Abstract
Major microtubules in epithelial cells are not anchored to the centrosome, in contrast to the centrosomal radiation of microtubules in other cell types. It remains to be discovered how these epithelial microtubules are generated and stabilized at noncentrosomal sites. Here, we found that Nezha [also known as calmodulin-regulated spectrin-associated protein 3 (CAMSAP3)] and its related protein, CAMSAP2, cooperate in organization of noncentrosomal microtubules. These two CAMSAP molecules coclustered at the minus ends of noncentrosomal microtubules and thereby stabilized them. Depletion of CAMSAPs caused a marked reduction of microtubules with polymerizing plus ends, concomitantly inducing the growth of microtubules from the centrosome. In CAMSAP-depleted cells, early endosomes and the Golgi apparatus exhibited irregular distributions. These effects of CAMSAP depletion were maximized when both CAMSAPs were removed. These findings suggest that CAMSAP2 and -3 work together to maintain noncentrosomal microtubules, suppressing the microtubule-organizing ability of the centrosome, and that the network of CAMSAP-anchored microtubules is important for proper organelle assembly.
Keywords: γ-tubulin, EB1
Animal cells organize two kinds of microtubules, centrosomal and noncentrosomal, during interphase of the cell cycle. Centrosomal microtubules grow radially from the centrosome, with their “minus ends” capped with a γ-tubulin ring complex (1). Their growing “plus ends” are subjected to dynamic instability, which is regulated by +TIPs (microtubule plus-end tracking proteins) (2). Noncentrosomal microtubules are produced by various mechanisms, and their minus ends are located in the cytoplasm without a focus on the centrosome (3, 4). The proportion of centrosomal and noncentrosomal microtubules in a cell varies depending on the cell type. For example, in fibroblast cells, centrosomal microtubules dominate, whereas in other cell types such as epithelial cells and neurons, noncentrosomal microtubules dominate (4). How such cell type-dependent differences are generated is unknown. It is also poorly understood how the minus ends of noncentrosomal microtubules are stabilized without their anchorage to the centrosome (5).
Only a few proteins have been identified as a cytoplasmic regulator of microtubules that can anchor their minus ends to noncentrosomal sites in animal cells. The centrosomal protein Ninein relocates to noncentrosomal sites, anchoring microtubule minus ends there (6, 7). Nezha [also known as calmodulin-regulated spectrin-associated protein 3 (CAMSAP3)] tethers noncentrosomal microtubules to the adherens junctions via its attachment to the minus ends of these microtubules (8). The Drosophila protein Patronin, which is related to CAMSAP3, stabilizes the minus ends of microtubules by protecting them against Kinesin 13-mediated depolymerization (9). Two other proteins, CAMSAP1 and CAMSAP2, are also related to CAMSAP3 (10), but their functions remain undetermined. In the present study, we investigated the roles of these proteins, focusing on CAMSAP2 and CAMSAP3, in microtubule organization in human Caco2 epithelial cells, whose microtubules are essentially noncentrosomal (11). Our results show that CAMSAPs play a key role in maintaining a population of noncentrosomal microtubules and that this population of microtubules is important for proper organelle assembly.
Results
Colocalization of CAMSAP2 and -3 at the Minus Ends.
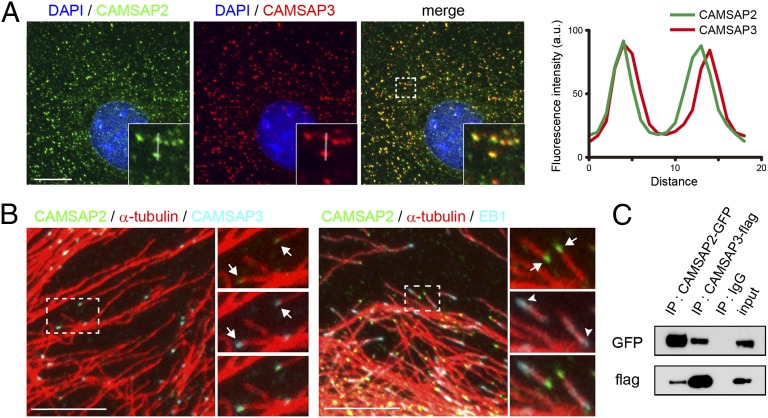
We used subconfluent cultures of Caco2 cells throughout the experiments, unless otherwise noted. CAMSAP3 protein is detected in small, distinct clusters, which are scattered through the cytoplasm, in addition to their accumulation along cell junctions, as reported previously (8). The number of these clusters per 100 μm2 ranged between 13.2 and 30.1 (n = 29 cells) depending on the subcellular positions. CAMSAP2 displayed a similar distribution pattern to that of CAMSAP3. Double immunostaining for these two proteins showed that their major immunofluorescence signals overlapped (Fig. 1A), although some faint signals of CAMSAP2 did not overlap with CAMSAP3. Closer observations of the coclusters of CAMSAP2 and -3 indicated that their outlines were slightly different (Fig. 1A, Right), suggesting that these two molecules were not evenly intermixed. CAMSAP2 and -3 coclusters were detected at an end of microtubules (Fig. 1B, Left). Coimmunostaining for CAMSAP2 and end-binding protein 1 (EB1), a microtubule plus-end tracking protein, showed that these two molecules were located at the opposite ends of a microtubule (Fig. 1B, Right), confirming that the CAMSAP2 and -3 bind the minus ends forming a cocluster. A biochemical sedimentation assay also confirmed that both CAMSAPs are associated with microtubules (Fig. S1A).
