Abstract
The plant hormone jasmonate (JA) plays an important role in regulating growth, development and immunity. A key step in JA signaling is ligand-dependent assembly of a coreceptor complex consisting of the F-box protein COI1 and JAZ transcriptional repressors. Assembly of this receptor complex results in proteasome-mediated degradation of JAZ repressors, which at resting state bind to and repress the MYC transcription factors. Although the JA receptor complex is believed to function within the nucleus, how this receptor complex enters the nucleus and, more generally, the cell biology of jasmonate signaling are not well understood. In this study, we conducted mutational analysis of the C termini (containing the conserved Jas motif) of two JAZ repressors, JAZ1 and JAZ9. These analyses unexpectedly revealed different subcellular localization patterns of JAZ1ΔJas and JAZ9ΔJas, which were associated with differential interaction of JAZ1ΔJas and JAZ9ΔJas with MYC2 and differential repressor activity in vivo. Importantly, physical interaction with MYC2 appears to play an active role in the nuclear targeting of JAZ1 and JAZ9, and the nuclear localization of JAZ9 was compromised in myc2 mutant plants. We identified a highly conserved arginine residue in the Jas motif that is critical for coupling MYC2 interaction with nuclear localization of JAZ9 and JAZ9 repressor function in vivo. Our results suggest a model for explaining why some JAZΔJas proteins, but not others, confer constitutive JA-insensitivity when overexpressed in plants. Results also provide evidence for a transcription factor-dependent mechanism for nuclear import of a cognate transcriptional repressor JAZ9 in plants.
Keywords: biotic stress, E3 ubiquitin ligase, salicylic acid, plant immunity
Plants, being sessile organisms subjected to ever-changing environmental stresses, have evolved complex mechanisms to properly balance their growth and development with appropriate defense responses. One of the major plant hormones involved in regulating the balance between growth and defense belongs to a class of lipid-derived molecules, collectively termed jasmonates (JAs) (1). In healthy plants, JA signaling plays a role in reproductive development, photomorphogenesis, and other growth responses (2–6). Defense responses that are regulated by JA signaling are activated by environmental stress factors including tissue damage from herbivorous insects, pathogen attack, drought, and UV irradiation (7–12). Induction of the JA response pathway results in significant transcriptional reprogramming and typically shifts the balance from growth to defense-related cellular processes, through inhibition of gene expression involved in cell cycle progression and photosynthesis, and activation of defense-related genes (13–16).
In the past five years, exciting progress has been made in the understanding of JA signaling and how this stress hormone influences plant growth, development and defense. The bioactive JA ligand, (3R,7S)-jasmonoyl-l-isoleucine (JA-Ile), is perceived by a receptor complex consisting of CORONATINE-INSENSITIVE1 (COI1), the F-box subunit of the SCFCOI1 ubiquitin ligase, and members of the JASMONATE-ZIM DOMAIN (JAZ) family of transcriptional repressors (17–23). As an elegant example of host-pathogen coevolution, several strains of the plant pathogen Pseudomonas syringae have developed the ability to produce coronatine (COR), a structural mimic of JA-Ile that increases susceptibility of the host plant (24–27). Arabidopsis has 12 JAZ proteins, which share two conserved functional motifs: the ZIM motif in the central part of the protein and the Jas motif at the C terminus (28). The ZIM motif mediates homo- and heteromeric interactions among JAZ repressors and JAZ interactions with the adapters/corepressors NINJA and TPL proteins (29–31). The C-terminal Jas motif is necessary for interaction with the LRR domain of COI1 (17, 19, 20, 32, 33). A short peptide of 21 amino acids within the Jas motif defines the minimal “degron” that is sufficient for ligand-dependent formation of COI1-JAZ1 receptor complexes (19). In addition, the Jas motif mediates interaction between JAZ proteins and their target transcription factors (TFs) that control downstream responses (17). Among the most well characterized TFs targeted by JAZ are members of a basic-helix–loop–helix family, including MYC2, -3, and -4, that are key regulatory components of diverse aspects of JA-mediated physiological responses (17, 33–38).
The core JA signaling components, including a multimeric transcriptional corepression complex consisting of JAZ, NINJA, and TPL proteins bound to JA-responsive TFs and the COI1-JAZ receptor complex, are believed to function in the nucleus (31). However, direct evidence of nuclear localization (and underlying nuclear targeting mechanisms) for many of the key JA signaling components is lacking and, more generally, the cell biology of jasmonate signaling is poorly understood. Previous studies have shown that JAZ1 variants (JAZ1∆Jas) lacking the entire Jas motif or carrying alanine substitutions at specific arginine residues in the Jas motif are unable to interact with COI1 in a ligand-dependent manner and are therefore resistant to SCFCOI1-dependent degradation through the proteasome (20, 33). As a result, transgenic Arabidopsis plants that express these JAZ1 variants exhibit JA insensitivity (20, 32, 33). A similar JA-insensitive phenotype was observed for JAZ3 (JAZ3∆Jas) and JAZ10 (JAZ10∆Jas) (17, 23, 30, 39). However, it has also been shown that the Jas motif is required for JAZ3, but not JAZ1 or JAZ10, interaction with TFs such as MYC2, which should be a key step in TF-specific transcriptional repression (29, 30). Therefore, although transgenic overexpression of JAZΔJas proteins has been used as the primary means in the discovery of the repressor function of JAZ proteins, how JAZ∆Jas proteins repress JA signaling has remained enigmatic (40). We report here that, in contrast to plants expressing JAZ1∆Jas, plants expressing JAZ9∆Jas do not exhibit obvious JA-insensitive phenotypes. This observation led us to conduct a series of experiments to understand the apparently different effects of the Jas motifs of JAZ1 and JAZ9 on JA signaling. Our results provide evidence for the existence of a TF-dependent mechanism for nuclear localization of cognate transcriptional repressor in plants, and show that nuclear localization of JAZ9 and interaction with MYC2 in the nucleus are coupled and are both required for repression of JA signaling.
