Abstract
The glucagon-like peptide 1 receptor (GLP1R) is a G protein-coupled receptor (GPCR) involved in insulin synthesis and regulation; therefore, it is an important drug target for treatment of diabetes. However, GLP1R is a member of the class B1 family of GPCRs for which there are no experimental structures. To provide a structural basis for drug design and to probe class B GPCR activation, we predicted the transmembrane (TM) bundle structure of GLP1R bound to the peptide Exendin-4 (Exe4; a GLP1R agonist on the market for treating diabetes) using the MembStruk method for scanning TM bundle conformations. We used protein–protein docking methods to combine the TM bundle with the X-ray crystal structure of the 143-aa N terminus coupled to the Exe4 peptide. This complex was subjected to 28 ns of full-solvent, full-lipid molecular dynamics. We find 14 strong polar interactions of Exe4 with GLP1R, of which 8 interactions are in the TM bundle (2 interactions confirmed by mutation studies) and 6 interactions involve the N terminus (3 interactions found in the crystal structure). We also find 10 important hydrophobic interactions, of which 4 interactions are in the TM bundle (2 interactions confirmed by mutation studies) and 6 interactions are in the N terminus (6 interactions present in the crystal structure). Thus, our predicted structure agrees with available mutagenesis studies. We suggest a number of mutation experiments to further validate our predicted structure. The structure should be useful for guiding drug design and can provide a structural basis for understanding ligand binding and receptor activation of GLP1R and other class B1 GPCRs.
Keywords: protein structure prediction, incretin receptors, peptide hormones
G protein-coupled receptors (GPCRs) are the largest family of integral membrane proteins within the human genome, and they are all characterized by seven transmembrane (TM) helices, with the N terminus on the extracellular (EC) side and the C terminus on the intracellular (IC). This family of proteins senses molecules outside of the cell and activates signal transduction pathways to cause cellular responses. Because of this vital role in cellular signaling networks, GPCRs are involved in many diseases, and they are the target of ∼40% of all prescription pharmaceuticals on the market (1). Because of the importance of GPCRs as drug targets, it is vital to gain structural information for aiding in drug design. Unfortunately, GPCRs, like other membrane proteins, are difficult to crystallize. There are now X-ray crystal structures for more than 12 distinct receptors of the (at least) 800 human GPCRs (2–16). All of the crystallized receptors belong to the class A (rhodopsin-like) family of GPCRs. However, a phylogenetic analysis of GPCRs classifies them into different subfamilies: class A (rhodopsin-like), class B1 (secretin-like), class B2 (adhesion-like), class C (glutamate-like), and Frizzled/Taste2 (12).
Class B1 (secretin-like) GPCRs are activated by peptide hormones. The large ectodomain of these receptors interacts strongly with the C-terminal halves of their endogenous polypeptide agonists. The N terminus of their ligands putatively binds to the TM bundle and EC loops. The current structural understanding of ligand binding and activation of class B1 GPCRs comes from functional and ligand binding studies as well as the crystallized ectodomains of several class B1 GPCRs (17–19). These studies have put forth speculations as to the plausible mechanisms for class B1 GPCR agonist binding and activation initiation, but in the absence of atomic-level structures of these receptors, it is difficult to understand, probe, and expand on these activation hypotheses.
In this work, we focus on the glucagon-like peptide 1 receptor (GLP1R). Activation of GLP1R by GLP1 stimulates the adenylyl cyclase pathway, which increases insulin synthesis and release of insulin in a glucose-dependent fashion (20). In addition, GLP1 reduces body weight by increasing satiety in the brain (21). Consequently, GLP1 would seem to be attractive for treating both type 2 diabetes and obesity. However, GLP1 is rapidly degraded by the serine protease dipeptidyl peptidase-IV in the body, resulting in its half-life of only 1–2 min (22, 23). Exendin-4 (Exe4), a 39-aa peptide isolated from the venom of the Gila monster, is a more stable analog of GLP1 with a half-life of 2.5 h in its marketed form (24–26). It has a 50% sequence homology with GLP1, and it is a full agonist with a stronger affinity and potency for GLP1R (27). Indeed, Exe4 is currently on the market for treatment of diabetes. Despite the success of Exe4 and its derivatives, there is still a need to develop small-molecule (nonpeptide) orally active agonists of GLP1R. This need is further supported by recent reports of oncogenic side effects of Exe4 (28). The process of novel drug design targeting GLP1R could be aided significantly if there was a structure of the full GLP1R bound to Exe4, which is the motivation of the research reported here. Our structure also provides testable hypotheses of GLP1R activation on ligand binding.
In the following sections, we present the predicted structure of the full membrane-bound GLP1R/Exe4 complex in the presence of water. The TM bundle was predicted using the MembStruk methodology (29). This bundle, which was attached to the GLP1R ectodomain crystallized with partial Exe4 (30), was inserted into a periodic membrane-water box and relaxed by molecular dynamics (MD). We find that this predicted structure is consistent with all available mutation data, and we suggest additional experimental tests to validate key aspects of our structure. We believe that this structural information presented should help the development of selective active small-molecule agonists for GLP1R and also aid in probing the activation of class B1 GPCRs.