Fig. 1.
CAMSAP2 and -3 form a complex at the minus end of noncentrosomal microtubules. (A) Triple staining for CAMSAP2, CAMSAP3, and DAPI in Caco2 cells. (Inset) Enlargement of boxed area. The immunofluorescence signals of CAMSAP2 and -3 are traced along the white line, and their relative intensities are plotted at Right. (B) Triple immunostaining for CAMSAP2, CAMSAP3, and α-tubulin (Left), and for CAMSAP2, α-tubulin, and EB1 (Right). (Right) Enlarged view of boxed areas, in which the stains for CAMSAP2 (green) and CAMSAP3 (light blue) are separately shown with their merged image at the bottom. The arrows point to clusters of CAMSAP2 and -3 (Left) or CAMSAP2 (Right) attaching to an end of a microtubule. The arrowheads indicate an EB1 comet. (Scale bars, 10 μm.) (C) Coimmunoprecipitation of CAMSAP2 and -3. HEK293T cells were transiently transfected with plasmids for GFP-tagged CAMSAP2 and Flag-tagged CAMSAP3, and their lysates were subjected to immunoprecipitation (IP) with the antibodies against each tag. The precipitates were analyzed by SDS/PAGE and Western blotting using these antibodies.
To corroborate these observations, we ectopically expressed EGFP-tagged CAMSAP2 (CAMSAP2-GFP) and Kusabira Orange-tagged CAMSAP3 (CAMSAP3-mKOR) in Caco2 cells. The distributions of ectopic CAMSAP2 and CAMSAP3 molecules were essentially identical with those found endogenously (Fig. S1B, Top): They coaggregated. This was also confirmed by live imaging of the two molecules (Movie S1): The majority of the coclusters stayed at a fixed position, displaying dynamic morphological changes at their peripheries, whereas a minor fraction showed rapid movement or migration. When CAMSAP2 and -3 were singly or doubly overexpressed, their immunostaining signals extended along the microtubule body, displaying a rod-like shape (Fig. S1B, Middle), and in extreme cases, they decorated the entire microtubule (Fig. S1B, Bottom), suggesting that these molecules can bind to the body of microtubules when present excessively. In the present experiments, we chose transfectants expressing minimal levels of ectopic CAMSAPs to avoid artificial effects from their overexpression.
As found for CAMSAP3 previously (8), CAMSAP2 signals became diffused when microtubules were depolymerized with nocodazole. Even the rod-shaped clusters of CAMSASP2 or -3, produced by their overexpression, disappeared when polymerized tubulins are removed by nocodazole treatments at 4 °C (Fig. S1C). These observations suggest that the clustered appearance of these CAMSAPs is established only through the association with polymerized tubulins.
The colocalization of the two CAMSAPs suggested that they might physically bind to one another. We tested this through coprecipitation experiments. Because the antibodies against CAMSAP2 showed certain levels of cross-reactivity to CAMSAP3 when used for immunoprecipitation of native molecules or Western blotting (see Materials and Methods for details), we used alternative methods: We transfected cells with tagged CAMSAP2 and/or -3, and precipitated these molecules from their lysates, using antibodies specific to the tags. Analysis of the precipitates indicated that CAMSAP2 and -3 can cosediment together (Fig. 1C and Fig. S1D). Thus, the interactions between CAMSAP2 and -3 may also contribute to the observed appearance of these molecules.
CAMSAP2 and -3 Cooperate to Support Microtubule Growth.
Next, we examined whether these two CAMSAPs have any distinct properties, by depleting each molecule using specific siRNAs (Fig. S2A). When CAMSAP2 was removed, CAMSAP3 clusters did not show any changes in morphology and localization. On the contrary, when CAMSAP3 was depleted, CAMSAP2 immunostaining signals began to elongate, covering the microtubule (Fig. S2 B and C), suggesting that the behavior of CAMSAP2 is regulated by CAMSAP3. We also noted that the band intensity in Western blots of CAMSAP2 was reproducibly up-regulated after CAMSAP3 depletion (Fig. S2A). This may explain why CAMSAP2 clusters were elongated in CAMSAP3-depleted cells, as similar elongation was observed when CAMSAP2 was overexpressed.