Results
Constitutive Expression of Different JAZΔJas Proteins Results in Disparate JA-Insensitive Phenotypes.
Several previous studies have reported the effects of ectopically expressing JAZ∆Jas proteins on JA signaling. Specifically, overexpression of JAZ1, JAZ3, and JAZ10 variants lacking the C-terminal Jas motif led to JA insensitivity, providing key evidence that JAZ proteins are transcriptional repressors (17, 20, 23, 30, 33). During investigation of the role of JAZ9 in JA signaling, we introduced an HA-tagged JAZ9 variant lacking the entire Jas motif (35S:3×HA:JAZ9∆Jas) into Arabidopsis, expecting a phenotypic effect on JA signaling similar to that in plants overexpressing JAZ1∆Jas (20, 33). Surprisingly, lines expressing this construct did not exhibit JA insensitivity as determined by root growth inhibition assays, in contrast to lines transformed with 35S:3×HA:JAZ1∆Jas (SI Appendix, Fig. S1). One explanation for the lack of phenotype in 35S:3×HA:JAZ9∆Jas lines is that JAZ9∆Jas maintains the ability to interact with COI1 in vivo, resulting in JAZ9∆Jas degradation in response to JA stimulus. However, in vivo degradation assays performed on 10-d-old transgenic seedlings revealed that both 3×HA:JAZ1∆Jas and 3×HA:JAZ9∆Jas were highly stable in comparison with the wild-type JAZ1 and JAZ9 fusion proteins (SI Appendix, Fig. S1). This finding is in agreement with previous yeast two-hybrid (Y2H) results indicating that JAZ1∆Jas and JAZ9∆Jas do not interact with COI1 in the presence of COR (33). Therefore, the observed phenotypic difference cannot be explained by differential stability of JAZ1∆Jas and JAZ9∆Jas.
Role of MYC2 Interaction in Nuclear Localization of JAZ1 and JAZ9.
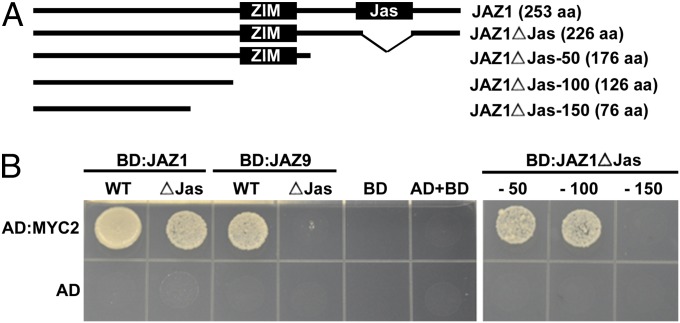
We next compared the ability of JAZ1∆Jas and JAZ9∆Jas mutants to interact with MYC2 using Y2H assay. Here, we found a difference between JAZ1∆Jas and JAZ9∆Jas. Whereas JAZ1∆Jas retained the ability to interact with MYC2, albeit to a lesser extent than the wild-type JAZ1, JAZ9∆Jas did not interact with MYC2 (Fig. 1B and SI Appendix, Fig. S2). Next, we deleted 50 amino acids at a time from the C terminus of JAZ1ΔJas (Fig. 1A) and tested the ability of each to interact with MYC2 in Y2H. JAZ1ΔJas −50 and −100 retained interaction with MYC2 (Fig. 1B). In contrast, JAZ1ΔJas −150 lost the ability to interact with MYC2, indicating that a second MYC2 interacting motif is located in the N terminus before the conserved ZIM motif (Fig. 1B). In addition, we found a difference in the subcellular localization patterns of YFP:JAZ1∆Jas and YFP:JAZ9∆Jas in Agrobacterium-mediated transient expression experiments in Nicotiana tabacum. Both YFP:JAZ1 and YFP:JAZ9 were found to be localized to the nucleus, mostly in undefined subnuclear bodies, whereas the YFP alone was localized in both the nucleus and cytoplasm in a diffuse manner (SI Appendix, Fig. S3). YFP:JAZ1∆Jas could be readily detected both in the nucleus and in the cytoplasm, but lost the localization to subnuclear bodies and resembled the diffuse localization of the YFP control (SI Appendix, Fig. S3). YFP:JAZ9∆Jas was also detected in both the nucleus and the cytoplasm, but the signal in the nucleus was weaker compared with YFP:JAZ1∆Jas, even though YFP:JAZ1∆Jas and YFP:JAZ9∆Jas proteins were expressed to similar levels (SI Appendix, Figs. S3 and S4). These results provided an initial indication that there might be a correlation between the ability of JAZ proteins to interact with MYC2 and an enhanced localization to the nucleus. We also determined the subcellular localization of COI1 and NINJA and found that both YFP:COI1 and YFP:NINJA localize diffusely within the nucleus in our transient expression experiments (SI Appendix, Fig. S5). The nuclear localization of YFP:NINJA was in agreement with previously reported localization results (31).
Fig. 1.