Methods
We have been developing methods for predicting the structures of GPCRs since the late 1990s. The earlier methodology, denoted as MembStruk, focused on sequential optimization of the seven TM helices starting from a homology template (29). More recently, we have developed a method, denoted as GEnSeMBLE, that aims at a combinatorially complete set of helix rotations and tilts (31). The structure that we report here was built entirely using MembStruk several years ago, but it was not published. We applied our GEnSeMBLE methodology to the older MembStruk structure, but we obtained essentially the same packing of the seven TM helices (more details are in SI Methods, section 8 and Tables S1 and S2). Therefore, we decided to continue with our previous structure for the seven TM helix bundle and its connection to the N terminus, but we replaced the previously homology-built ectodomain of the N terminus of GLP1R (nGLP1R)/Exe4 part (where nGLP1R is the N terminus of GLP1R) with its crystal structure, which appeared recently (30).
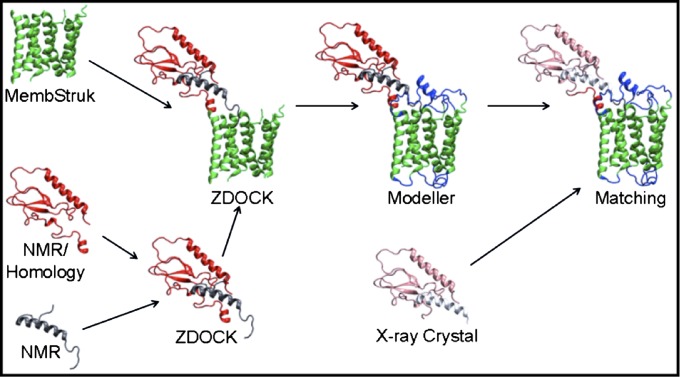
A summary of the full procedures used to generate the GLP1R/Exe4 can be given in the following nine steps (the procedure is depicted in Fig. 1).
i) The initial step in MembStruk (29) is to use comparative hydrophobicity analysis of GLP1R and related GPCRs to identify the seven likely TM domains and then, position (x, y, z) the hydrophobic centers of these helices on a common plane with preselected tilts (θ, φ) and axis rotations (η) based on some template (in this case, we used the predicted prostaglandin D2 (PGD2) receptor structure) (32).
ii) This step is followed by sequential optimization of each TM domain by varying η over 360° in 30° increments and side chain optimization (SCREAM with modest minimization) (33) in a sequence that considers all TM domains multiple times.
iii) Because of the large 143-aa N terminus of GLP1R, we originally built the structure for nGLP1R from homology to the NMR structure of the mouse Corticotropin-releasing factor (CRF) receptor 2 (Protein Data Bank ID code 2JND). This 131-aa region then underwent optimization of the side chains (34).
iv) We used the ZDOCK procedure (35) to dock the NMR structure of the full Exe4 ligand (Protein Data Bank ID code 1JRJ) to the nGLP1R structure from step iii (36).
v) We manually docked the nGLP1R/Exe4 complex from step iv to the TM bundle in such a way that the N terminus of the ligand could interact with the TM region from step i.
vi) We connected the N terminus to the TM region (residues 131–145) and built the three EC and three IC loops using Modeler; this process was followed by SCREAM to position the side changes and then, energy minimization (37).
vii) Then, we inserted the full GLP1R protein into a periodic membrane and water box (75 × 75 × 117 Å), eliminating overlapping species to obtain a system with 61,119 atoms. This system was equilibrated at 300 K (first, the water and membrane and then, all atoms for 20 ns) using NAMD 2.6, with CHARMM22 charges for the protein and CHARMM27 charges for the lipids (38–40). The waters were modeled using the TIP3P potential function (41).
viii) In the meantime, an X-ray crystal structure had appeared for nGLP1R bound to part of Exe4 (9–39; Protein Data Bank ID code 3C5T) (42). We matched this structure to our predicted structure and reoptimized (SCREAM, minimization, and MD for 18 ns).
ix) After 18 ns of full-atom and full-solvent MD, we performed simulated annealing of the TM portion of the ligand binding site, and then, we carried 10 ns of full-atom and full-solvent MD at 300 K. A representative snapshot of the final 10 ns of MD was chosen for the discussion below.
Fig. 1.
Creating the GLP1R/Exe4 bundle. The steps used to create our GLP1R/Exe4 structure are depicted along with the methods used at each point. After the steps shown here, the entire complex was optimized through 28 ns of MD.
Experimental information was not used during any of the above steps, except that information known about GLP1 loop structures was used to select loops from Modeler in step vi. The specific information used is discussed in SI Methods, section 4.
Results and Discussion
Intraprotein Interactions.
For class A GPCRs, several conserved interhelical interactions, such as the 1-2-7 or 2-3-4 hydrogen bond networks, are present in most crystal structures. The amino acids involved in the 1-2-7 and 2-3-4 interactions are not conserved in class B GPCRs. However, we do find many interhelical interactions, some of which occur between residues conserved in class B1 GPCRs, which may be important for their structure and function. The GLP1R/Exe4 complex has 16 hydrogen bonds within its TM bundle (excluding helix-forming hydrogen bonds) or connecting loops, which is shown in Table S3 and discussed in the following sections. The TM bundle interactions are pictured in Fig. 2.
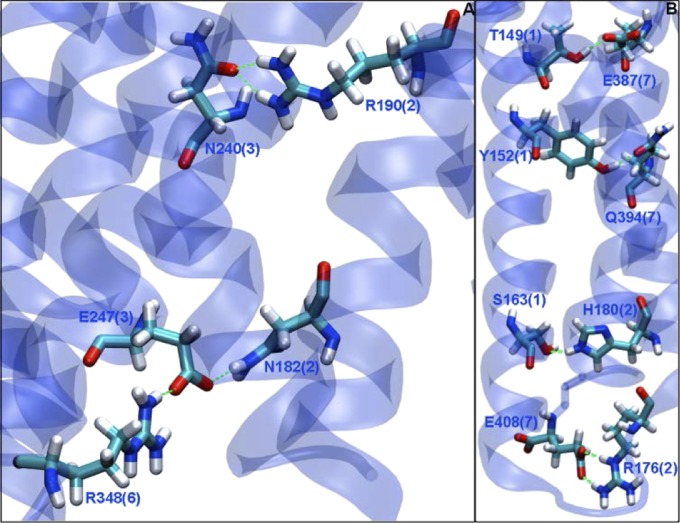
Fig. 2.
