Abstract
Inducible degradation is a powerful approach for identifying the function of a specific protein or protein complex. Recently, a plant auxin-inducible degron (AID) system has been shown to degrade AID-tagged target proteins in nonplant cells. Here, we demonstrate that an AID-tagged protein can functionally replace an endogenous protein depleted by RNAi, leading to an inducible null phenotype rapidly after auxin addition. The AID system is shown to be capable of controlling the stability of AID-tagged proteins that are in either nuclear or cytoplasmic compartments and even when incorporated into protein complexes. Induced degradation occurs rapidly after addition of auxin with protein half-life reduced to as little as 9 min and proceeding to completion with first-order kinetics. AID-mediated instability is demonstrated to be rapidly reversible. Induced degradation is shown to initiate and continue in all cell cycle phases, including mitosis, making this system especially useful for identifying the function(s) of proteins of interest during specific points in the mammalian cell cycle.
Keywords: proteasome, SCF, ubiquitin
Conditional inactivation or depletion of proteins is a powerful method for determining gene function and central to our understanding of complex biological systems. Protein expression is frequently controlled at the DNA level by disruption of the coding sequence of the gene (1) or at the level of the mRNA by using methodologies for suppressing mRNA accumulation, especially with RNAi (2, 3). However, in both cases, protein depletion is indirect and the rapidity of loss is dependent on the stability of the protein, resulting in very slow loss for long-lived proteins. This serves as a major limitation for all but the shortest lived proteins, because different severities of phenotypes are typically observed at differing levels of depletion, especially in the case of proteins that have multiple functions (4). Additionally, neither gene inactivation nor RNAi silencing is readily reversible (in the latter case, this is an obligate consequence of the relatively long half-life of guide RNAs once incorporated into the RNAi-induced silencing complex) (5). RNAi-mediated mRNA degradation often also suffers from incomplete silencing and/or off-target effects.
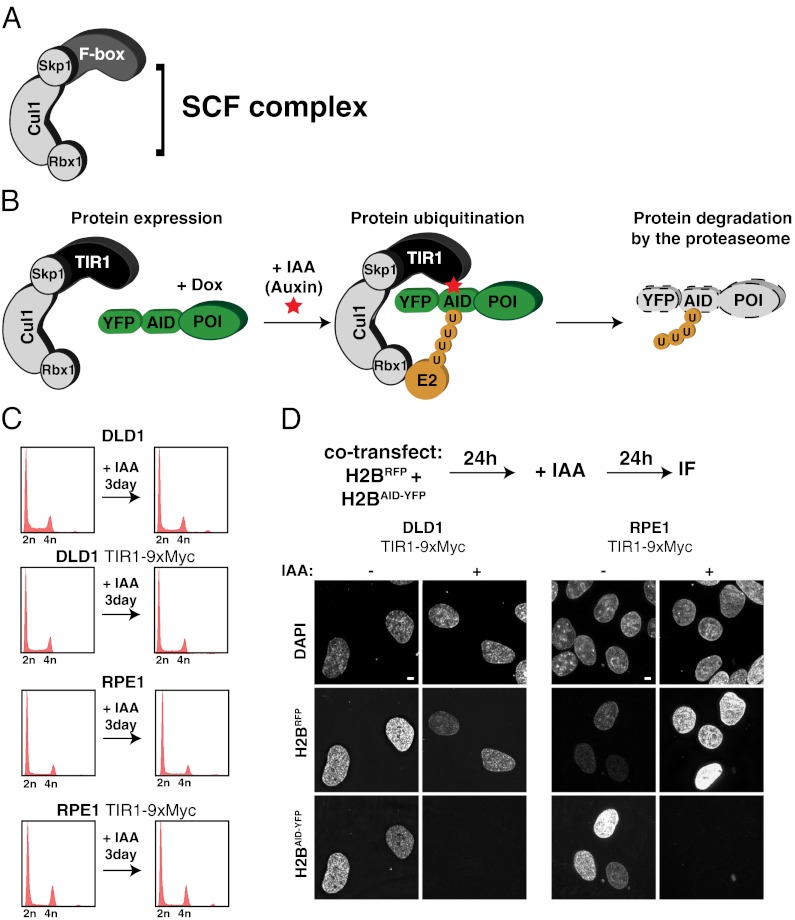
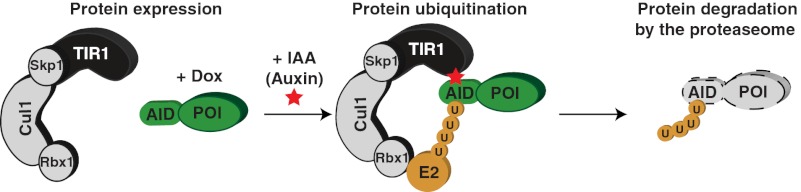
To overcome these limitations, a variety of systems have been developed that allow the posttranslational degradation of proteins using cell-permeable small molecules (4, 6–10). Recently, a system for inducible-protein depletion was developed that relies on the transplantation of a ligand-induced degradation system found in plants (4). All eukaryotes possess Skp1, Cullin, and F-Box protein (SCF) ubiquitin ligases, which are composed of three core subunits and a variable F-box that is responsible for substrate recruitment to the SCF complex (11, 12) (Fig. 1A). F-box proteins associate with the SCF complex through an interaction of their F-box domain with the Skp1 protein (13). In plants, auxin hormones promote the interaction between the F-box protein TIR1 and proteins containing an auxin-inducible degron (AID) (14–16). Orthologs of TIR1 and AID are only found in plant species; however, due to the high degree of conservation of Skp1 in eukaryotes, ectopically expressed TIR1 can associate with Skp1 in animal cells and form an SCFTIR1 complex (Fig. 1B).
Fig. 1.
Auxin-inducible degradation system for controlling protein stability in human cells. (A) Schematic illustration of the SCF ubiquitin ligase. The variable F-box protein is involved in substrate recruitment. (B) Schematic illustration of the auxin-induced degradation system. Ectopically expressed TIR1 F-box protein incorporates into the SCF complex in human cells. In the presence of the IAA, TIR1 associates with the AID fused to the protein of interest (POI). SCFTIR1 recruits an E2 ligase and polyubiquitinates the AID, resulting in the degradation of the POI by the proteasome. (C) Parental and TIR1-9Myc expressing DLD-1 and RPE1 were treated with or without IAA for 3 d and processed for flow cytometry. Note expression of TIR1-9Myc and/or treatment with IAA does not cause alterations in the cell cycle profile. (D) DLD-1 or RPE1 cells stably expressing TIR1-9Myc were cotransfected with histone H2BmRFP or histone H2BAID-YFP; after 24 h, they were treated with (+) or without (−) IAA for a further 24 h. Fluorescent images show the presence or absence of histone H2BmRFP and histone H2BAID-YFP. (Scale bars = 5 μm.)