Deletion of the Jas domain differentially affects interaction with MYC2. (A) A diagram of the coding sequences of JAZ1, JAZ1ΔJas, and the C-terminal truncations used to create yeast two-hybrid (Y2H) constructs. (B) MYC2 interaction with JAZ1, JAZ9, JAZ1ΔJas, JAZ9ΔJas, and C-terminal deletions of JAZ1ΔJas in Y2H assays. Yeast cultures cotransformed with BD:JAZ and AD:MYC2 (Upper) or GAL4AD (Lower) spotted on −LWH drop-out media. Protein interactions are indicated by colony growth.
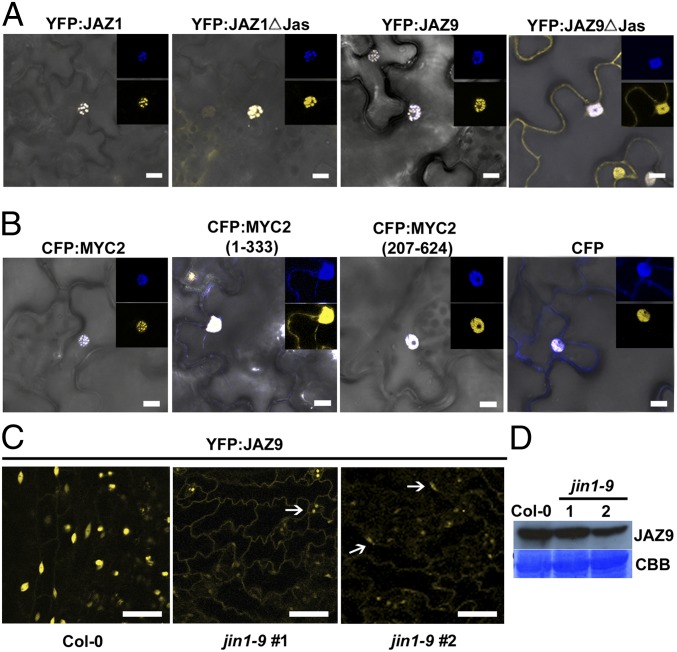
The apparent correlation between the ability of JAZ proteins to interact with MYC2 and an enhanced localization to the nucleus prompted us to examine the possibility that MYC2 might play an active role in targeting JAZ repressors to the nucleus. MYC2 was transiently coexpressed together with either JAZ1 or JAZ9 in N. tabacum leaves. Both YFP:JAZ1 and YFP:JAZ9 colocalized with CFP:MYC2 in subnuclear bodies (Fig. 2A). Interestingly, coexpression of YFP:JAZ1∆Jas with CFP:MYC2 changed the localization of this truncated JAZ from both nuclear and cytoplasmic (SI Appendix, Fig. S3) to predominantly nuclear (Fig. 2A). In contrast, coexpression of YFP:JAZ9∆Jas with CFP:MYC2 did not have the same effect on the cytoplasmic location of YFP:JAZ9∆Jas (Fig. 2A). Thus, overexpression of CFP:MYC2 was sufficient to drive complete nuclear localization of YFP:JAZ1∆Jas (which retains interaction with MYC2), but not YFP:JAZ9∆Jas (which does not interact with MYC2).
Fig. 2.
Nuclear localization of JAZ is influenced by interaction with MYC2. Overlay of bright-field and fluorescent images of YFP:JAZ and CFP:MYC2 (A), or YFP:JAZ9 and CFP:MYC2 or truncated derivatives of CFP:MYC2 (B), transiently coexpressed in N. tabacum epidermal cells. (Insets) Portions of images enlarged to show nuclei with CFP channel alone (Upper) and YFP channel alone (Lower). (Scale bar: 10 µm.) (C) Fluorescent images of leaf epidermal cells expressing YFP:JAZ9 in transgenic Col-0 or jin1-9 (myc2) mutants. Arrows indicate nuclei. (Scale bar: 50 µm.) (D) Full-length YFP:JAZ9 protein was detected from each line by Western blot using a polyclonal antibody against GFP. CBB, Coomassie brilliant blue staining of PVDF membrane.
MYC2 has been shown to interact with JAZs via its N terminus (37). Protein sequence analysis using ProteinPredict software, which is based on the LOCtree and PredictNLS algorithms (41, 42), showed that the C-terminal half of MYC2 contains a monopartite nuclear localization signal (NLS), KRPKKRGRK433-441. We examined the localization of the N-terminal (1–333) and C-terminal (207–624) halves of MYC2 and the effects of the N-terminal and C-terminal halves of MYC2 on the localization of JAZ9. Consistent with the presence of a NLS in the C terminus, the C-terminal half of MYC2 was localized exclusively in the nucleus. In contrast, the N-terminal half was localized partially in the cytoplasm and partially in the nucleus (Fig. 2B). YFP-JAZ9 nuclear localization was not affected when coexpressed with the C terminus of MYC2, which does not interact with JAZ9 in Y2H (Fig. 2B and SI Appendix, Fig. S6). However, the cytoplasmically localized N terminus of MYC2, which interacts with JAZ9, trapped a portion of YFP:JAZ9 in the cytoplasm (Fig. 2B and SI Appendix, Fig. S6). This result further indicates that physical interaction with MYC2 affects JAZ9 localization in the cell.
We next asked the question about whether endogenous MYC2 is necessary for nuclear localization of full-length JAZ9. For this purpose, we transformed the 35S:YFP:JAZ9 construct into the Arabidopsis jin1-9 (myc2) mutant (43). Confocal microscopy of the resulting lines revealed partial mis-localization of YFP:JAZ9 to the cytoplasm, in addition to the expected nuclear localization (Fig. 2C); this localization pattern contrasted with that of YFP:JAZ9 in wild-type Col-0 plants, where YFP:JAZ9 signal was located entirely in the nucleus. There are at least three MYC-family transcription factors (MYC2, MYC3, and MYC4) that have been shown to interact with JAZs and are involved in JA signaling (37, 44). Quantitative RT-PCR analysis showed that, under our growth conditions, MYC2 and MYC3 are highly expressed in leaves, whereas MYC4 was expressed at a lower level (SI Appendix, Fig. S7). Taken together, the results provide genetic evidence that MYC2 is partially required for proper nuclear localization of JAZ9 and suggest that MYC3, and potentially MYC4, may also contribute to JAZ9 nuclear localization.