The (A) TM2-TM3-TM6 and (B) TM1-TM2-TM7 conserved hydrogen bond networks. (A) We believe that the E247(TM3)-R348(TM6) ionic lock may be associated with the unactivated GPCR structure (analogous to the R3.50-D/E6.30 interaction in class A GPCRs). TM3 is additionally coupled to TM2 through the conserved N182(TM2) to form a 2-3-6 interaction. This interaction is further stabilized by the R190(TM2)-N240(TM3) hydrogen bond, which may be analogous to the TM2.45(S/N/T)-3.42(S/N/T)-4.50(W) conserved interaction of class A GPCRs. (B) We also see several more transient couplings between TM1, -2, and -7, which are shown here. These four interactions help solidify the local structure of TMs 1, 2, and 7, and their disruption may be involved in activation.
TM3-TM6 ionic lock.
We consider that the conserved E247(TM3)-R348(TM6) ionic lock is analogous to the R(3.50)-D/E(6.30) [using Ballesteros numbering of TM residues (43)] ionic lock of class A GPCRs known to stabilize the GPCR in an inactive form, although these donor and acceptor residue types are swapped in the class B1 version of the TM3-TM6 ionic lock (44). We find this ionic lock to be maintained through most of the dynamics. This interaction could play the role of maintaining the inactive form of class B1 GPCRs. Indeed, E247 is fully conserved among class B1 GPCRs, whereas R348 is either an R or K in all class B1 GPCRs. This hypothesis could be tested by mutations that break the ionic lock, which might lead to a constitutively active receptor (GLP1R does not exhibit constitutive activity) (45). This hypothesis is complicated in the light of GLP1R studies like the work by Heller et al. (46), which looked at the R348G mutant of GLP1R and found no activation for all concentrations of GLP1; in contrast, the work by Takhar et al. (47) found R348A to have no effect on binding or activation of GLP1. Perhaps, these large changes to alanine or glycine led to a modified TM bundle that changed the binding site, and/or this residue is critical for G-protein coupling and activation. A more subtle R348Q mutation would test the intricacies of this ionic lock in more detail.
Because the TM3-TM6 ionic lock is present in our structure, it is likely that our complex is not yet fully activated. However, our structure is bound to an agonist, and TM6 makes almost no interactions other than this ionic lock. Thus, it may be at least partially activated. This lack of TM6 interactions would allow TM6 (on breaking of the ionic lock) to immediately move away from TM3 to interact with the Gα-subunit of the G protein, which it does in the active conformation of the G protein-bound β2-adrenergic receptor structure (48).
Coupling of TM2-TM3-TM6.
The 3-6 ionic lock is additionally coupled to TM2 by the conserved N182(TM2)-E247(TM3) hydrogen bond, resulting in a TM2-TM3-TM6 (2-3-6) hydrogen bond network [N182(TM2)-E247(TM3)-R348(TM6)]. This N182(TM2)-E247(TM3) has only minor fluctuations through the course of dynamics. Because N182 is an N, H, or Q in class B1 GPCRs, this 2-3-6 hydrogen bond network may be conserved in class B1 GPCRs. The 2-3-6 conserved network may be further stabilized by the similarly conserved R190(TM2)-N240(TM3) hydrogen bond, which is shown in conjunction with the 2-3-6 network in Fig. 2. We suggest that this interaction might be analogous to the 2.45(S/N/T)-3.42(S/N/T)-4.50(W) conserved interaction of class A GPCRs. Indeed, N240(TM3) is just 7 aa away from E247(TM3) compared with the 3.42 residue in class A GPCRs, which is 8 aa away from the 3.50 residue. We find the R190(TM2)-N240(TM3) interaction to be very constantly maintained throughout the dynamics. N240 is fully conserved among class B1 GPCRs, whereas R190 is R, K, or N, and therefore, this interaction is possible in any class B1 GPCR and may be a feature of this receptor class.
TM1-TM2-TM7 interaction network.
The remaining two strong and conserved interactions between TM1 and TM7 [Y152(TM1)-Q394(TM7)] and TM2 and TM7 [R176(TM2)-E408(TM7)] might play an analogous role to the TM1.50(N)-TM2.50(D)-TM7.49(N) interactions conserved among class A GPCRs. Perhaps it is important to keep the TM1-TM2-TM7 (1-2-7) region rigid to control activation. Both interactions are stable during the course of the dynamics simulation. These residues are also conserved in class B1 GPCRs: Y152 may be a Y or H, whereas Q394 may be a Q or H. R176 and E408 are fully conserved in all class B1 GPCRs. Note that the R176(TM2)-E408(TM7) interactions are located in the IC end of the TMs, and as such, they could also play a role in GPCR activation.