SCFTIR1 has been demonstrated to promote auxin-inducible degradation of proteins tagged with an AID in a variety of animal cells (4, 8) (Fig. 1B), including inducible degradation of a nuclear localized GFP in human cells. However, not yet established is whether auxin-inducible degradation of AID-tagged proteins is (i) active against a diverse array of both nuclear and cytoplasmic substrates, (ii) capable of controlling the stability of substrates incorporated in macromolecular complexes, (iii) sufficiently robust to achieve rapid and complete degradation of all target proteins, (iv) initiated and continuously active in all phases of the cell cycle, and (v) rapidly reversible within minutes of auxin removal. Here, we provide a detailed characterization of the AID system in human cells (4, 8). We demonstrate that the AID system possesses all five of these properties, and thus represents a powerful tool for studying protein function in mammalian cells.
Results
AID System Allows Protein Degradation in Both Transformed and Nontransformed Human Cells.
We used retroviral transduction to construct a monoclonal colorectal cancer cell line (DLD-1) and a diploid nontumor cell line (RPE-1) stably expressing TIR1-9xMyc. Stable expression of TIR1-9xMyc with or without 3 d of incubation with 500 μM auxin hormone indole-3-acetic acid (IAA) did not affect growth or cell cycle distribution of either cell type (Fig. 1C), consistent with an earlier report in chicken DT40 cells (4). To test if rapid proteolysis of substrates could be induced by addition of auxin, TIR1-9Myc expressing DLD-1 and RPE-1 cells was cotransfected with mRFP-tagged histone H2B (H2BmRFP) and AID-YFP-tagged histone H2B (H2BAID-YFP; the AID tag is 228 aa or ∼25 kDa). In the absence of IAA, histone H2BmRFP and histone H2BAID-YFP accumulated in the nucleus of both cell lines (Fig. 1D). However, addition of IAA led to degradation of histone H2BAID-YFP but not histone H2BmRFP, demonstrating that the AID system can be used for induced proteolysis of target proteins in both transformed and nontransformed human cells (Fig. 1D). All additional experiments were performed using TIR1-9Myc expressing DLD-1 cells.
AID System Can Quantitatively Degrade Divergent Protein Substrates in Human Cells.
To determine whether rapid, auxin-induced degradation was a general property of AID-tagged protein substrates, five different transgenes encoding carboxy-terminally AID-YFP–tagged substrates were integrated at the same genomic locus of DLD-1 TIR1-9Myc cells using Flp-mediated recombination (17). Two of these transgenes encoded cytosolic proteins: polo-kinase 4 (Plk4), which regulates centriole duplication (18–20), and cyclin B1, the regulatory subunit of the cyclin-dependent kinase Cdk1 (21). In addition, we analyzed three nuclear proteins: histone H2B, a subunit of the nucleosome; centromere protein A (CENP-A), the centromere-specific histone H3 variant that assembles into centromeric chromatin (22); and Telomeric Repeat-Binding Factor 2 (TRF2), a component of the shelterin complex required for the protection of telomere ends (23, 24).
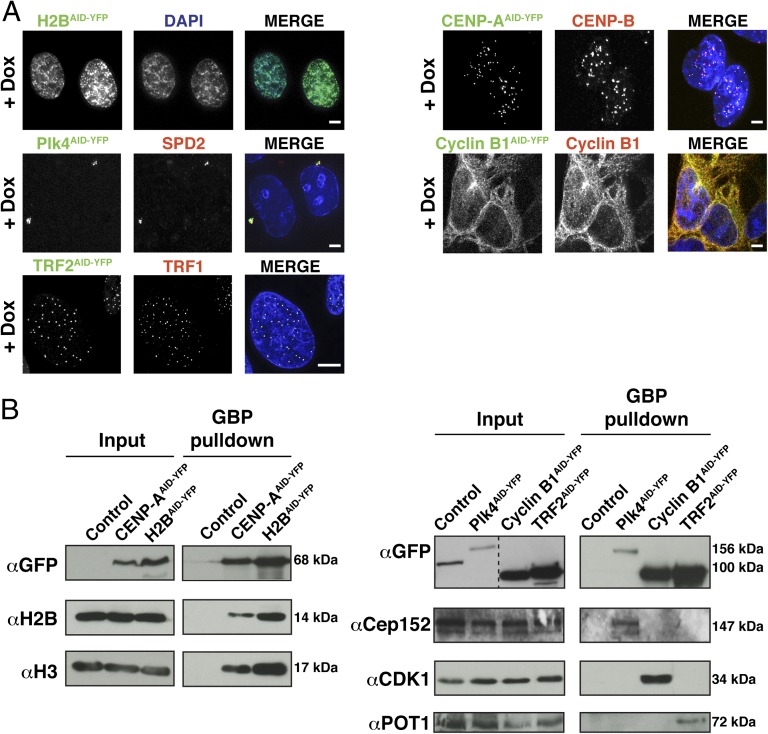
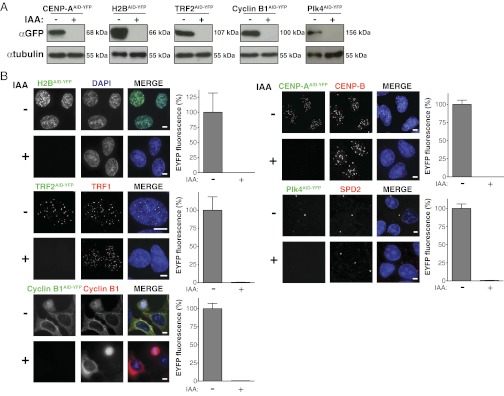
Transgene expression was under doxycycline control, and following induction, all five substrates localized as expected: Plk4AID-YFP localized at the centrosome, cyclin B1AID-YFP localized diffusely in the cytoplasm, histone H2BAID-YFP localized within the nucleus, CENP-AAID-YFP localized to centromeres, and TRF2AID-YFP associated with telomeres (Fig. 2A). Purifications using the GFP-binder protein demonstrated that all substrates were incorporated into protein complexes, with histone H2BAID-YFP and CENP-AAID-YFP both associating with endogenous histone H2B and histone H3, Plk4AID-YFP binding to CEP152, TRF2AID-YFP associating with POT1, and cyclin B1AID-YFP binding to Cdk1 (Fig. 2B). All five proteins were quantitatively destroyed within 24 h of addition of auxin, as seen with immunoblotting (Fig. 3A) and quantitative immunofluorescence (Fig. 3B).