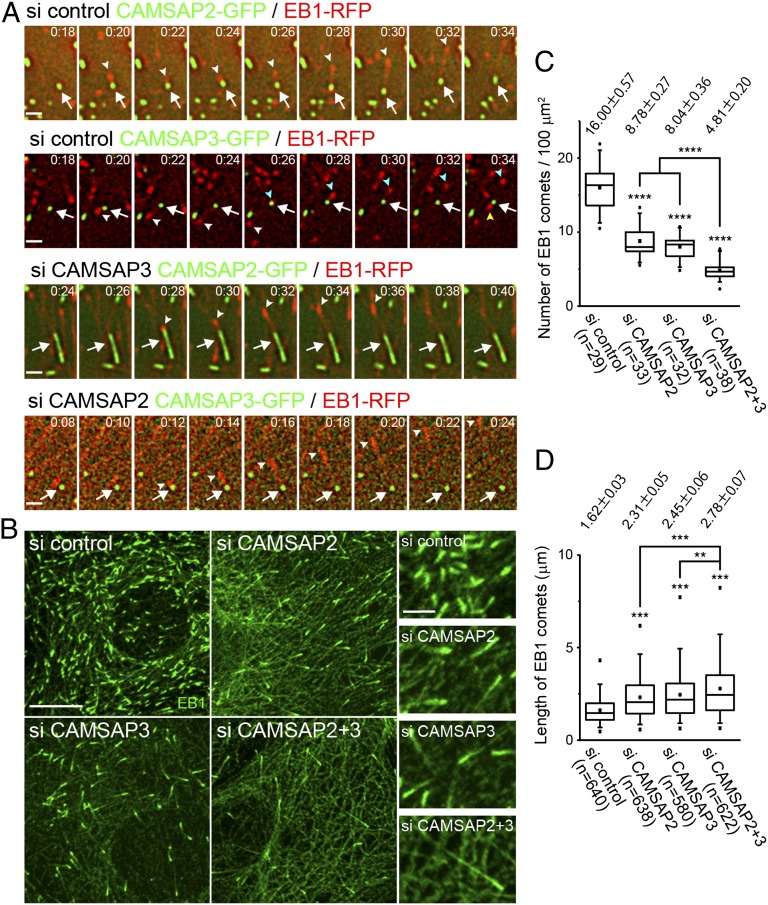
Then, we compared their ability to support microtubule growth. We doubly transfected Caco2 cells with the plus-end tracking protein EB1 and CAMSAP2 or CAMSAP3. EB1 was tagged with red fluorescent protein (EB1-RPF) and the CAMSAPs were tagged with green fluorescent protein (CAMSAP2-GFP or CAMSAP3-GFP). Time-lapse live imaging of these molecules showed that EB1 comets emanated from both CAMSAP2 and CAMSAP3 clusters (Fig. 2A and Movie S2, Upper). The frequency of EB1 emanation from each cluster varied in a range between zero and four times per minute, and the direction of EB1 emanation from a single CAMSAP cluster either kept constant or changed from time to time. These features of EB1 behavior were indistinguishable between the ectopic CAMSAP2 and -3 clusters. In these experiments, however, we could not determine which CAMSAP in their coclusters was responsible for EB1 emanation. We therefore depleted endogenous CAMSAPs from these transfectants. We found that, even when CAMSAP2 was removed, EB1 emanation still occurred from the ectopic CAMSAP3 clusters, and vice versa (Fig. 2A and Movie S2, Lower). This suggested that each CAMSAP can independently support microtubule plus-end polymerization. In the case of ectopic CAMSAP2 clusters, which show a rod-like shape due to the absence of CAMSAP3, EB1 comets emanated from only one end of the rod, indicating that this elongated cluster has a polarity.
Fig. 2.
CAMSAP2 and -3 can nucleate microtubules independently. (A) Time-lapse images of EB1-RFP and CAMSAP2-GFP or CAMSAP3-GFP coexpressed in Caco2 cells, which were transfected with control (si control), CAMSAP2 (si CAMSAP3), or CAMSAP3 (si CAMSAP3) siRNA. The arrows indicate CAMSAP2 or -3 clusters, and the arrowheads point to EB1 comets. In the control CAMSAP3 images, the second and third EB1 comets emerge as pointed with blue and yellow arrowheads, respectively. Montage images of Movie S2 are shown. (Scale bar, 1 μm.) (B) Cells transfected with the indicated siRNAs were immunostained for EB1. (Scale bar, 10 μm.) (C) The number of EB1 comets was quantified. Data were collected from three independent experiments, and more than nine cells were analyzed in each experiment. Values indicate average ± SEM. ****P < 1.0E-5. (D) The length of EB1 comets was measured. Data were collected from three independent experiments, and more than seven cells were analyzed in each experiment. Values indicate average ± SEM. **P < 0.001, ***P < 0.0001.
In the above observations, we could not determine whether the overall efficiency of microtubule growth from CAMSAP clusters remained unchanged or not, when one CAMSAP was depleted. To clarify this point, we compared the total number of EB1 comets between control and CAMSAP-depleted cells, using a fixed specimen. The number of EB1 comets decreased when either CAMSAP2 or -3 was depleted (Fig. 2 B and C). This suggested that, although each CAMSAP can independently support microtubule plus-end growth, these molecules do not seem to exhibit their full potential unless both CAMSAPs are present. Then, we depleted the two CAMSAPs together, and found that the number of EB1 comets was further decreased, suggesting that their coexistence is required for maintaining the normal number of microtubules with polymerizing plus ends. We also noted that EB1 comets exhibited longer tails in the absence of CAMSAPs and were longest when both CAMSAPs were codepleted (Fig. 2D).