We find two additional interactions in the 1-2-7 region that involve nonconserved amino acids—T149(TM1)-E387(TM7) and H180(TM2)-S163(TM1)—that further stabilize the 1-2-7 coupling. We find that the T149(TM1)-E387(TM7) interaction is only transient during dynamics, being often mediated by the H1 residue of the ligand. Perhaps the agonist will eventually break this interaction as part of activation. The H180(TM2)-S163(TM1) interaction forms/breaks/reforms several times during the course of dynamics, indicating that it is less stable than the hydrogen bonds discussed previously. Despite their transience, these hydrogen bonds do help to stabilize the coupling of the 1-2-7 helices, and in conjunction with the with the more stable conserved interactions discussed earlier, they form a solid grouping of TMs 1, 2, and 7. The two conserved interactions discussed above, along with these two nonconserved interactions, are pictured in Fig. 2.
EC loop couplings and N-terminal interactions.
The remaining four interhelical interactions are between the EC loops. The first three interactions are with D222(EC1) and adjoining EC2 residues R299, W297, and C296. In addition, there is a helical region present in EC1 from residue 215 to 225. It is the base of this helix that interacts with EC2. The final interloop hydrogen bond is between H374(EC3) and M303(EC2). The EC loops are clearly closely coupled, and they provide order to the flexible loop regions, which has been seen in other GPCR crystal structures (49). These stabilizing interactions would play a role in peptide binding, because they need to accommodate a peptide reaching from the N terminus past EC1 and into the TM bundle interior. Finally, GLP1R also has the TM3-EC2 disulfide coupling (C296-C226) conserved among class A GPCRs and class B1. There are no other Cys residues in the EC loops.
The overall architecture of the N terminus from the crystal structure remains stable during the course of the dynamics. There are still the three conserved disulphide bonds, two regions of antiparallel β-sheets, and an α-helix adjacent to the ligand, which all occur in all of the class B1 GPCR N-terminal crystal structures (50). No significant deviations from the crystal are observed.
Overall, our study of GLP1R shows that there are several conserved hydrogen bond networks that mimic those networks of class A GPCRs. The 3-6 ionic lock is similar to the lock of class A GPCRs, although it may play a somewhat different structural and functional role in G-protein activation. The 1-2, 1-7, and 2-7 networks mimic the 1-2-7 interaction motif of class A GPCRs. Finally, the loop structures, which may have direct bearing on ligand binding, are stabilized by several interloop interactions.
Protein–Ligand Interactions.
Structure overview.
The GLP1R/Exe4 binding site involves interactions throughout the N terminus, TM regions, and EC loops with the primary interactions occurring with the N terminus, TMs 1, 2, and 7, and EC1 (Fig. 3). We find Exe4 to be helical from residues 9–29. The strongest interactions with the N terminus include six polar interactions (three of which were present in the crystal structure) and six hydrophobic interactions (all were present in the crystal structure). The TM bundle features eight polar interactions (two of which were confirmed by mutation studies) and four hydrophobic interactions (two of which were confirmed by mutation studies). The full-energy analysis for these interactions is shown in Table S4. We will discuss the binding site in three parts: hydrogen bonds, nonpolar interactions, and detailed comparison with mutation data.
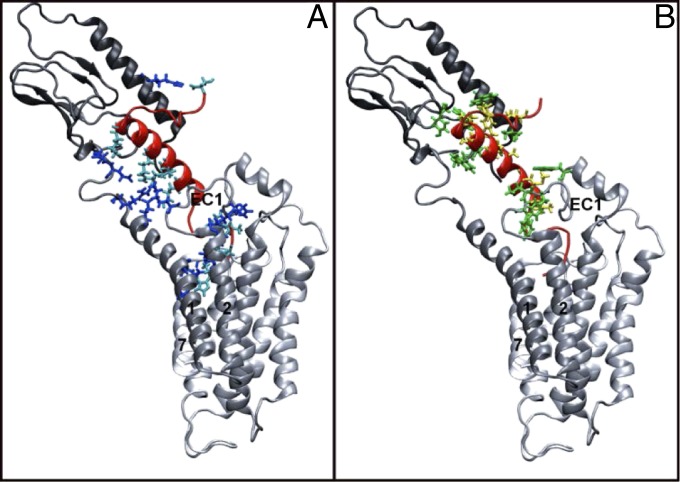
Fig. 3.
Overview of the ligand binding site’s hydrophobic (A) and hydrophilic (B) interactions. GLP1R is shown with a color transition from black to white as the protein goes from the N terminus to the C terminus. Exe4 is shown in red. Hydrophilic interactions are shown in A, with protein residues in blue and ligand residues in turquoise. Hydrophobic interactions are shown in B, with protein residues in green, and ligand residues in yellow.
Polar interactions.
We find 14 polar interactions (hydrogen bonds or salt bridges) between GLP1R and Exe4 (Fig. 4 and Table 1). There are eight polar interactions within the TM region, which reflects the primary areas of interaction between the ligand and TM bundle: TMs 1, 2, and 7 as well as EC1. The TM region polar interactions are particularly focused at the first few residues of the ligand—specifically H1 and E3 but also, T5 and F6. Our TM bundle interactions are also validated by site-directed mutagenesis studies for residues T149, E387, T391, and K197, which we find to interact with H1 and E3 of the ligand (14, 51) as shown in Table 1.