Fig. 2.
Auxin-induced degradation system is capable of controlling the stability of substrates localized to different regions of cells. (A) AID-YFP–tagged proteins were induced for 8–24 h. Fluorescence images show the appropriate subcellular localization of doxycycline (Dox)-induced transgenes, histone-H2BAID-YFP (nucleus), Plk4AID-YFP (centrosome), TRF2AID-YFP (telomere), CENP-AAID-YFP (centromere), and cyclin B1AID-YFP (cytoplasm). (Scale bars = 5 μm.) (B) AID-YFP–tagged proteins were induced for 24 h and subsequently purified from cell lysates using GFP-binder protein (GBP)-coupled beads. Control purifications were performed in parallel from parental cells that do not contain an inducible AID-YFP transgene. GBP-purified protein complexes were analyzed by immunoblotting.
Fig. 3.
Complete depletion of target proteins can be achieved with the auxin-induced degradation system. (A) AID-YFP–tagged proteins were induced for 24 h, and cells were treated with (+) or without (−) IAA for a further 24 h. Remaining protein levels were analyzed by immunoblotting. (B) AID-YFP–tagged proteins were induced for 8–24 h and treated with (+) or without (−) IAA for a further 24 h. Fluorescence images show the localization and level of AID-YFP–tagged target proteins. Bar graphs show the quantification of EYFP levels in fluorescence images. Bars represent the mean of >20 cells per condition. Error bars represent the SEM. (Scale bars = 5 μm.)
Auxin-Induced Degradation Reduces Protein t1/2 to <20 Min.
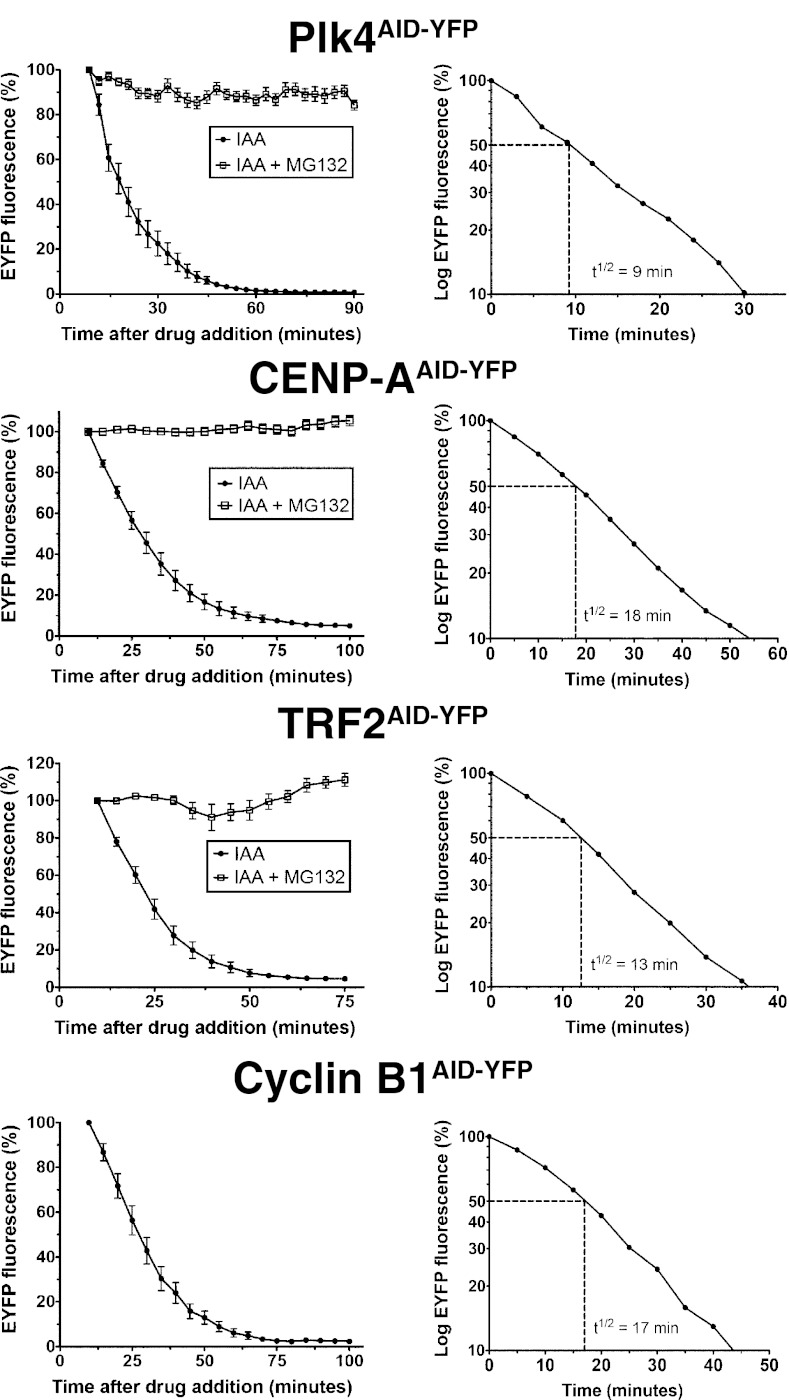
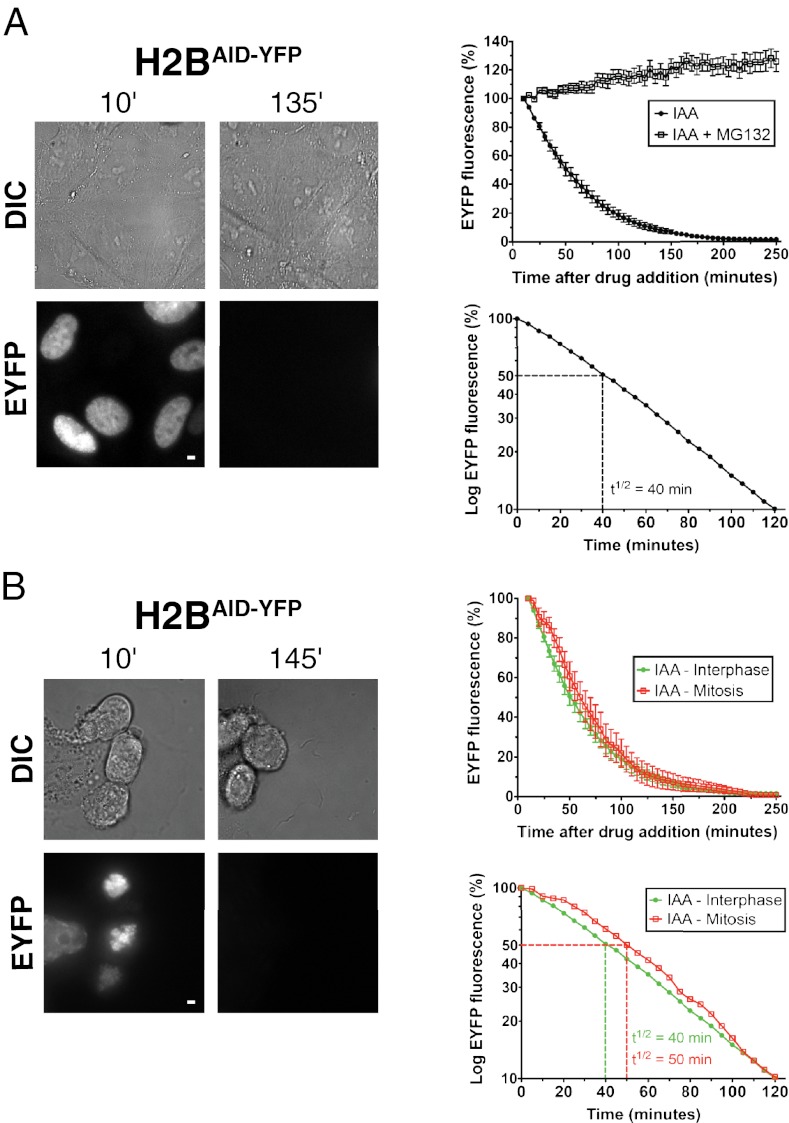
The rapidity of induced degradation of each of the five differentially localized substrates incorporated into different protein complexes was quantified with fluorescence time-lapse microscopy after addition of IAA. Substrate degradation began rapidly after the addition of IAA and proceeded with first-order kinetics (Fig. 4). For Plk4AID-YFP, CENP-AAID-YFP, TRF2AID-YFP, and cyclin B1AID-YFP, the t1/2 for each was <20 min (9, 18, 13, and 17 min, respectively), essentially yielding quantitative loss within 80 min (Fig. 4 and Movies S1, S2, S3, and S4). Degradation of histone H2BAID-YFP occurred more slowly (initial t1/2 = 40 min; Fig. 5A and Movie S5); however, even here, loss to undetectable levels was reached within 3 h. For all five substrates, t1/2 was constant during the degradation of >90% of the protein (Figs. 4 and 5A). For both CENP-AAID-YFP and TRF2AID-YFP, induction of expression for >8 h resulted in a proportion of YFP-tagged protein localizing diffusely throughout the nucleus (Movies S2 and S3). Nevertheless, IAA addition led to the precipitous degradation of the entire pool of CENP-AAID-YFP and TRF2AID-YFP. CENP-A present at centromeres has been demonstrated to have a very long t1/2 in cells (25); however, remarkably, centromeric and diffusely localized pools of CENP-AAID-YFP were destroyed with identical kinetics (Fig. S1 and Movie S2). This demonstrates that even protein incorporated into a stable complex with a very long t1/2 can be rapidly depleted using the AID system. The rapid decline in fluorescence observed after IAA addition was a result of proteolysis and not photobleaching; addition of the proteasome inhibitor MG132 prevented the degradation of each YFP-tagged target protein tested (Figs. 4 and 5A).