To assess the role of CAMSAPs in microtubule polymerization, we decided to observe how microtubules would respond to the acute loss of CAMSAPs. To this end, we cotransfected Caco2 cells with EB1-RPF and DD-tagged CAMSAP3-GFP; the DD tag facilitates the degradation of CAMSAP3-GFP (12). Live imaging of a pair of CAMSAP3 and EB1-decorated microtubule in these transfectants revealed that, when CAMSAP3 was degraded, the partner microtubule also retracted from the original attachment site (Fig. S2D and Movie S3). This observation suggested that CAMSAP3 loss induced subsequent depolymerization of the associated microtubule, presumably at its minus ends. In this experiment, we could not visualize endogenous CAMSAP2, and therefore it remains unclear how or whether CAMSAP2 participated in the process observed. However, our finding is consistent with the observation that depletion of CAMSAP3 alone could reduce the number of EB1 comets.
CAMSAP Depletion Alters the Assembly Pattern of Microtubules.
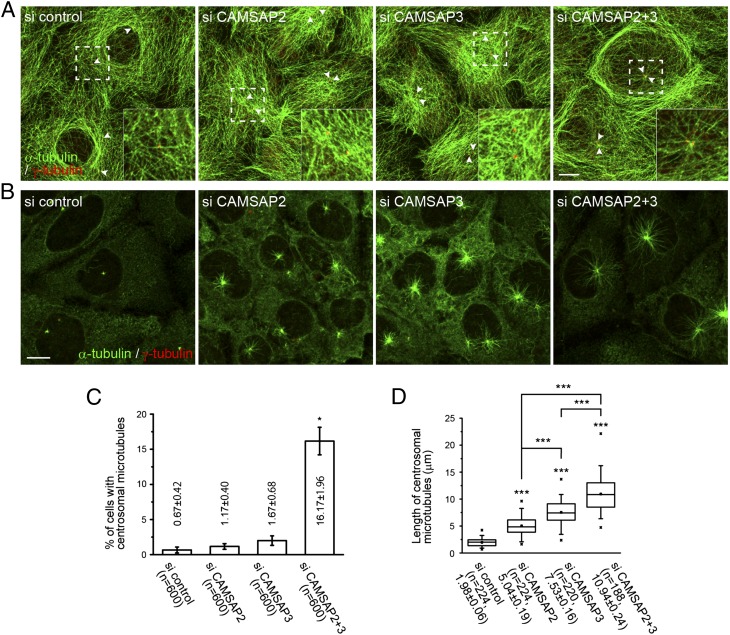
We next looked at the effect of CAMSAP depletion on the overall microtubule assembly pattern. In subconfluent cultures of Caco2 cells, the majority of microtubules are arranged in a pattern surrounding the nucleus, with microtubules only sparsely detected over the nucleus. In these cells, the centrosomes were located at random positions, and they rarely nucleated radial microtubules. However, when CAMSAP2 or -3 were depleted, microtubules became redistributed so as to densely cover the nucleus, and the centrosomes also redistributed around the center of these reorganized microtubule arrays (Fig. 3A). Surprisingly, when both CAMSAP2 and -3 were simultaneously knocked down, a certain population of cells exhibited the centrosomal radiation of microtubules (Fig. 3 A and C). Another set of microtubules also emerged in these cells, running along the cell periphery. To confirm whether these changes in microtubule assembly pattern were due to the specific effects of siRNA-mediated removal of CAMSAPs, we performed rescue experiments. We cotransfected Caco2 cells with cDNAs encoding mouse CAMSAP2 and -3 and siRNAs for CAMSAP2 and -3. This coexpression of mouse cDNAs completely abolished the RNAi-induced phenotypes (Fig. S3, Top). In these cells, centrosomal microtubules were no longer detected (nearly 100% efficiency). Expression of cDNA encoding either one of the two CAMSAPs in the double-knockdown cells also resulted in the complete removal of centrosomal microtubules, but condensed microtubules tended to be detectable near the nucleus in these cells (Fig. S3, Middle and Bottom), suggesting that single CAMSAPs may not be able to cover all of the cooperative functions of the two CAMSAPs.
Fig. 3.