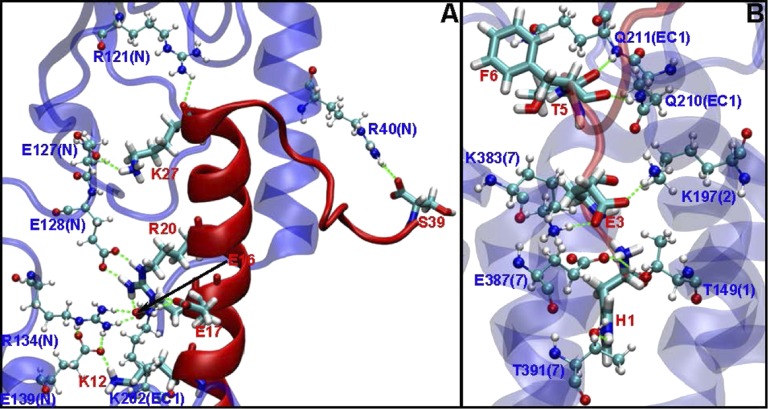
Fig. 4.
Exe4 hydrogen bonds with the N terminus (A) and TM region of GLP1R (B). All of the receptor–ligand hydrogen bonds are depicted here and quantified in Table S3. Protein residues are shown in blue [Corey-Pauling-Koltun (CPK) drawing method], whereas ligand residues are shown in red (licorice drawing method). We find a total of 14 important polar interactions of Exe4 with GLP1R, of which 6 interactions involve the N terminus (A; including 3 interactions found in the X-ray crystal structure) and 8 interactions are in the TM bundle or loops (B; including 2 interactions that have been confirmed by mutation studies).
Table 1.
Polar interactions between GLP1R and Exe4
| GLP1R residue | Location | Exe4 residue | Energy | Mutation | Reduces binding |
| TM region interactions | |||||
| T149 | TM1 | H1 | −1.0 | T149M (47) | 5.0-fold |
| E387 | TM7 | H1 | −6.2 | E387A (48) | 3.9-fold |
| T391 | TM7 | H1 | −2.8 | T391A (48) | 2.8-fold |
| K197 | TM2 | E3 | −39.6 | K197A (48) | 3.0-fold |
| K383 | TM7 | E3 | −37.9 | ||
| Q211 | EC1 | T5 | −1.0 | ||
| Q210 | EC1 | F6 | −3.1 | ||
| K202 | EC1 | E17 | −48.8 | ||
| N-terminal interactions | |||||
| R134 | N | E16 | −47.6 | ||
| E128 | N | R20 | −44.8 | E128A (29) | 2.4-fold* |
| E139 | N | K12 | −38.4 | ||
| E127 | N | K27 | −33.4 | E127A (29) | 6.8-fold* |
| R40 | N | S39 | −23.3 |
Energies are given in kilocalories per mole and are provided to show the relative strength of the interactions.
*Present in the crystal structure of nGLP1R/Exe4.
Our five N-terminal interactions include the two crystal salt bridges: E128(N)-R20 and E127(N)-K27 (42). These two residues’ importance has also been shown through mutation studies on E127 and E128 (52). In addition, we find three very strong interactions of the N terminus, of which two interactions are in the flexible region between the structured N terminus and TM1 and the third interaction is at the C terminus of the ligand. The remaining crystal hydrogen bond between R121(N) and the backbone of K27 alternates during the MD from water-mediated interaction to a weak hydrogen bond. This weak interaction is consistent with a study by Underwood et al. (53), which found that mutating R121 to alanine (R121A) decreased ligand binding by only 1.6-fold.
Hydrophobic interactions.
We predict 21 strong hydrophobic interactions between GLP1R and Exe4. The 10 strongest interactions (cutoff of −3 kcal/mol for the van der Waals (VDW) energy; 1 kcal = 4.18 kJ) are shown in Fig. 5 and Table S5. We find two main clusters of hydrophobic interactions.
Fig. 5.
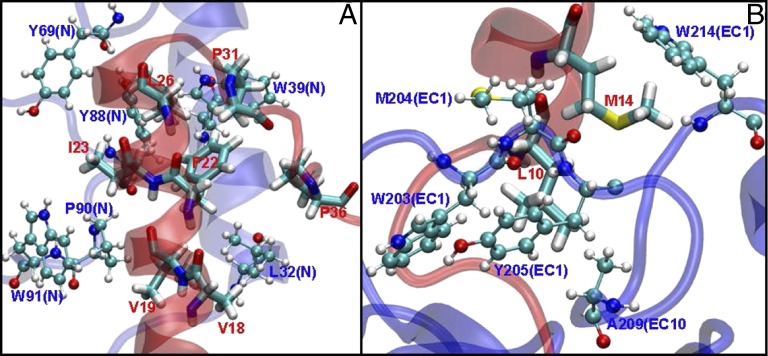
Hydrophobic interactions between GLP1R and Exe4 in the N terminus (A) and EC1 (B). Protein residues are shown in blue (CPK drawing method), whereas ligand residues are shown in red (licorice drawing method). We find 10 important hydrophobic interactions, of which 6 interactions are in the N terminus (A; all confirmed by X-ray crystal structure) and 4 interactions are in the TM bundle (B; of which 2 interactions have been confirmed by mutation studies).
The first cluster of hydrophobic interactions occurs in EC1. GLP1R residues W203, M204, Y205, A209, W214, and L217 interact with Exe4 residues L10 and M14. Indeed, experiments on residues M204A/Y205A found a 2.7-fold decrease in binding of Exe4 (54). We find that M204 has a −7.0 kcal/mol interaction energy with Exe4, whereas the interaction energy of Y205 with Exe4 is −2.6 kcal/mol.