Fig. 4.
Protein degradation occurs rapidly following IAA addition. AID-YFP–tagged proteins were induced for 8–24 h, and cells were treated with IAA in the presence or absence of the proteasome inhibitor MG132. EYFP levels were monitored by fluorescence time-lapse microscopy beginning 9–10 min after drug addition. (Left) Graphs show EYFP fluorescence at various times after drug addition. (Right) Graphs show the logarithmic plot of EYFP fluorescence at various time points after filming began. Graphs represent the mean of 7–20 cells per condition. Error bars represent the SEM.
Fig. 5.
Protein degradation occurs during all phases of the cell cycle. (A) Histone H2BAID-YFP was induced for 8 h, and cells were treated with IAA in the presence or absence of the proteasome inhibitor MG132. EYFP levels were monitored by fluorescence time-lapse microscopy beginning 10 min after drug addition. (Upper) Graph shows EYFP fluorescence at various times after drug addition. (Lower) Graph shows the logarithmic plot of EYFP fluorescence at various time points after filming began. Graphs represent the mean of 19 (IAA + MG132) or 32 (IAA) cells per condition, and error bars represent the SEM. Representative images show the level of histone H2BAID-YFP and corresponding differential interference contrast (DIC) images at the indicated times. (B) Same as in A except that in one condition, cells were arrested in mitosis with nocodazole for 6 h before the beginning of filming. Destruction was quantified in mitotically arrested cells, which can be distinguished by their highly condensed chromosomes. Graphs represent the mean of 10 (mitosis) or 32 (interphase) cells per condition. Error bars represent the SEM. (Scale bars = 5 μm.)
To establish how continued synthesis affected the kinetics of AID-induced degradation, all cellular translation in histone H2BAID-YFP-expressing cells was inhibited by addition of cycloheximide before addition of IAA. Absence of new synthesis of histone H2BAID-YFP modestly decreased the time required for quantitative depletion, with reduction in the initial t1/2 from 40 min to 33 min (Fig. S2), indicating that even for the most stable of our five AID-tagged proteins, the rapid kinetics of auxin-induced degradation are largely unaffected by continued protein synthesis.
Auxin-Induced Proteolysis Occurs in All Phases of the Cell Cycle.
An initial view of when during the cell cycle auxin-induced degradation was active came from comparing the kinetics of loss of Plk4AID-YFP, CENP-AAID-YFP, TRF2AID-YFP, cyclin B1AID-YFP, and histone H2BAID-YFP within individual cells of a randomly cycling population. Similar kinetics were found in all cells within each population (Fig. S3 and Movies S1, S2, S3, S4, and S5). Because these cells were distributed in G1, S, and G2 phases of the cell cycle in relative abundances of ∼5:2:1 [based on FACS profiles (Fig. S4) and the typical length of each portion of the cycle (26)], auxin-induced degradation of substrates must be able to initiate and proceed in all three portions of the interphase cell cycle.