Alteration of microtubule assembly pattern in the absence of CAMSAPs. (A) Double immunostaining for α-tubulin (green) and γ-tubulin (red) in Caco2 cells transfected with the indicated siRNA. (Inset) Enlargement of boxed area. The arrowheads point to centrosomes, which are generally detected as a doublet in each cell. (B) Microtubule growth after nocodazole washout. Confluent cells transfected with the indicated siRNAs were treated with 10 μM nocodazole for 1 h at 4 °C, incubated for another 7 min after removing nocodazole, and then fixed and immunostained for α-tubulin and γ-tubulin. The relative differences in the degree of centrosomal radiation of microtubules between the samples are maintained during further incubation periods, although noncentrosomal microtubule fragments gradually increase after 10 min. (Scale bars, 10 μm.) (C) The number of cells with an array of microtubules radiating from the centrosomes in A was quantified. Date represent the mean ± SEM from six independent experiments, in which 100 cells were analyzed per experiment. *P < 0.01. (D) The extent of microtubule radiation from the centrosomes in B was quantified by measuring the length of the four longest microtubules in each cell. Data were collected from three independent experiments. Values indicate mean ± SEM. ***P < 0.0001.
These observations suggest that the microtubule-organizing ability of the centrosomes was restored or up-regulated in CAMSAP-depleted cells. To verify this idea, we treated confluent Caco2 cells with nocodazole to depolymerize microtubules and observed the microtubule regrowth processes after nocodazole washout. Although Caco2 cells normally have no centrosomal microtubules, the initial repolymerization of microtubules occurred from the centrosome under this specific condition. As anticipated, this centrosomal radiation of microtubules was enhanced in CAMSAP2- or CAMSAP3-depleted cells, where CAMSAP3 loss was more effective than that of CAMSAP2, and this effect was maximized when both CAMSAPs were codepleted (Fig. 3 B and D). These observations imply that CAMSAPs or CAMSAP-associated microtubules suppress centrosomal microtubule growth in normal cells.
γ-Tubulin plays a central role in the nucleation of microtubules. This tubulin was, however, not particularly concentrated on CAMSAP2 clusters (Fig. S4A), as we reported for CAMSAP3 (8). To confirm γ-tubulin dependency or independency of microtubule growth from CAMSAPs, we looked at the effect of γ-tubulin depletion on microtubule patterning. Western blot analysis showed that γ-tubulin was greatly reduced in the cells treated with specific siRNAs (Fig. S4B). Immunostaining experiments confirmed that γ-tubulin signals were strongly down-regulated in the cytoplasm of siRNA-treated cells, although some γ-tubulin remnants were still detectable in the centrosomes. In these cells, the microtubule assembly pattern was not particularly altered (Fig. S4C), supporting the idea that γ-tubulin is dispensable for the CAMSAP-dependent organization of microtubules.
During these observations, we noticed that at least one of the two centrosomes present in each cell, detected by immunostaining for γ-tubulin, overlapped CAMSAP2 or -3 signals (Fig. S4A). Similar results were obtained by observing exogenously introduced tagged CAMSAP2 and -3, suggesting that these CAMSAPs could also associate with the centrosome, although the mechanism for their asymmetrical distribution in each pair of centrosomes remains unknown. In our previous studies, we missed the centrosomal localization of CAMSAP3 (8): this is probably due to the higher sensitivity of our newly prepared anti-CAMSAP3 antibodies in antigen detection.
Alterations in Microtubule Dynamics After CAMSAP Depletion.
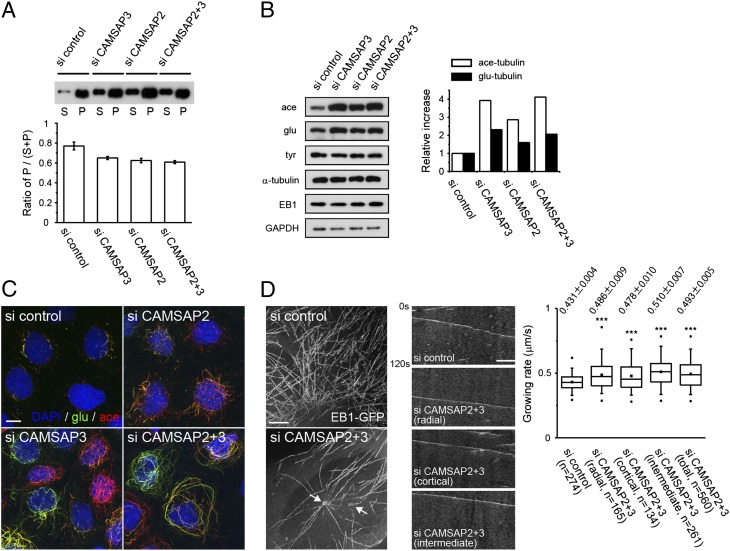
Analysis of EB1-tracked microtubules suggested that the microtubules with growing plus ends were decreased in CAMSAP-depleted cells (Fig. 2B). Consistently, tubulin fractionation assays showed that the ratio of insoluble α-tubulin to total α-tubulin decreased in CAMSAP-depleted cells (Fig. 4A), confirming that the dynamic equilibrium between tubulin monomers and polymers shifted toward the monomer phase in these cells. We also examined posttranslational modifications of microtubules and found that the level of tyrosination slightly decreased in CAMSAP-depleted cells, whereas that of acetylation and detyrosination increased in these cells (Fig. 4 B and C). These results suggest that the overall microtubule dynamics was changed by CAMSAP depletion.