The second cluster of hydrophobic interactions occurs on one face of the helical portion of Exe4, interacting with the hydrophobic face of a helix of GLP1R in the N terminus. These interactions include the interactions between GLP1R residues L32, T35, V36, W39, Y69, Y88, L89, P90, W91, and L123 and Exe4 residues V18, V19, F22, L23, L26, P31, and P36. These interactions include some of the strongest hydrophobic interactions in our entire structure: L32 at −8.4, W39 at −6.6, P90 at −4.2, and W91 at −4.0 kcal/mol. These interactions were all present in the crystal structure of nGLP1R/Exe4, and they have been confirmed by mutation studies (42, 53). Specifically, the L32A mutation had a 7.1-fold effect on Exe4 binding and 9.5-fold effect on activation, whereas P90A had a 2.1-fold effect on binding and 5.5-fold effect on activation.
We also find a final small cluster of nonpolar residues in the middle of TM3 (L232, M233, and V229; not pictured), which form a hydrophobic pocket around the G2 residue of Exe4. These hydrophobic interactions, plus the K202-E17 salt bridge, and the weaker polar interactions with EC1 in Table S4 (to Q210 and Q211) indicate clearly that EC1 is extremely important for Exe4 binding.
Comparison with mutation data.
Several mutation studies have been carried out on Exe4 with the intent of determining the residues that are important for ligand binding (51–55). These studies are summarized in Table S6. Of the 26 mutations leading to a decrease in binding or activity, 24 mutations are consistent with our predicted equilibrium structure, whereas the remaining 2 mutations appear transiently during the course of the dynamics. Of these 26 residues, 13 residues involve the N terminus (and were in the X-ray structure), whereas 13 residues involve the TM helices and EC1; 11 of these 13 residues interact with six of seven TMs (all but TM6) plus EC1. The remaining two residues on TM6 (H363 and E364) are transiently involved in a hydrogen bond network that spans the middle of the TM bundle and extends to H1 of the ligand. Our prediction agrees with the conclusion of mutation studies that D198 is not crucial for ligand binding (56). It is important to emphasize that the GLP1R structure (except for the N terminus) was derived strictly from our MembStruk method without any use of mutation data (except in the loop growing, which used a distance constraint between Y205(EC1)-F6 and required M204, Y205, D215, and R227 to be close to some part of Exe4).
Finally, we note that our structure preserved all interactions found in the crystal structure—both hydrophilic and hydrophobic—over the course of the dynamics. This finding provides additional validation of our structure, because these interactions would be expected to be stable. In addition, all residues indicated in the literature to be potentially important for binding or activation are found in our structure to point into the TM bundle, or they are otherwise accessible to the ligand. As such, they can either interact with the ligand itself or have structural effects. Overall, our GLP1R structure strongly agrees with the available experimental data, making it valuable for further structural and activation studies.
Testing the Predicted Binding Site.
Our predicted structure of GLP1R and its binding site for Exe4 suggest many mutation studies for its further validation. Indeed, we constructed structures for 14 such mutations (Table S7 shows their effects on energies). The procedure was to use SCREAM to introduce the desired mutation and then minimize the protein to 0.5 kcal/mol per angstrom rms force (33). These calculations assumed that the overall backbone structure remains intact. Of 14 mutations, 2 mutations are predicted to improve binding, whereas 12 mutations are predicted to decrease binding.
The first set of mutations was chosen with the goal of validating our predictions of the strongest interactions between GLP1R and Exe4. Ten cases were aimed at disrupting the binding site by breaking interactions discussed previously: R134A/Q, K202A/Q, and K383A/Q break salt bridges or hydrogen bonds, whereas W203N/Y and W214N/Y disrupt the EC1–ligand hydrophobic interactions. We also suggest two ligand mutations that would decrease binding: K12A and M14Q. The K12A mutation would lose the E139 salt bridge, whereas the M14Q would disrupt the hydrophobic network that the M14 has in the EC1 area. In each of the 12 cases, our predicted change in binding agrees with expectation. Finally, we predicted two mutations to improve ligand binding. The first mutation was S11W of Exe4, which allows a new interaction to be formed with W214 on the receptor. The second change is Q213K of GLP1R, which forms a new hydrogen bond with D9 on Exe4. Both of these changes improve our predicted interaction energies between the receptor and ligand.
Discussion of Ligand Binding and Protein Activation.
Our GLP1R/Exe4 structure suggests several general features of ligand binding to GLP1R and potentially, class B1 proteins as a whole. One feature is that we find that the TM region polar interactions are particularly focused at the first few residues of the ligand—specifically, H1 and E3 but also, T5 and F6. This finding is in accordance with the known importance of the N terminus (specifically residues 1–7) of class B1 agonists for protein activation (50). In the specific case of Exe4, if the ligand is truncated by eight residues, it becomes a competitive antagonist for GLP1R, because it can still bind the receptor but no longer cause activation (27).
Next, we find that the binding pocket of Exe4 shows strong polar and hydrophobic interactions with EC1. Experimental studies showed that mutations of EC2 residues to alanine dramatically decreased binding of GLP1 but had no effect on the binding of Exe4 (57), consistent with our structure. We suggest that the reason for the importance of the loops in the peptide binding is to align the ligand in the correct conformation for TM bundle entry. In the two-domain model of class B1 GPCR protein binding, the N-terminal ectodomain plays the role of recognizing the ligand and supports the initial binding (58). We believe that the next step is for the flexible N terminus/ligand complex to align itself to the TM bundle by loop interactions followed by final insertion of the head region of the ligand into the TM bundle itself.
Also, we note that the flexible N terminus of the ligand is nestled in the TM1-TM2-TM7 binding pocket, which leaves the TM3-TM4-TM6 region largely open, making this area available for binding of small molecules serving as ago-allosteric modulators (59). In addition, the residues of the ligand inserted themselves between hydrogen bonds of the apo GLP1R [for example, T149(TM1)-H1-E387(TM7)]. This finding suggests that part of the effect of Exe4 binding may be to break some of the TM1-TM2-TM7 strong interactions, giving the structure the flexibility to achieve its active conformation.