To test whether substrate degradation could be initiated during mitosis, histone H2BAID-YFP-expressing cells were arrested in mitosis by treatment for 6 h with the microtubule assembly poison nocodazole. Following addition of IAA, histone H2BAID-YFP degradation occurred rapidly with kinetics similar to those observed in interphase cells (t1/2 = 50 min in interphase vs. t1/2 = 60 min in mitosis; Fig. 5B and Movie S6), demonstrating that induced degradation is almost as efficient in mitosis as it is in the other three cell cycle phases.
AID-Mediated Protein Degradation Is Quickly Reversible, Enabling Inducible Protein Expression in Human Cells.
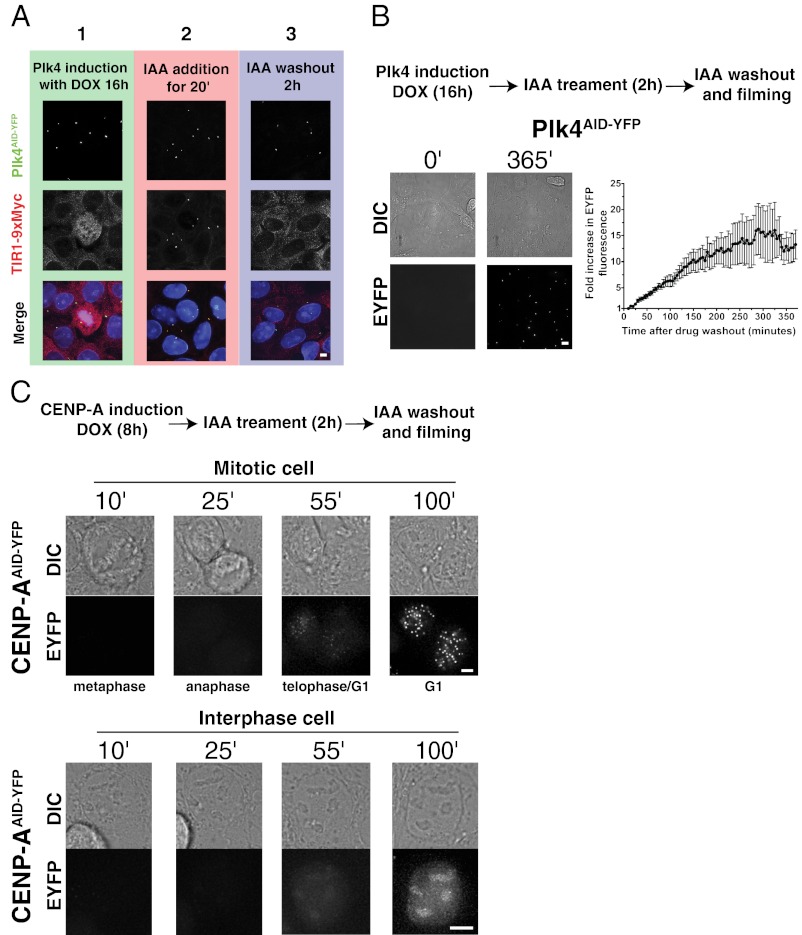
To determine the reversibility of AID-targeted degradation after removal of IAA, we first examined the localization of TIR1-9xMyc in cells expressing Plk4AID-YFP. Within 20 min of IAA addition, TIR1-9xMyc became enriched at the centrosome, colocalizing with the substrate Plk4AID-YFP (Fig. 6A). This recruitment of TIR1-9xMyc to the centrosome was reversible; 2 h after IAA washout, TIR-9xMyc localized diffusely in the nucleus/cytoplasm and was undetectable at the centrosome (Fig. 6A), consistent with rapid reversibility of AID-targeted degradation.
Fig. 6.
Protein depletion is readily reversible on removal of IAA. (A) Plk4AID-YFP expression was induced for 16 h, and cells were treated with IAA for 20 min. Cells were washed to remove IAA and supplemented with doxycycline (DOX) for a further 2 h. Cells were fixed at the indicated times after drug addition, and the localization of Plk4AID-YFP and TIR1-9Myc was determined by fluorescent microscopy. In the presence of IAA, TIR1-9Myc is reversibly recruited to the centrosome. (B) Plk4AID-YFP expression was induced for 16 h, and cells were treated with IAA for a further 2 h. Cells were washed to remove IAA and supplemented with doxycycline, and levels of Plk4AID-YFP fluorescence were monitored by fluorescence time-lapse microscopy. The graph represents the mean of 10 cells, and error bars represent the SEM. Representative images show the level of Plk4AID-YFP and corresponding differential interference contrast (DIC) images at the indicated times. (C) Expression of CENP-AAID-YFP was induced for 8 h, and cells were treated with IAA for a further 2 h. Cells were washed to remove IAA and supplemented with doxycycline, and recruitment of CENP-AAID-YFP to the centromere was monitored by fluorescence time-lapse microscopy. Note that although newly synthesized CENP-AAID-YFP accumulates diffusely in the nuclei of cells, loading of CENP-AAID-YFP onto centromeres is delayed until late telophase/early G1. (Scale bars = 5 μm.)
To establish the kinetics with which AID-tagged target proteins reaccumulate after removal of IAA, cells were treated with IAA for 2 h to degrade Plk4AID-YFP quantitatively (>99%); IAA was then removed, and quantitative time-lapse fluorescence microscopy was used to follow Plk4AID-YFP reaccumulation. Newly made Plk4AID-YFP was detectable almost immediately after IAA washout (within <10 min), and Plk4AID-YFP protein levels increased linearly for 200 min, reaching steady state by ∼350 min after IAA removal (Fig. 6B and Movie S7).
The rapid reversibility of the AID system raised the possibility that it could be exploited to achieve inducible protein expression. To monitor the incorporation of newly synthesized CENP-A in living cells, we treated cells expressing CENP-AAID-YFP with IAA for 2 h to degrade previously synthesized CENP-AAID-YFP, removed the IAA, and followed cells by time-lapse fluorescence microscopy. Newly synthesized CENP-AAID-YFP accumulated diffusely in the nuclei of interphase cells; however, consistent with our previous report (25), it incorporated into centromeres soon after cells exited mitosis and entered G1 phase (Fig. 6C and Movie S8). This demonstrates that the rapid reversibility of the AID system is useful for providing inducible control of protein expression in human cells.