Fig. 4.
Microtubule dynamics in CAMSAP-depleted cells. (A) Changes in the ratio of polymerized and unpolymerized α-tubulins after CAMSAP depletion. Cells were incubated with a microtubule-stabilizing buffer, and the lysates were fractionated into the supernatant (S) and pellet (P). α-Tubulin was detected by Western blotting. The ratio of P to S+P was quantified below. Data represent the mean ± SEM from three independent experiments. (B) Posttranslational modification of microtubules. Lysates of Caco2 cells treated with the indicated siRNAs were subjected to Western blotting using antibodies against acetylated (ace), detyrosinated (glu), and tyrosinated (tyr) tubulin. Band intensities were measured and normalized using the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) bands at Right. A typical result is shown. (C) Triple immunostaining for detyrosinated and acetylated tubulins, and DAPI. (Scale bar, 10 μm.) (D) Maximum projection images (Left) and kymograph (Center) of Movie S4. The arrow points to the putative position of centrosomes; EB1 radiation mainly occurs from the left one. (Scale bars: Left, 5 μm; Right, 2.5 μm.) The velocity of EB1 comets was measured and shown at Right. For CAMSAP2/3 codepleted cells, the EB1 comets were classified into three groups: “radial,” the comets nucleating from the centrosome; “cortical,” those growing in the cortical region that was defined as the area spanning 5 μm from the cellular edge; and “intermediate” for all other comets. For control cells, the measurement was performed without classification. Data were compiled from at least five cells per experiment in four independent experiments. Values show average ± SEM. ***P < 0.0001.
To further characterize the dynamic behavior of the microtubule arrays reorganized by CAMSAP depletion, we compared the plus-end dynamics of microtubules between control and CAMSAP2/3 codepleted cells by analyzing the EB1 comets. Time-lapse images of EB1-GFP (Movie S4) showed that EB1 comets migrated in variable directions in control cells, whereas those in CAMSAP-depleted cells seemed to radiate either from the centrosome or from unidentified origins. The latter was most evident at the cortical area of the cell. When radial EB1 comets reached the cortical zone, they occasionally turned to migrate along the cell border, whereas others underwent “catastrophe” or “capture” before the turn. Therefore, the cortical microtubules might be a mixture of centrosomal and noncentrosomal microtubules. Then, we found that the average velocity of EB1 comets increased for any population of microtubules in CAMSAP-depleted cells (Fig. 4D), suggesting that the speed of plus-end polymerization was generally enhanced in these cells. This feature of microtubule dynamics is probably linked to the other profiles of microtubule behavior observed in CAMASAP-depleted cells, including the increase of free tubulins and that of the EB1 comet length, because similar relations were found through in vitro study of the yeast EB1 homolog Mal3 (13). That is, the increased concentration of tubulins leads to the enhanced velocity as well as elongation of Mal3 comets. To summarize, CAMSAP depletion caused a reduction in the number of microtubules with growing plus ends, as well as in the amount of polymerized tubulins. The microtubules remaining in CAMSAP-depleted cells exhibit altered properties, including enhanced posttranslational modifications.
Depletion of CAMSAPs Affects Organelle Assembly.
We wondered whether the alterations of microtubule pattern and dynamics affected any other intracellular structures. Immunostaining for various organelles revealed that early endosomes and the Golgi apparatus showed altered distributions in CAMSAP-depleted cells. In control Caco2 cells, early endosomes were detected as dispersed vesicles by immunostaining for the early endosome antigen 1 (EEA1). After CAMSAP depletion, however, these vesicles accumulated around the centrosome, on which radial microtubules converge (Fig. S5A). In control Caco2 cells, the Golgi apparatus is distributed as discontinuous clusters at the perinuclear regions, and the centrosomes did not show any spatial relations to the Golgi clusters. When CAMSAPs were depleted, however, the Golgi complex became fragmented and scattered over wider areas of the cytoplasm, as revealed by immunostaining for giantin, a cis and medial Golgi marker (Fig. S5 B and C). In the cells having acquired the centrosomal radiation of microtubules, a fraction of these Golgi fragments came to accumulate in the centrosomal zone (Fig. S5D). These effects of CAMSAP removal on Golgi redistribution were enhanced most by codepletion of CAMSAP2 and -3 (Fig. S5C). The specificity of these RNAi effects was confirmed by rescue experiments (Fig. S5E). As it is well known that endosomes (14) and endoplasmic reticulum–Golgi membranes (15, 16) use microtubules for their trafficking, these findings suggest that the microtubule network organized by CAMSAPs is important for proper assembly of these organelles.
Discussion
Our results demonstrate that CAMSAP2 and -3 play a critical role in maintaining noncentrosomal microtubules and in determining the overall growth pattern of microtubules in epithelial cells. Although each protein can independently cap the minus ends, the properties and functions of the two CAMSAPs appeared not identical; cells required the presence of both molecules for maintaining the normal organization of microtubules. The two CAMSAPs probably work together through their partly redundant and partly complementary properties.