Any discussion of GPCR activation would be incomplete without mention of the TM3-TM6 ionic lock, of which we find a variation in our structure. Instead of the conserved R(3.50)-D/E(6.30) ionic lock of class A GPRCs, we find an analogous conserved E247(TM3)-R348(TM6) ionic lock. Breaking this interaction may be crucial for GLP1R activation. To test this hypothesis, one could mutate one of the charged residues to a polar residue, such a glutamine; and therefore, the overall hydrophilicity of the region could be preserved, but the interaction would be broken.
Finally, it has been suggested that an N-terminal helix capping motif of a peptide agonist is a key element underlying class B1 GPCR activation (18). This structural feature is theorized to consist of a hydrophobic interaction between residues 6 and 10, and it is stabilized by a salt bridge between residues 7 and 10 of the ligand. The result of these interactions is that the ligand forms an L-shape at its N terminus. Although we do not see the exact 7–10 and 6–10 interactions—instead, we find that residues Phe6 and Ser8 form a backbone hydrogen bond—this alternate interaction causes the ligand to adopt a slightly more loose L-conformation. This structural constraint may be important for the rational drug design of peptide agonists targeting class B1 receptors.
Conclusion
We present here the predicted TM bundle for GLP1R (residues 146–408), which we combined with the crystal structure for the N terminus (residues 28–145) and Exe4. The resulting structure was then equilibrated in an explicit membrane and water environment for 28 ns. We find strong agreement with available experimental data, most of which played no role in the predictions. We suggest 14 mutations to provide strong tests of our predicted binding site. This structure can now form the basis for the rational design of other peptide ligands and the greatly needed small-molecule ligands.
The model that we present here can be used to explore the mechanisms of class B1 GPCR binding and activation (52). In addition, we expect that this structure will provide a basis for the design and optimization of new small-molecule ligands that bind selectively and specifically to GLP1R.
Finally, one of the grand challenges in understanding GPCRs is to elucidate the mechanism of activation. This study does not address this issue directly; however, we do identify a TM3-TM6 ionic lock that is conserved across class B1 GPCRs that we believe may play a very similar or more complex role in GLP1R activation compared with the TM3-TM6 ionic lock conserved across a subset of class A GPCRs. Mutation studies on this ionic lock will help test such speculations and provide structural signatures of active and inactive receptor conformations.
Supplementary Material
Acknowledgments
The MembStruk studies by J.H. were funded by Allozyne (Ken Grabstein). The final optimization of the structure and analysis (A.K.) was funded by Sanofi-Aventis (Ken Wertman).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218051109/-/DCSupplemental.
References
- 1.Filmore D. It's a GPCR world. Modern Drug Discovery. 2004;7(11):24–28. [Google Scholar]
- 2.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 3.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330(6007):1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475(7354):65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482(7386):552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335(6070):851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7(4):339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 13.Wu HX, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485(7398):327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manglik A, et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485(7398):321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granier S, et al. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485(7398):400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson AA, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485(7398):395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parthier C, Reedtz-Runge S, Rudolph R, Stubbs MT. Passing the baton in class B GPCRs: Peptide hormone activation via helix induction? Trends Biochem Sci. 2009;34(6):303–310. doi: 10.1016/j.tibs.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Neumann JM, et al. Class-B GPCR activation: Is ligand helix-capping the key? Trends Biochem Sci. 2008;33(7):314–319. doi: 10.1016/j.tibs.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Pal K, Melcher K, Xu HE. Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. Acta Pharmacol Sin. 2012;33(3):300–311. doi: 10.1038/aps.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt WE, Siegel EG, Creutzfeldt W. Glucagon-like peptide-1 but not glucagon-like peptide-2 stimulates insulin release from isolated rat pancreatic islets. Diabetologia. 1985;28(9):704–707. doi: 10.1007/BF00291980. [DOI] [PubMed] [Google Scholar]
- 21.Kieffer TJ, Habener JF. The glucagon-like peptides. Endocr Rev. 1999;20(6):876–913. doi: 10.1210/edrv.20.6.0385. [DOI] [PubMed] [Google Scholar]
- 22.Kieffer TJ, McIntosh CH, Pederson RA. Degradation of glucose-dependent insulinotropic polypeptide and truncated glucagon-like peptide 1 in vitro and in vivo by dipeptidyl peptidase IV. Endocrinology. 1995;136(8):3585–3596. doi: 10.1210/endo.136.8.7628397. [DOI] [PubMed] [Google Scholar]
- 23.Mentlein R, Gallwitz B, Schmidt WE. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7-36)amide, peptide histidine methionine and is responsible for their degradation in human serum. Eur J Biochem. 1993;214(3):829–835. doi: 10.1111/j.1432-1033.1993.tb17986.x. [DOI] [PubMed] [Google Scholar]
- 24.Eng J, Kleinman WA, Singh L, Singh G, Raufman JP. Isolation and characterization of exendin-4, an exendin-3 analogue, from Heloderma suspectum venom. Further evidence for an exendin receptor on dispersed acini from guinea pig pancreas. J Biol Chem. 1992;267(11):7402–7405. [PubMed] [Google Scholar]