ArginineJas17 Is Critical for JAZ9 Nuclear Localization and Interaction with MYC2.
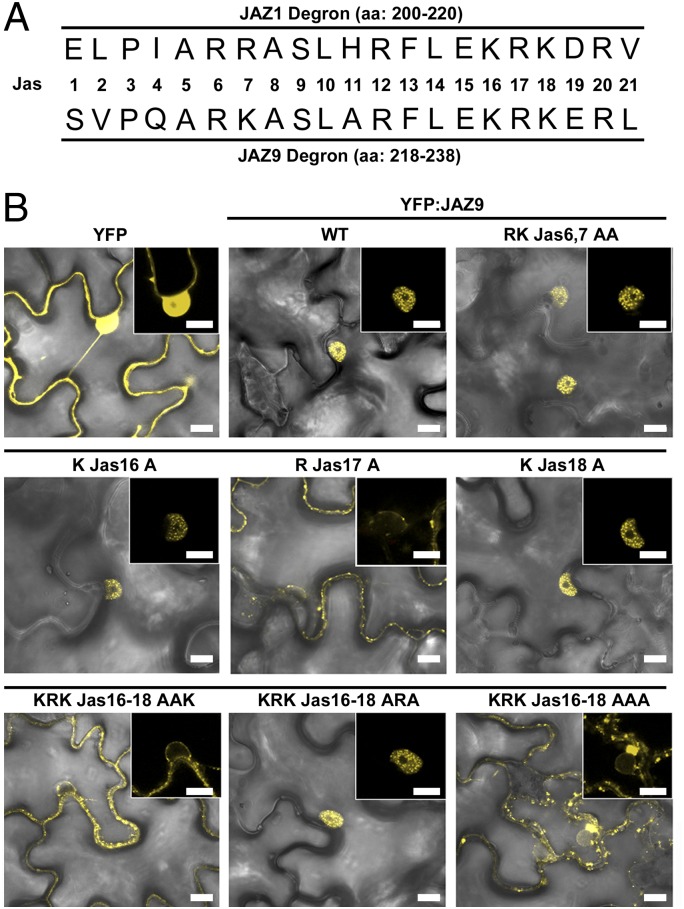
Having established an active role for MYC2 in nuclear localization of JAZ9, we next sought to identify specific residues within the Jas motif that are required for this process. The minimal JAZ1 degron was previously identified in the crystal structure of the COI1-JAZ1 coreceptor complex (19). Because different JAZs are of different lengths, the specific amino acid positions within the conserved Jas motif vary greatly among different JAZs. We use a simplified nomenclature to describe amino acid positions within the Jas motif (Fig. 3A). The Jas motif contains clusters of basic amino acid residues that resemble the classic mono- and bipartite NLS (45–47). In addition, the highly conserved KRK(E/D)RX5PY sequence in the C-terminal end of the Jas motif resembles a nonclassical NLS (46, 48), and has been implicated as a nuclear localization signal for JAZ1 (49). However, alternative splice variants of JAZ10 that lack the PY motif retain the ability to enter the nucleus (30, 39). To determine which, if any, of the motifs are involved in nuclear localization of JAZ9, we transiently expressed the wild-type JAZ9 and the RKJas6,7AA or KRKJas16-18AAA mutants as YFP fusions in N. tabacum leaf cells and analyzed their subcellular localization by confocal microscopy. YFP:JAZ9-RKJas6,7AA retained the ability to accumulate in the nucleus and to form subnuclear bodies, whereas YFP:JAZ9-KRKJas16-18AAA exhibited a cytoplasmic localization (Fig. 3B). These nuclear and cytoplasmic localization patterns were further confirmed in transgenic Arabidopsis expressing the same YFP:JAZ9 fusions (SI Appendix, Fig. S8), thus implicating the KRK motif as a critical element for nuclear localization of JAZ9. We noticed that the mis-localization of YFP:JAZ9-KRKJas16-18AAA to the cytoplasm was more complete than YFP:JAZ9ΔJas (SI Appendix, Fig. S3), indicating that the residual nuclear localization of YFP:JAZ9ΔJas may be caused by some nonspecific diffusion because YFP:JAZ9ΔJas is smaller than full-length YFP:JAZ9-KRKJas16-18AAA.
Fig. 3.
ArginineJas17 in the Jas domain is critical for nuclear localization of JAZ9. (A) Amino acid sequences of the Jas degron from JAZ1 and JAZ9. Jas1 is the first amino acid in sequence. Subsequent amino acids are numbered sequentially. (B) Bright-field and fluorescent image overlay of YFP:JAZ9 variants transiently expressed in N. tabacum epidermal cells. Single, double, or triple alanine substitutions in the Jas motif of JAZ9 are indicated in each panel. (Insets) Portions of images enlarged to shown nuclei. (Scale bar: 10 µm.)