Three important questions remain unanswered. First, why are polymerized tubulins reduced in CAMSAP-depleted cells? One likely possibility is that CAMSAP2 and -3 are the major minus-end stabilizers in Caco2 cells, and thus their depletion resulted in a simple shortage of cytoplasmic factors that can stabilize the minus ends. This would have naturally reduced the microtubule number. The second question is the following: How is the centrosomal nucleation of microtubules induced by CAMSAP depletion? A plausible idea is that CAMSAPs and centrosomes compete for free tubulins. In normal cells, CAMSAPs may dominate in this competition, and only in their absence are centrosomes able to nucleate their own microtubules. Alternatively, CAMSAPs might directly inhibit the function of centrosomes, as these proteins were detected on some of the centrosomes. On the other hand, it is of note that the centrosome could not fully compensate for the reduction of polymerized tubulins in CAMSAP-depleted cells. This suggests that the microtubule-organizing ability of the centrosome might be suppressed by additional mechanisms in Caco2 epithelial cells. Third, how did CAMSAP depletion lead to the increased posttranslational modification of microtubules? This would be possible if CAMSAP clusters could supply some biochemical modulators onto the microtubules growing from themselves.
In the present study, we purposely used a subconfluent culture of Caco2 cells to obtain high-resolution optical images of microtubules and associated proteins. In vivo, however, cells are tightly packed together, epithelial cells are apicobasally polarized, and microtubule minus ends are oriented toward the apical membrane (11, 17, 18). Therefore, it will be interesting to determine whether CAMSAPs can also regulate the orientation of microtubules in polarized epithelial cells; such studies are actually under way. In addition, it should be reemphasized that, in many cell types, the majority of microtubules focus on the centrosome, resulting in the formation of radial microtubule arrays, in contrast with the pattern seen in epithelial cells. Future studies will be needed to determine whether CAMSAPs are involved in such cell type-specific microtubule patterning.
Materials and Methods
Plasmid Construction.
cDNA of mouse Camsap2 was cloned from an E16 mouse brain cDNA library by PCR and inserted into a pCA-sal-EGFP or pCA-sal-Flag vector (19). To obtain DD-tagged CAMSAP3-GFP, CAMSAP3-mKOR, CAMSAP3-Flag, and CAMSAP3-HA, mouse Camsap3 (8) was subcloned into a pPTuner vector (Clontech), pmKO1-MC1 vector (MBL), pCMV-Tag 2B (Stratagene), and a pHA vector, in which the GFP tag of the pGFP vector (Clontech) was replaced with an HA tag, respectively. To construct EB1-RFP, EB1 was subcloned into the pCANw-RFP vector with a RFP-tag sequence on its 3′ end. cDNA of EB1 (20) was a gift from Y. Mimori-Kiyosue (RIKEN Center for Developmental Biology, Kobe, Japan). Stealth siRNAs and Mission siRNAs were purchased from Invitrogen and Sigma, respectively. Sequence information for siRNAs can be found in SI Materials and Methods.
Antibodies Preparation and Specificity.
Rabbit polyclonal antibody (pAb) against CAMSAP3 was raised by immunizing rabbits with GST-tagged mouse CAMSAP3 (596–1076 aa) and affinity-purified with the antigen. The rat monoclonal antibody (mAb) against CAMSAP2 (clone 37C2) was generated by injection of Donryu rats (Japan SLC) with GST-tagged mouse CAMSAP2 (313–755 aa) and subsequent hybridoma screening. The mAbs were purified through ammonium sulfate precipitation and DEAE column chromatography. Rabbit pAb against CAMSAP2 (Proteintech) was also used. We confirmed the specificity and availability of these antibodies using RNAi for each molecule: The anti-CAMSAP3 pAb exclusively recognized CAMSAP3 under any experimental conditions. The rat anti-CASAMP2 mAb specifically reacted with CAMSAP2 in immunostaining, but this mAb could not be used for other immunological methods. The rabbit anti-CASAMP2 pAb recognized CAMSAP2, but it cross-reacted with mouse CAMSAP3 in Western blotting and both mouse and human CAMSAP3 in immunoprecipitation; these cross-reactions did not occur in immunostaining. All animal studies have been approved by the Animal Care and Use Committee at RIKEN Center for Developmental Biology. Other antibodies used are listed in SI Materials and Methods.
Cell Culture and Transfection.
Caco2 and HEK293T cells were cultured as described previously (8). Unless otherwise mentioned, all experiments were performed with 40–70% confluence. Cells were transfected using Lipofectamine 2000 (Invitrogen), FuGENE HD, X-tremeGENE 9, or X-tremeGENE HP (Roche), according to the manufacturer’s instructions. For siRNA treatments, cells were transfected using Lipofectamine RNAi MAX (Invitrogen). In nocodazole washout experiments, cells were treated with 10 μM nocodazole at 4 °C for 60 min. After removing nocodazole, cells were incubated in a 1:1 mixture of DMEM and Ham’s F-12 supplemented with 10% (vol/vol) FBS, and subjected to live-cell imaging or fixation.