Functional Replacement of BubR1 with an AID-Tagged BubR1 Transgene Allows Temporal Control of Mitotic Checkpoint Activity.
The mitotic checkpoint is an essential cell cycle control mechanism that operates during every division to prevent the irreversible transition into anaphase until all chromosomes attach to microtubules of the mitotic spindle (27). BubR1 is an essential component of the mitotic checkpoint, but previous studies of the protein’s function have relied on strategies that chronically deplete BubR1 levels over multiple cell cycles (28, 29). To establish if the AID system can be used to achieve the rapid, functional inactivation of BubR1, we established a method to replace endogenous BubR1 protein with an AID-tagged version capable of rapid, inducible destruction.
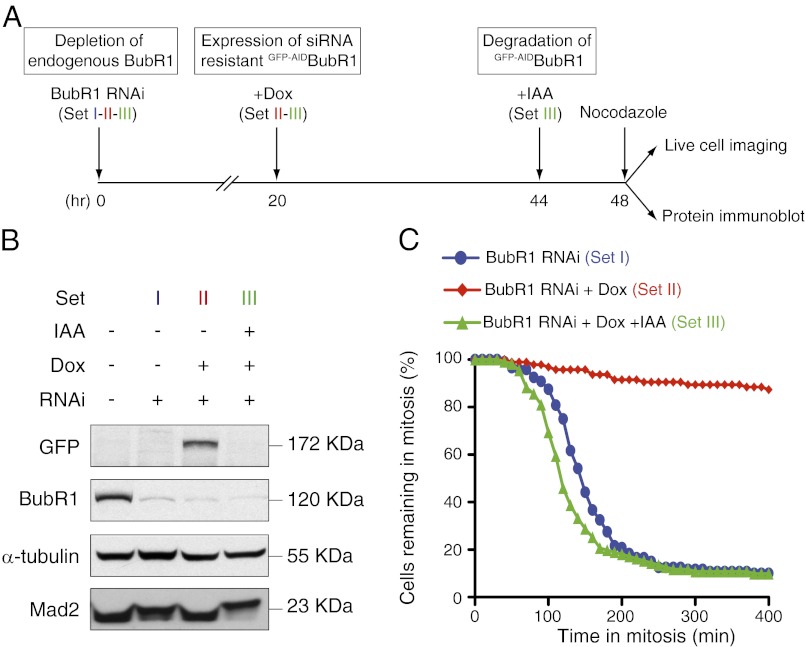
Endogenous BubR1 was depleted by >90% in cells using siRNA targeting the 3′ UTR of the BubR1 mRNA and was replaced by expression of siRNA-resistant, amino-terminally GFP-AID-tagged BubR1 (GFP-AIDBubR1) encoded by a transgene containing a distinct 3′ UTR (Fig. 7 A and B). Cells were treated with nocodazole to block spindle microtubule assembly, thereby chronically activating the mitotic checkpoint. The duration of the mitotic arrest was then determined by time-lapse microscopy. As expected, suppression of endogenous BubR1 resulted in premature mitotic exit in the presence of nocodazole, and this was rescued by expression of GFP-AIDBubR1: ∼80% of BubR1-depleted cells exited mitosis in 200 min compared with <10% of the cells expressing GFP-AIDBubR1 (Fig. 7C, sets I and II). Addition of IAA for 4 h before mitotic entry led to the complete destruction of GFP-AIDBubR1 (Fig. 7 A and B, set III) and a rapid mitotic exit in the presence of nocodazole (Fig. 7C, set III). Surprisingly, following destruction of GFP-AIDBubR1, cells exited mitosis even more rapidly than cells depleted of endogenous BubR1 (Fig. 7C, compare sets II and III). A likely explanation for this effect is that cells displaying the most complete depletion of BubR1 will undergo catastrophic mitotic divisions and/or cease cycling during the 48-h period required for the siRNA-mediated reduction in endogenous BubR1. By contrast, functional complementation with GFP-AIDBubR1 allows cells to survive complete loss of endogenous BubR1. Therefore, following destruction of GFP-AIDBubR1, these cells will more rapidly exit mitosis than cells only partially depleted in endogenous BubR1. Thus, rapid depletion of BubR1 using the AID system can produce a more complete null phenotype than is achieved by depletion of the BubR1 mRNA.
Fig. 7.
Functional replacement of siRNA-depleted BubR1 with an AID-tagged BubR1 transgene. (A) Schematic outlines the experimental strategy used to replace endogenous BubR1 with GFP-AIDBubR1. (B) Immunoblot shows the levels of various proteins at 48 h after siRNA depletion of endogenous BubR1. (C) Graph shows the fraction of cells remaining in mitosis at various times after addition of nocodazole. There were >95 cells per condition from two independent experiments. Dox, doxycycline.
Discussion
Systems that permit the posttranslational regulation of protein stability with small molecules are valuable tools for interrogating biological systems. Here, we have provided an extensive characterization of the auxin-inducible degradation system in human cells and demonstrated that this system (i) is active against multiple substrates, (ii) is capable of targeting degradation of both nuclear and cytoplasmic substrates that are monomeric or in macromolecular complexes, (iii) proceeds rapidly and to completion with a diverse array of substrates, (iv) is active in all phases of the cell cycle, (v) is rapidly reversible within minutes of auxin removal, and (vi) is able to be used in combination with a functional replacement of an endogenous gene. For all five substrates we tested using the AID system, protein degradation proceeded to completion and occurred rapidly, with a t1/2 as short as 9 min and with an average time of 19 min. The time required for depletion of >90% of the substrate ranged between 30 and 120 min (Figs. 4 and 5). The speed and level to which substrates are depleted from cells make the AID system an ideal choice for studying phenotypes resulting from acute loss of protein function.
The AID system offers a method to overcome limitations of previous approaches (e.g., siRNA) in which gradual depletion of a protein may produce a different phenotype compared with when a protein is rapidly removed from cells (4). Indeed, different phenotypes may be observed at differing levels of protein depletion; thus, to determine the true “null’ phenotype, the protein in question should be rapidly and completely depleted from cells. This caveat is especially relevant when examining long-lived proteins (e.g., CENP-A), which may require days to be fully depleted from cells (25, 30, 31).