To further delineate the KRKJas16-18 motif in nuclear localization of JAZ9, we next analyzed single, double and triple alanine mutations of KRKJas16-18. Single alanine point mutations of KJas16 or KJas18 had no effect on nuclear localization, as evidenced by the formation of subnuclear bodies (Fig. 3B). However, RJas17A completely excluded the fusion protein from the nucleus; all double and triple amino acid substitutions that included the RJas17A mutation had the same effect (Fig. 3B). Moreover, with only RJas17 present in the KRK motif (i.e., KRKJas16-18ARA), the fusion protein became properly localized to subnuclear bodies (Fig. 3B). The results from these experiments revealed a key role of RJas17 in the correct nuclear localization of YFP:JAZ9.
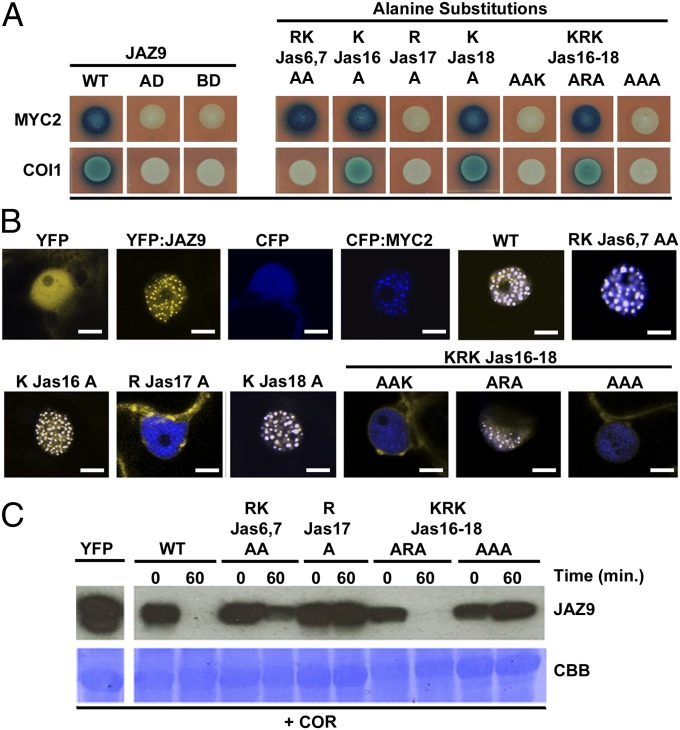
The identification of RJas17 as a critical determinant of JAZ9 nuclear localization provided us with another opportunity to test the hypothesis that JAZ9 nuclear localization is coupled to the physical interaction with MYC2. Results from Y2H assays revealed that KRKJas16-18 triple alanine mutations disrupted interaction with MYC2. However, JAZ9-KRKJas16-18ARA had normal interaction with MYC2. Therefore, within the KRK sequence, RJas17 was necessary and sufficient for mediating interaction with MYC2 (Fig. 4A).
Fig. 4.
ArginineJas17 is critical for JAZ9 interaction, stability, and colocalization with MYC2. (A) Yeast two-hybrid assay. Yeast cultures cotransformed with AD:MYC2 and BD:JAZ9 variants (Upper), or BD:COI1 and AD:JAZ9 variants (Lower), were spotted on −UWH drop-out media supplemented with X-Gal (and 10µM COR for COI1 interaction). Blue color indicates protein interaction. (B) Colocalization of YFP:JAZ9 variants and CFP:MYC2 transiently expressed in N. tabacum epidermal cells. Fluorescent images of YFP:JAZ9, CFP:MYC2, and vector controls, and dual channel overlay of YFP:JAZ9 carrying the indicated alanine substitutions coexpressed with CFP:MYC2. (Scale bar: 5 µm.) (C) In vivo JAZ protein degradation assay. Stability of YFP:JAZ9 variants expressed in Arabidopsis was determined by Western blot 1 h after 1 µM COR treatment. CBB, Coomassie brilliant blue staining of PVDF membrane.
Next, we examined the ability of various mutants to colocalize with CFP:MYC2 upon transient expression in N. tabacum. Again, we observed a strong correlation between nuclear localization of YFP:JAZ9 and the ability of these fusion proteins to interact with MYC2 and to colocalize with CFP:MYC2. Specifically, YFP:JAZ9-RKJas6,7AA, which interacted with MYC2 in Y2H assays, was colocalized with CFP:MYC2 in subnuclear bodies, similar to YFP:JAZ9 and CFP:MYC2 (Fig. 4B). Single lysine mutations in the KRKJas16-18 motif had no effect on nuclear import or colocalization with CFP:MYC2. However, any variation of YFP:JAZ9 lacking the critical amino acid RJas17 was excluded from the nucleus, whereas the CFP:MYC2 signal remained nuclear. Furthermore, within the KRK sequence RJas17 was sufficient for JAZ9 colocalization with CFP:MYC2 to subnuclear bodies (Fig. 4B). Consistent with findings reported in a previous study (33), we found that RKJas6,7 are not required for JAZ9 interaction with MYC2 (Fig. 4A). This result can now be explained by the fact that, of the basic amino acids in the Jas motif, only RJas17 is critical for JAZ9 interaction with MYC2.
We also determined the ability of JAZ9 mutants to interact with COI1 in Y2H experiments. Consistent with previous results (33), JAZ9-RKJas6,7AA did not interact with COI1, and again, we found that the RJas17A mutation disrupted interaction (Fig. 4A). These results indicated that alanine substitutions of both JAZ9-RKJas6,7 and -RJas17 would result in increased stability. As expected, in vivo protein degradation experiments in transgenic Arabidopsis seedlings showed that whereas YFP:JAZ9 and YFP:JAZ9-KRKJas16-18ARA are completely degraded within 1 h after treatment with coronatine, YFP:JAZ9-RKJas6,7AA and YFP:JAZ9-RJas17A were resistant to coronatine-mediated degradation (Fig. 4C). This finding is consistent with the ability of JAZ9 and JAZ9-KRKJas16-18ARA, but not JAZ9-RKJas6,7AA or JAZ9-RJas17A, to interact with COI1 (Fig. 4C) (33).