Image Acquisition and Live-Cell Imaging.
Methods for immunostaining of fixed cells are described in SI Materials and Methods. Time-lapse imaging for Movies S1, S2, and S4 was performed using a Deltavision microscope (Applied Precision) as described earlier (19). Cells were transiently transfected with indicated expression vectors. When necessary, sequential images were deconvoluted using SoftWoRx (Applied Precision). Live cell imaging for Movie S3 was performed on an Olympus IX71-ZDC spinning disk confocal microscope. Details for image processing and quantification are described in SI Materials and Methods.
Biochemical Assays.
Methods for immunoprecipitation are described in SI Materials and Methods. Measurement of the soluble and polymerized tubulin fractions was performed as described previously with slight modifications (21). Briefly, Caco2 cells transfected with indicated siRNAs were incubated with a microtubule stabilization buffer [80 mM K–1,4-piperazinediethanesulfonic acid, pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 μM Taxol, 0.5% (vol/vol) Nonidet P-40, and protease and phosphatase inhibitor mixture] at 37 °C for 5 min in the dark. Lysates were collected and centrifuged at 17,400 × g for 15 min at 30 °C. After the supernatant was separated, the pellet was washed once with microtubule stabilization buffer without detergents, protease and phosphatase inhibitors, and then the supernatant and pellet were mixed with an equal volume of a sample buffer, respectively. An equivalent volume of each fraction was subjected to SDS/PAGE and analyzed by Western blotting probed with the antibody against α-tubulin. A microtubule cosedimentation assay was carried out as described earlier (8).
Statistical Analysis.
We performed the same experiments at least three times to confirm reproducibility. In the histograms, data are represented as mean ± SEM, except for Fig. 4A. The box-and-whisker plots show the mean (small square), median (line), 1st and 99th (crosses), 5th and 95th (whisker), and 25th and 75th (boxes) percentiles. The results were analyzed by Student t test, Welch’s t test [two-tailed, unpaired; Excel (Microsoft)], or Mann–Whitney U test [Prism 5 (Graphpad)]. Values of P < 0.01 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Y. Mimori-Kiyosue for reagents; M. Nomura, H. Saitou, H. Abe-Ishigami, and C. Uemura-Yoshii for technical support; and Optical Image Analysis Unit for microscopical support. This work was supported by the programs Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (to W.M.) and Grants-in-Aid for Specially Promoted Research of the Ministry of Education, Science, Sports, and Culture of Japan (to M.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218017109/-/DCSupplemental.
References
- 1.Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12(11):709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhmanova A, Steinmetz MO. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9(4):309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 3.Keating TJ, Borisy GG. Centrosomal and non-centrosomal microtubules. Biol Cell. 1999;91(4–5):321–329. [PubMed] [Google Scholar]
- 4.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119(Pt 20):4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 5.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13(15):R614–R624. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- 6.Lechler T, Fuchs E. Desmoplakin: An unexpected regulator of microtubule organization in the epidermis. J Cell Biol. 2007;176(2):147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss DK, et al. Ninein is released from the centrosome and moves bi-directionally along microtubules. J Cell Sci. 2007;120(Pt 17):3064–3074. doi: 10.1242/jcs.010322. [DOI] [PubMed] [Google Scholar]
- 8.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135(5):948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143(2):263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baines AJ, et al. The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol Biol Evol. 2009;26(9):2005–2014. doi: 10.1093/molbev/msp115. [DOI] [PubMed] [Google Scholar]
- 11.Meads T, Schroer TA. Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motil Cytoskeleton. 1995;32(4):273–288. doi: 10.1002/cm.970320404. [DOI] [PubMed] [Google Scholar]
- 12.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450(7172):1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 14.Apodaca G. Endocytic traffic in polarized epithelial cells: Role of the actin and microtubule cytoskeleton. Traffic. 2001;2(3):149–159. doi: 10.1034/j.1600-0854.2001.020301.x. [DOI] [PubMed] [Google Scholar]
- 15.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: Regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7(4):631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246(2):263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 17.Bacallao R, et al. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109(6 Pt 1):2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert T, Le Bivic A, Quaroni A, Rodriguez-Boulan E. Microtubular organization and its involvement in the biogenetic pathways of plasma membrane proteins in Caco-2 intestinal epithelial cells. J Cell Biol. 1991;113(2):275–288. doi: 10.1083/jcb.113.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichii T, Takeichi M. p120-catenin regulates microtubule dynamics and cell migration in a cadherin-independent manner. Genes Cells. 2007;12(7):827–839. doi: 10.1111/j.1365-2443.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- 20.Mimori-Kiyosue , et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168(1):141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minotti AM, Barlow SB, Cabral F. Resistance to antimitotic drugs in Chinese hamster ovary cells correlates with changes in the level of polymerized tubulin. J Biol Chem. 1991;266(6):3987–3994. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.