- 25.Parkes DG, Pittner R, Jodka C, Smith P, Young A. Insulinotropic actions of exendin-4 and glucagon-like peptide-1 in vivo and in vitro. Metabolism. 2001;50(5):583–589. doi: 10.1053/meta.2001.22519. [DOI] [PubMed] [Google Scholar]
- 26.Bray GM. Exenatide. Am J Health Syst Pharm. 2006;63(5):411–418. doi: 10.2146/ajhp050459. [DOI] [PubMed] [Google Scholar]
- 27.Göke R, et al. Exendin-4 is a high potency agonist and truncated exendin-(9-39)-amide an antagonist at the glucagon-like peptide 1-(7-36)-amide receptor of insulin-secreting beta-cells. J Biol Chem. 1993;268(26):19650–19655. [PubMed] [Google Scholar]
- 28.Elashoff M, Matveyenko AV, Gier B, Elashoff R, Butler PC. Pancreatitis, pancreatic, and thyroid cancer with glucagon-like peptide-1-based therapies. Gastroenterology. 2011;141(1):150–156. doi: 10.1053/j.gastro.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaidehi N, et al. Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA. 2002;99(20):12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Runge S, Thøgersen H, Madsen K, Lau J, Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J Biol Chem. 2008;283(17):11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 31.Abrol R, Griffith AR, Bray JK, Goddard WA., 3rd Structure prediction of g protein-coupled receptors and their ensemble of functionally important conformations. Methods Mol Biol. 2012;914:237–254. doi: 10.1007/978-1-62703-023-6_14. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, et al. Prediction of the 3D structure and dynamics of human DP G-protein coupled receptor bound to an agonist and an antagonist. J Am Chem Soc. 2007;129(35):10720–10731. doi: 10.1021/ja070865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kam VWT, Goddard WA. Flat-bottom strategy for improved accuracy in protein side-chain placements. J Chem Theory Comput. 2008;4(12):2160–2169. doi: 10.1021/ct800196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grace CR, et al. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci USA. 2007;104(12):4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R, Li L, Weng ZP. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 36.Neidigh JW, Fesinmeyer RM, Prickett KS, Andersen NH. Exendin-4 and glucagon-like-peptide-1: NMR structural comparisons in the solution and micelle-associated states. Biochemistry. 2001;40(44):13188–13200. doi: 10.1021/bi010902s. [DOI] [PubMed] [Google Scholar]
- 37.Eswar N, et al. Current Protocols in Bioinformatics. 2006. Comparative protein structure modeling using Modeller. (Wiley, New York), Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 42.Runge VM. Notes on “Characteristics of gadolinium-DTPA complex: A potential NMR contrast agent”. AJR Am J Roentgenol. 2008;190(6):1433–1434. doi: 10.2214/AJR.07.3549. [DOI] [PubMed] [Google Scholar]
- 43.Ballesteros A, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 44.Vogel R, et al. Functional role of the “ionic lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380(4):648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Fortin JP, Schroeder JC, Zhu Y, Beinborn M, Kopin AS. Pharmacological characterization of human incretin receptor missense variants. J Pharmacol Exp Ther. 2010;332(1):274–280. doi: 10.1124/jpet.109.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller RS, Kieffer TJ, Habener JF. Point mutations in the first and third intracellular loops of the glucagon-like peptide-1 receptor alter intracellular signaling. Biochem Biophys Res Commun. 1996;223(3):624–632. doi: 10.1006/bbrc.1996.0945. [DOI] [PubMed] [Google Scholar]
- 47.Takhar S, et al. The third cytoplasmic domain of the GLP-1[7-36 amide] receptor is required for coupling to the adenylyl cyclase system. Endocrinology. 1996;137(5):2175–2178. doi: 10.1210/endo.137.5.8612565. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33(1):17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couvineau A, Laburthe M. The family B1 GPCR: Structural aspects and interaction with accessory proteins. Curr Drug Targets. 2012;13(1):103–115. doi: 10.2174/138945012798868434. [DOI] [PubMed] [Google Scholar]
- 51.Beinborn M, Worrall CI, McBride EW, Kopin AS. A human glucagon-like peptide-1 receptor polymorphism results in reduced agonist responsiveness. Regul Pept. 2005;130(1–2):1–6. doi: 10.1016/j.regpep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Coopman K, et al. Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: Modelling the ligand-bound receptor. Mol Endocrinol. 2011;25(10):1804–1818. doi: 10.1210/me.2011-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Underwood CR, et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J Biol Chem. 2010;285(1):723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López de Maturana R, Treece-Birch J, Abidi F, Findlay JB, Donnelly D. Met-204 and Tyr-205 are together important for binding GLP-1 receptor agonists but not their N-terminally truncated analogues. Protein Pept Lett. 2004;11(1):15–22. doi: 10.2174/0929866043478491. [DOI] [PubMed] [Google Scholar]
- 55.Al-Sabah S, Donnelly D. A model for receptor-peptide binding at the glucagon-like peptide-1 (GLP-1) receptor through the analysis of truncated ligands and receptors. Br J Pharmacol. 2003;140(2):339–346. doi: 10.1038/sj.bjp.0705453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.López de Maturana R, Donnelly D. The glucagon-like peptide-1 receptor binding site for the N-terminus of GLP-1 requires polarity at Asp198 rather than negative charge. FEBS Lett. 2002;530(1–3):244–248. doi: 10.1016/s0014-5793(02)03492-0. [DOI] [PubMed] [Google Scholar]
- 57.Koole C, et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J Biol Chem. 2012;287(6):3642–3658. doi: 10.1074/jbc.M111.309328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov Today. 2005;10(6):417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 59.Knudsen LB, et al. Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc Natl Acad Sci USA. 2007;104(3):937–942. doi: 10.1073/pnas.0605701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.