RJas17 Is Required for JAZ9 to Repress JA Responses in Planta.
Our results suggest a model in which the repressor function of JAZ9 protein in planta requires two inseparable RJas17-mediated processes: (i) MYC2-assisted nuclear localization of JAZ9 protein and (ii) physical interaction with MYC2. If this model is correct, we expect that (i) directing JAZ9∆Jas (SCFCOI1 degradation-resistant, but not MYC2-interacting) to the nucleus may not be sufficient to confer JA insensitivity, but (ii) JAZ9-RKJas6,7AA (SCFCOI1 degradation-resistant, MYC2-interacting, and nuclear-localized) should confer JA insensitivity when transgenically expressed in wild-type plants. To test these predictions, we fused JAZ9∆Jas to mCherry (mCH) and VirD2NLS, a well characterized nuclear localization signal from the Agrobacterium tumefaciens VirD2 protein (50, 51), and expressed these constructs in Col-0 plants. As expected, mCH:JAZ9ΔJas:NLS was localized in the nucleus (SI Appendix, Fig. S9). However, Col-0 plants expressing mCH:JAZ9ΔJas:NLS remained sensitive to JA, as indicated by inhibition of root elongation in seedlings grown on MS agar supplemented with 10 µM methyl-JA (MeJA; SI Appendix, Fig. S9). This result demonstrates that redirecting JAZ9∆Jas to the nucleus with a classical NLS is not sufficient to repress JA signaling.
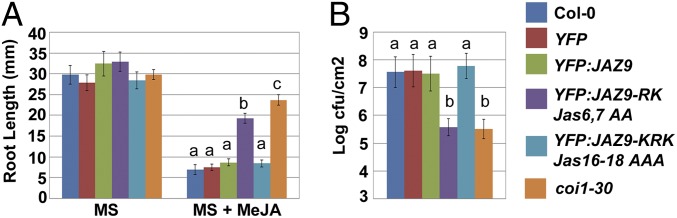
In contrast, transgenic Col-0 seedlings expressing YFP:JAZ9-RKJas6,7AA were partially insensitive (26% root growth inhibition, compared with 62–65% root growth inhibition for Col-0), whereas transgenic expression of YFP:JAZ9-KRKJas16-18AAA did not affect root or rosette growth sensitivity to JA (Fig. 5A and SI Appendix, Fig. S10). We also performed disease assays to evaluate whether these plants are altered in response to the coronatine-producing strain Pseudomonas syringae pv tomato (Pst) DC3000. We found typical symptom development on wild-type Col-0 and transgenic plants expressing YFP, YFP:JAZ9, or YFP:JAZ9-KRKJas16-18AAA (SI Appendix, Fig. S10). In contrast, plants overexpressing YFP:JAZ9-RKJas6,7AA showed significant resistance to Pst DC3000, comparable to that seen in the coi1-30 mutant (Fig. 5B and SI Appendix, Fig. S10). Bacterial counts were as much as 100 times lower than that in Col-0, and leaves retained a disease-free appearance (Fig. 5B and SI Appendix, Fig. S10). These findings indicate that the ability of JAZ9 to strongly repress JA responses in planta requires nuclear localization, MYC2 interaction, and resistance to SCFCOI1-dependent degradation.
Fig. 5.
Trangenic expression of JAZ9-RKJas6,7AA confers JA-insensitive phenotypes. (A) Quantification of root growth. Seedlings were grown in the absence or presence of 10 µM MeJA for 10 d, and root length was quantified using ImageJ software. Data are the means from 15 plants; error bars represent SD. Letters on columns indicate statistically significant differences (P < 0.01, Tukey’s HSD test). (B) Five-week-old plants were syringe-infiltrated with Pst DC3000 at a concentration of 106 cfu/mL. Bacterial enumeration was conducted 3 d postinoculation. Data are the mean cfus counted from four individual leaves per genotype; error bars represent SD. Letters on columns indicate statistically significant differences (P < 0.01, Tukey’s HSD test).
Discussion
JA is an important hormone that regulates diverse physiological processes in plants, ranging from growth and development to immunity against biotic and abiotic stresses. Recent studies from many laboratories have contributed to several significant advances in the dissection of the COI1-JAZ-MYC core signaling module and molecular connections to different downstream cellular pathways (7, 52–58). Although the COI1-JAZ-MYC signaling module is thought to perceive ligands and function inside the nucleus, direct evidence supporting this model and the underlying mechanisms for the nuclear entry of various signaling components has remained largely enigmatic. In this study, we addressed this important issue and show that COI1, JAZ1, JAZ9, MYC2, and NINJA are constitutively localized in the nucleus. In particular, our results revealed a TF-dependent mechanism for the nuclear localization of JAZ9 and suggest a model for explaining why some JAZΔJas proteins, but not others, confer constitutive JA-insensitivity when overexpressed in plants, a puzzle that has remained unresolved since the discovery of JAZ repressors.
