidation reaction. The eEF-2 concentration was 0.5 μM and the purified YLR143W concentration was 3 nM. The reaction buffer contained 80 mM Tris⋅HCl (pH 8.0), 15 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM ATP, 10 mM NH4Cl, and 0.1 mg/mL BSA. Glutamine was used at a 10-mM concentration as the potential alternative nitrogen source. The reaction was carried out at 30 °C for 30 min. The formation of diphthamide was visualized by Rh-NAD labeling as described previously.
Additional methods are available as supplementary information online.
Supplementary Material
Acknowledgments
We thank Dr. J. Collier for providing pLMY101. This work was supported by National Institutes of Health/National Institute of General Medical Sciences Grant GM088276.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.A.v.d.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214346109/-/DCSupplemental.
References
- 1.Robinson EA, Henriksen O, Maxwell ES. Elongation factor 2. Amino acid sequence at the site of adenosine diphosphate ribosylation. J Biol Chem. 1974;249(16):5088–5093. [PubMed] [Google Scholar]
- 2.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J Biol Chem. 1980;255(22):10710–10716. [PubMed] [Google Scholar]
- 3.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255(22):10717–10720. [PubMed] [Google Scholar]
- 4.Collier RJ. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39(11):1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 5.Moehring JM, Moehring TJ. The post-translational trimethylation of diphthamide studied in vitro. J Biol Chem. 1988;263(8):3840–3844. [PubMed] [Google Scholar]
- 6.Moehring JM, Moehring TJ, Danley DE. Posttranslational modification of elongation factor 2 in diphtheria-toxin-resistant mutants of CHO-K1 cells. Proc Natl Acad Sci USA. 1980;77(2):1010–1014. doi: 10.1073/pnas.77.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moehring TJ, Danley DE, Moehring JM. In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol Cell Biol. 1984;4(4):642–650. doi: 10.1128/mcb.4.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Leppla SH. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell. 2003;12(3):603–613. doi: 10.1016/j.molcel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24(21):9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465(7300):891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, et al. Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol Biosyst. 2011;7(1):74–81. doi: 10.1039/c0mb00076k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Kim J, Su X, Lin H. Reconstitution of diphthine synthase activity in vitro. Biochemistry. 2010;49(44):9649–9657. doi: 10.1021/bi100812h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326(5957):1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 14.Su X, et al. YBR246W is required for the third step of diphthamide biosynthesis. J Am Chem Soc. 2012;134(2):773–776. doi: 10.1021/ja208870a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillenmeyer ME, et al. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010;11(3):R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattheakis LC, Shen WH, Collier RJ. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(9):4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22(25):5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jørgensen R, Wang Y, Visschedyk D, Merrill AR. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008;9(8):802–809. doi: 10.1038/embor.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll SF, Collier RJ. Amino acid sequence homology between the enzymic domains of diphtheria toxin and Pseudomonas aeruginosa exotoxin A. Mol Microbiol. 1988;2(2):293–296. doi: 10.1111/j.1365-2958.1988.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 21.Bork P, Koonin EV. A P-loop-like motif in a widespread ATP pyrophosphatase domain: Implications for the evolution of sequence motifs and enzyme activity. Proteins. 1994;20(4):347–355. doi: 10.1002/prot.340200407. [DOI] [PubMed] [Google Scholar]
- 22.Demeshkina N, Hirokawa G, Kaji A, Kaji H. Novel activity of eukaryotic translocase, eEF2: Dissociation of the 80S ribosome into subunits with ATP but not with GTP. Nucleic Acids Res. 2007;35(14):4597–4607. doi: 10.1093/nar/gkm468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb TR, et al. Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J Cell Sci. 2008;121(Pt 19):3140–3145. doi: 10.1242/jcs.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C-M, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18(3):320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, et al. Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol Cell Biol. 2006;26(10):3835–3841. doi: 10.1128/MCB.26.10.3835-3841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281(43):32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci USA. 2012;109(34):13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei H, et al. Immunotoxin resistance via reversible methylation of the DPH4 promoter is a unique survival strategy. Proc Natl Acad Sci USA. 2012;109(18):6898–6903. doi: 10.1073/pnas.1204523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10):M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48(13):2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.