A remarkable feature of the AID system is its ability to destroy substrates that are incorporated into large protein complexes, such as TRF2AID-YFP (part of the six-subunit shelterin complex) and CENP-AAID-YFP and Histone H2BAID-YFP (components of the eight-subunit nucleosome complex) (Fig. 2 B and C). This feature makes the AID system suitable for the analysis of single-protein components of multisubunit complexes. It was notable that the AID system promoted the depletion of chromatin-bound TRF2AID-YFP, CENP-AAID-YFP, and histone H2BAID-YFP, as well as cytosolic components, such as Plk4AID-YFP and cyclin B1AID-YFP (Fig. 3), indicating that proteins localized to many different regions of the cell will be susceptible to AID-mediated degradation.
Our evidence that the AID system is capable of destroying substrates at all phases of the cell cycle demonstrates that SCFTIR1 is active throughout the cell cycle, with the recruitment of SCFTIR1 to the substrate being a rate-limiting step for proteolysis. The ability to destroy substrates at all stages of the cell cycle forms a major advantage for studying cell cycle-regulated processes and, to our knowledge, has not been demonstrated with any other system that allows regulation of protein stability with small molecules.
A final feature of the auxin-mediated degradation system is that it is rapidly reversible because new protein begins accumulating almost immediately after IAA removal (Fig. 6). Protein accumulation after removal of auxin does not require transcription, and is thus faster than would occur following activation of an inducible promoter (e.g., doxycycline-inducible promoters). The extensive depletion of target proteins with the AID system raises the possibility that this technology may be useful (either alone or in combination with inducible promoters) in temporally regulating the induction of toxic genes.
Like other methods for posttranslational regulation of protein stability (4, 6–10), the auxin system does not control stability of endogenous, untagged proteins. Nevertheless, advances in the use of adeno associated viruses (32), zinc finger (33), and transcription activator-like effector endonucleases (34) open the possibility of carrying out homologous recombination in somatic human cells to create endogenous proteins fused to the AID. A second possibility is to deplete an endogenous protein using siRNA or gene-targeting technologies and reconstitute cells with an siRNA-resistant, AID-tagged transgene, as we have done for the mitotic checkpoint protein BubR1 (35) (Fig. 7). Third, a dominantly acting gene product can be tagged with the AID and rapidly removed from cells.
Finally, an interesting extension of this work will be to determine whether the AID system will also prove to be useful for modulating the expression levels of proteins in animals. This could be particularly valuable in genetically engineered mouse models of human disease, where the AID system can be used to provide validation of potential therapeutic targets in an animal before specific inhibitors are available for use. [A potential caveat, however, is whether IAA can be administered at levels sufficient to induce protein degradation in animals without causing significant adverse toxicity (36, 37).] Further studies will be required to address the applicability of the AID system in animals.
Materials and Methods
Constructs.
The osTIR1-9Myc and AID ORF was a kind gift from Masato Kanemaki (National Institute of Genetics, Mishima, Japan). Full-length ORFs were cloned into a pcDNA5/FRT/TO-based vector (Invitrogen) modified to contain an amino-terminus or carboxy-terminus AID and a YFP tag.
Cell Biology and Generation of Stable Cell Lines.
Cell culture and flow cytometry were performed as previously described (38). osTIR1-9Myc was introduced into Flp-In TRex–DLD-1 parental cells or into RPE1 cells using retroviral delivery as described previously (39). Stable integrates were selected in 2 μg/mL (DLD-1) or 5 μg/mL (RPE1) puromycin, and single clones were isolated using single-cell sorting (FACS-Vantage; Becton Dickinson). Stable, isogenic cell lines expressing AID-tagged transgenes were generated using FRT/Flp-mediated recombination, as described previously (40). Expression of the AID-tagged transgenes was induced with 1 μg/mL doxycycline (SIGMA) for 8–24 h. Flp-In TRex–DLD-1, TIR1-9Myc cells were used for all the experiments, except for the experiments in Fig. 1 C and D, where RPE1 TIR1-9Myc cells were also used. Cell transfection was performed using FuGENE (Invitrogen). Small molecules were used at the following final concentrations: IAA, 500 μM (I5148; SIGMA); nocodazole, 2 μg/mL (SIGMA); MG132, 20 μM (Calbiochem); and cycloheximide, 50 μg/mL (SIGMA). siRNA directed against the 3′ UTR of BubR1 (5′-CUGUAUGUGCUGUAAUUUA-3′) was purchased from Thermo Fisher Scientific. Cells were transfected with 100 nM oligonucleotides using Oligofectamine (Invitrogen). Twenty-four hours after transfection, tetracycline was added to express GFP-AID-BubR1 for another 24 h before collecting cells for immunoblotting or analysis by time-lapse microscopy.
Protein Purification and Western Blotting.
To purify AID-YFP-tagged transgenes, cells were lysed in lysis buffer [10 mM Tris (pH 7.5); 0.1% Triton X-100; 100 mM NaCl; 1 mM EDTA; 1 mM EGTA; 50 mM NaF; 20 mM β-glycerophosphate; 0.1 mM DTT; 200 nM microcystin; 1 mM PMSF; and 1 μM leupeptin, pepstatin, and chymostatin] and sonicated, and soluble extracts were prepared. The supernatant was incubated with beads alone or beads coupled to GFP binding protein (41). Beads were washed five times in lysis buffer, and protein complexes were analyzed by immunoblotting. For immunoblot analysis, the following antibodies were used: BubR1 (rabbit, 1:5,000 covance), Mad2 (rabbit, 1:1,000; Bethyl Laboratories), DM1A (mouse anti–α-tubulin, 1:5,000), YFP (mouse anti-GFP, 1:500; Roche), POT1 (rabbit, 1:1,000; Abcam), Cep152 (rabbit, 1:1,000; Bethyl Laboratories), CDK1 (mouse, 1:500; Santa Cruz Biotechnology), Histone H3 (rabbit, 1:5,000; SIGMA), and Histone H2B (mouse, 1:1,000; Upstate Biotechnology).
Immunofluorescence.