All JAZ proteins studied to date have been shown to be localized in the nucleus and contain a highly conserved Jas motif at the C terminus (20, 23, 30, 49, 59). How the Jas motif mediates nuclear import, however, is not understood. The Jas motif contains clusters of basic amino acids conserved among the JAZ proteins (RKJas6,7 and KRKERJas16-20, separated by eight amino acids in JAZ9), which superficially resembles a putative bipartite NLS. Importantly, this stretch of basic amino acids, plus five additional nonbasic amino acids in the Jas motif of JAZ1, was capable of driving GFP into the nucleus (60). Unexpectedly, our results show that none of the basic amino acids, except for RJas17, in the Jas motif of JAZ9, is required for nuclear localization (Fig. 3B). Because clustered, basic amino acids are essential for the function of an NLS (45, 61), our results cast doubt on the idea that the Jas motif functions as an NLS per se. Instead, we found that the requirement of RJas17 for JAZ9 nuclear entry is correlated with its requirement for JAZ9 interaction with MYC2. First, overexpression of nuclear-localized CFP:MYC2 in N. tabacum cells is sufficient to drive complete nuclear localization of YFP:JAZ1∆Jas (which retains interaction with MYC2), but not YFP:JAZ9∆Jas (which does not interact with MYC2) (Fig. 2A). Second, CFP:MYC2 lacking its C-terminal NLS (KRPKKRGRK433-441) not only was partially localized in the cytoplasm, but also trapped a substantial portion of YFP:JAZ9 in the cytoplasm (Fig. 2B and SI Appendix, Fig. S6). Third, the nuclear localization of YFP:JAZ9 is compromised in the myc2 mutant plants (Fig. 2C). Altogether, these results suggest that MYC2, but not JAZ9, plays an active role in determining the nuclear localization of both MYC2 and JAZ9 in the cell. We therefore hypothesize that the Jas motif (in particular, RJas17) is necessary for the nuclear localization of JAZ9 because it is essential for physical interaction with MYC2. This hypothesis is further supported by protein sequence analysis using ProteinPredict, which shows a monopartite NLS, KRPKKRGRK433-441, in the C-terminal half of MYC2, but failed to identify a predictable NLS in JAZ9.
It remains to be determined whether the TF-dependent mechanism for nuclear localization would be generally applicable to other JAZ repressors, although interaction with MYC2 and related TFs is a common property of all characterized JAZ repressors. Furthermore, although our results indicate the physical interaction with MYC2 (and presumably other TFs) being a major factor in directing the nuclear localization of JAZ9, it is possible that other MYC2-dependent processes also influence this process. For example, MYC2 may positively regulate the expression of the components of the nuclear import machinery as an additional mechanism of assisting the nuclear localization of JAZs. However, examination of publicly available Arabidopsis gene expression databases (36, 62) did not reveal obvious MYC2-dependent regulation of the expression of such genes.
Transgenic overexpression of JAZΔJas proteins has been used as the primary means in the discovery of the repressor function of JAZ proteins because these derivatives are resistant to SCFCOI1-dependent degradation and confer JA-insensitive phenotypes (17, 30, 33, 39). However, the mechanism by which JAZΔJas confers the dominant-negative repressor function is still unresolved (17, 20, 29, 40). Our results now provide an explanation for why some, but not other, JAZΔJas proteins confer constitutive JA-insensitivity when overexpressed in plants, and suggest a model in which the ability of a JAZΔJas to confer JA insensitivity is linked to three features: TF-interacting, nuclear-localized, and resistant to SCFCOI1-dependent degradation. First, we found that JAZ9ΔJas failed to repress JA signaling (SI Appendix, Figs. S1 and S9). This result was in contrast to strong JA insensitivity caused by overexpression of JAZ1ΔJas (20, 33). By investigating the capability of JAZ1ΔJas and JAZ9ΔJas to interact with MYC2, we have determined that interaction with MYC2 is a key feature that is associated with eliciting JA-insensitivity, as JAZ1ΔJas, but not JAZ9ΔJas, is still able to interact with MYC2 (Fig. 1B and SI Appendix, Fig. S2). Second, confocal microscopic examination revealed that YFP:JAZ9 is colocalized with CFP:MYC2 in the nucleus, whereas JAZ9ΔJas and those Jas motif mutants that do not confer JA insensitivity were not found to be colocalized with CFP:MYC2, but instead localized mostly in the cytosol, despite being resistant to degradation in response to coronatine (Figs. 3 and 4; and SI Appendix, Fig. S8). Third, we were able to identify two COI1-interacting residues, RKJas6,7, that when mutated to alanine, created a JAZ9 derivative that is nuclear localized, resistant to degradation, still interacts with MYC2, and is colocalized with MYC2 in the nucleus (Figs. 3 and 4; and SI Appendix, Fig. S8). This JAZ9 derivative now exerts JA insensitive phenotypes when transgenically overexpressed (Fig. 5 and SI Appendix, Fig. S10). On the other hand, we found that simply directing JAZ9ΔJas (resistant to SCFCOI1-dependent degradation, but not interacting with MYC2) into the nucleus by fusion to a NLS was not sufficient to create jasmonate insensitive phenotypes (SI Appendix, Fig. S9). The TF interaction-based model of repression not only could explain the lack of dominant-negative effect of JAZ9ΔJas, but also the ability of JAZ1ΔJas and JAZ10ΔJas to confer JA-insensitivity because both of them still interact with MYC2, indicating more than one MYC2-interacting region in these particular JAZs (30). However, this model could not yet explain the ability of JAZ3ΔJas to confer JA-insensitive phenotypes because the Jas motif is required for JAZ3 interaction with MYC2, but JAZ3ΔJas confers JA insensitivity (17, 29). It is possible that TFs other than MYC2 are biologically relevant targets of JAZ3, or that JAZ3ΔJas confers JA-insensitivity through heterodimerization with other nuclear-localized and MYC2-interacting JAZ repressors, as proposed (29).
To our knowledge, TF-dependent nuclear localization of cognate transcriptional repressors has not been reported in plants or other eukaryotic systems. However, Pfeiffer et al. (63) recently reported that phytochrome-interacting factor (PIF) TFs facilitate the nuclear import of a light receptor protein, phytochrome B, as a critical step in regulating light signaling. Thus, at least two major signaling systems in plants, light signaling and JA signaling, use the TF-dependent nuclear localization mechanism. It is possible that plants have evolved this mechanism to enable immediate targeting of TFs for repression by cognate repressors, possibly right after they are synthesized in the cytoplasm. This mechanism may be important if the TPL transcriptional repression complex is rate-limiting in the nucleus because of its involvement in many signaling processes (31). TF-dependent import of JAZs could efficiently target the NINJA/TPL transcriptional repression complex to only TF-bound JAZs, but not “nonproductive” TF-free JAZs, which could occur if JAZs were imported independently. Regardless, the TF-dependent nuclear import mechanism may be needed to ensure maximal and immediate repression of JA signaling to minimize unnecessary growth inhibition and other undesirable side effects that are known to accompany the activation of JA signaling (1).
Materials and Methods
All experiments reported in this work were performed three or more times with similar results. Confocal microscopy was conducted on leaf tissues from N. tabacum (transient expression) and Arabidopsis thaliana (stable expression). Dual-channel sequential imaging was used to capture images during colocalization experiments. Detailed experimental protocol and information about creating gene constructs and transgenic Arabidopsis can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Melinda Frame at the Center for Advanced Microscopy at Michigan State University for advice on microscopy. This work was supported by a Michigan State University Plant Science Fellowship and Dissertation Completion Fellowship (to J.W.), and National Institutes of Health Grant R01AI068718, Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science) Grant DE–FG02–91ER20021 for confocal microscopy, and a grant from the Gordon and Betty Moore Foundation (to S.Y.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210054109/-/DCSupplemental.
References
- 1.Yang DL, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109(19):E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6(5):751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16(1):126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandaokar A, et al. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46(6):984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- 5.Robson F, et al. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell. 2010;22(4):1143–1160. doi: 10.1105/tpc.109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song S, et al. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23(3):1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 8.Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol. 2008;146(3):832–838. doi: 10.1104/pp.107.115683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conconi A, Smerdon MJ, Howe GA, Ryan CA. The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature. 1996;383(6603):826–829. doi: 10.1038/383826a0. [DOI] [PubMed] [Google Scholar]
- 10.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 11.Kim EH, Park SH, Kim JK. Methyl jasmonate triggers loss of grain yield under drought stress. Plant Signal Behav. 2009;4(4):348–349. doi: 10.4161/psb.4.4.8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo JS, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011;65(6):907–921. doi: 10.1111/j.1365-313X.2010.04477.x. [DOI] [PubMed] [Google Scholar]
- 13.Goossens A, et al. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA. 2003;100(14):8595–8600. doi: 10.1073/pnas.1032967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pauwels L, et al. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA. 2008;105(4):1380–1385. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uppalapati SR, et al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005;42(2):201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE. 2008;3(11):e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 18.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 19.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468(7322):400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 21.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280(5366):1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14(8):1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19(8):2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126(5):969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 25.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46(1):34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36(4):485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 27.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105(19):7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsir L, Chung HS, Koo AJ, Howe GA. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr Opin Plant Biol. 2008;11(4):428–435. doi: 10.1016/j.pbi.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009;59(1):77–87. doi: 10.1111/j.1365-313X.2009.03852.x. [DOI] [PubMed] [Google Scholar]
- 30.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauwels L, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464(7289):788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146(3):952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55(6):979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe H, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Z, et al. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant. 2011;4(2):279–288. doi: 10.1093/mp/ssq073. [DOI] [PubMed] [Google Scholar]
- 36.Dombrecht B, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19(7):2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16(7):1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung HS, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010;63(4):613–622. doi: 10.1111/j.1365-313X.2010.04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009;70(13-14):1539–1546. doi: 10.1016/j.phytochem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Nair R, Carter P, Rost B. NLSdb: Database of nuclear localization signals. Nucleic Acids Res. 2003;31(1):397–399. doi: 10.1093/nar/gkg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol. 2005;348(1):85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 43.Anderson JP, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16(12):3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011;62(6):2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange A, et al. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8(3):195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 47.Wagstaff KM, Jans DA. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic. 2009;10(9):1188–1198. doi: 10.1111/j.1600-0854.2009.00937.x. [DOI] [PubMed] [Google Scholar]
- 48.Lee BJ, et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126(3):543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grunewald W, et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009;10(8):923–928. doi: 10.1038/embor.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee L-Y, Fang M-J, Kuang L-Y, Gelvin SB. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods. 2008;4(1):24. doi: 10.1186/1746-4811-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citovsky V, et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362(5):1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol. 2008;11(5):486–494. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009;276(17):4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- 54.Chung HS, Niu Y, Browse J, Howe GA. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry. 2009;70(13-14):1547–1559. doi: 10.1016/j.phytochem.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonseca S, Chico JM, Solano R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12(5):539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012;17(1):22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Pauwels L, Goossens A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci. 2008;13(2):66–71. doi: 10.1016/j.tplants.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 59.Shyu C, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24(2):536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grunewald W, et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009;10(8):923–928. doi: 10.1038/embor.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94(2):193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 62.Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. CressExpress: A tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 2008;147(3):1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfeiffer A, et al. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA. 2012;109(15):5892–5897. doi: 10.1073/pnas.1120764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.