Immunofluorescence was performed as previously described (42) using the following antibodies: 4A6 (mouse anti-Myc, 1:1,000; Upstate Biotechnology), TRF1 (rabbit, 1:1,000; a kind gift from Titia De Lange, The Rockefeller University, New York, NY), CENP-B (rabbit, 1:1,000; Abcam), Cyclin B1 (mouse, SC-245, 1:50; Santa Cruz Biotechnology), and SPD2-Cy3 (rabbit, 1:1,500; a kind gift from Karen Oegema, Ludwig Institute for Cancer Research, University of California at San Diego). YFP was visualized directly using a GFP filter set. Immunofluorescence images were collected using a Deltavision Core system (Applied Precision) controlling an interline CCD camera (Cooldsnap; Raper). Images were collected using a 100× 1.4-N.A. or 60× 1.4-N.A. oil objective at 0.2-μM z sections and subsequently deconvolved. Maximum intensity 2D projections were assembled for each image using softWoRx (Applied Precision). For quantification of YFP signal intensity, undeconvolved 2D maximum intensity projections were saved as unscaled 16-bit tagged image file format (TIFF) images and signal intensities were determined using MetaMorph (Molecular Devices). For Plk4AID-YFP, a 20 × 20 pixel box and a larger 25 × 25 pixel box were drawn around the centrosome (marked with γ-tubulin staining). Integrated YFP intensity in the smaller box was calculated by subtracting the mean fluorescence intensity in the area between the two boxes (mean background) from the mean YFP intensity in the smaller box and multiplying by the area of the smaller box. For CENP-AAID-YFP, a 15 × 15 pixel circle was drawn around a centromere (marked by CENP-B staining) and an identical circle was drawn adjacent to the structure (background). The integrated signal intensity of each individual centromere was calculated by subtracting the fluorescence intensity of the background from the intensity of the adjacent centromere. Approximately 25 centromeres were averaged to provide the average fluorescence intensity for each individual cell. The signal intensity of TRF2AID-YFP was determined using an identical method, except TRF1 staining was used to determine the localization of the telomere.
To determine Histone H2BAID-YFP intensity, a 50 × 50 pixel box was drawn in the nucleus or in the cytoplasm (background) and the integrated signal intensity of each cell was calculated by subtracting the fluorescence intensity of the background from the intensity of the nucleus. An identical procedure was used to calculate the intensity of Cyclin B1AID-YFP, except background readings from the nucleus were subtracted from the cytoplasmic signal.
Live Cell Microscopy.
Cells were seeded into 35-mm, poly-l-lysine–coated, glass-bottomed culture dishes (MatTek) and transferred to CO2 independent media supplemented with doxycycline 24 h later. Cells were maintained at 37 °C in an environmental control station, and images were collected using a Deltavision RT system (Applied Precision) with a 40× 1.35-N.A. oil lens at 3- or 5-min time intervals. IAA was added at the microscope stage. For each time point, 4 × 3-μM z sections were acquired for YFP, and maximum intensity projections were created using softWoRx. Movies were assembled and analyzed using MetaMorph software (Molecular Devices). For quantification of YFP signal intensity, undeconvolved 2D maximum intensity projections were saved as unscaled 16-bit TIFF images and signal intensity was determined for each frame of the movie using MetaMorph. The procedure for quantifying the levels of Plk4AID-EYFP, Histone H2BAID-EYFP, and Cyclin B1AID-EYFP was as described in the section on immunofluorescence. CENP-AAID-EYFP and TRF2AID-EYFP were quantified using a procedure identical to that used for Histone H2BAID-EYFP. To compare the degradation kinetics of centromeric and diffusely localized CENP-AAID-EYFP, a circle 10 pixels in diameter was drawn around an individual centromere (centromeric pool) and an identical circle was drawn adjacent to the structure (diffuse pool). The integrated signal intensity for six cells was averaged to provide the average fluorescence intensity at each time point.
Supplementary Material
Acknowledgments
The authors thank Masato Kanemaki, Titia de Lange, Eros Lazzerini Denchi, and Stephen Taylor for providing reagents. We gratefully acknowledge the University of California at San Diego Neuroscience Microscopy Shared Facility (P30 NS047101). This work was supported by Grant GM29513 from the National Institutes of Health (to D.W.C, who receives salary support from the Ludwig Institute for Cancer Research). A.J.H. is supported by a Leukemia and Lymphoma Society special fellowship, and D.F is supported by a European Molecular Biology Organization long-term fellowship.
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 19886 (volume 109, number 49).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216880109/-/DCSupplemental.
References
- 1.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85(14):5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34(1):322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17(9):981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonger KM, Chen LC, Liu CW, Wandless TJ. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat Chem Biol. 2011;7(8):531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanke M, et al. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011;12:8. doi: 10.1186/1471-2121-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neklesa TK, et al. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7(8):538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1(5):REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho MS, Tsai PI, Chien CT. F-box proteins: The key to protein degradation. J Biomed Sci. 2006;13(2):181–191. doi: 10.1007/s11373-005-9058-2. [DOI] [PubMed] [Google Scholar]
- 13.Schulman BA, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408(6810):381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 14.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 15.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 16.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 17.O’Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251(4999):1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 18.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15(24):2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 19.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 20.Holland AJ, Lan W, Cleveland DW. Centriole duplication: A lesson in self-control. Cell Cycle. 2010;9(14):2731–2736. doi: 10.4161/cc.9.14.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12(6):658–665. doi: 10.1016/s0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 22.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104(4):805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 24.Bilaud T, et al. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17(2):236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 25.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176(6):795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon RE, Lane BP. Duration of cell cycle and its phases measured in synchronized cells of squamous cell carcinoma of rat trachea. Cancer Res. 1980;40(12):4467–4472. [PubMed] [Google Scholar]
- 27.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 28.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7(1):45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101(23):8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemmerich P, et al. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180(6):1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151(5):1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Büning H, Perabo L, Coutelle O, Quadt-Humme S, Hallek M. Recent developments in adeno-associated virus vector technology. J Gene Med. 2008;10(7):717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- 33.Carroll D. Zinc-finger nucleases: A panoramic view. Curr Gene Ther. 2011;11(1):2–10. doi: 10.2174/156652311794520076. [DOI] [PubMed] [Google Scholar]
- 34.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14(1):47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142(3):444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furukawa S, Usuda K, Abe M, Hayashi S, Ogawa I. Indole-3-acetic acid induces microencephaly in mouse fetuses. Exp Toxicol Pathol. 2007;59(1):43–52. doi: 10.1016/j.etp.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 37.John JA, Blogg CD, Murray FJ, Schwetz BA, Gehring PJ. Teratogenic effects of the plant hormone indole-3-acetic acid in mice and rats. Teratology. 1979;19(3):321–324. doi: 10.1002/tera.1420190307. [DOI] [PubMed] [Google Scholar]
- 38.Holland AJ, et al. Polo-like kinase 4 controls centriole duplication but does not directly regulate cytokinesis. Mol Biol Cell. 2012;23(10):1838–1845. doi: 10.1091/mbc.E11-12-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah JV, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14(11):942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 40.Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117(Pt 26):6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 41.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7(2):282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188(2):191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]