Abstract
A common strategy among microbes living in iron-limited environments is the secretion of siderophores, which can bind poorly soluble iron and make it available to cells via active transport mechanisms. Such siderophore–iron complexes can be thought of as public goods that can be exploited by local communities and drive diversification, for example by the evolution of “cheating.” However, it is unclear whether bacterial populations in the environment form stable enough communities such that social interactions significantly impact evolutionary dynamics. Here we show that public good games drive the evolution of iron acquisition strategies in wild populations of marine bacteria. We found that within nonclonal but ecologically cohesive genotypic clusters of closely related Vibrionaceae, only an intermediate percentage of genotypes are able to produce siderophores. Nonproducers within these clusters exhibited selective loss of siderophore biosynthetic pathways, whereas siderophore transport mechanisms were retained, suggesting that these nonproducers can act as cheaters that benefit from siderophore producers in their local environment. In support of this hypothesis, these nonproducers in iron-limited media suffer a significant decrease in growth, which can be alleviated by siderophores, presumably owing to the retention of transport mechanisms. Moreover, using ecological data of resource partitioning, we found that cheating coevolves with the ecological specialization toward association with larger particles in the water column, suggesting that these can harbor stable enough communities for dependencies among organisms to evolve.
Keywords: microbial diversity, ocean bacteria, population structure, genome dynamics
Ecological populations of bacteria, comprising clusters of coexisting close relatives with similar resource preference in the environment, are often composed of organisms that can have hundreds of unique genes per genome (1). This enormous diversity among organisms with similar function and niche association remains one of the most puzzling phenomena in microbiology. Identifying the selective pressures that drive the evolution of variable gene content—the so-called flexible genome—is essential to understand the functional impact of genomic diversity on the ecological function of microbial populations in the environment. Functional gene annotations across close relatives in different bacterial species show that variable gene content is relatively enriched in genes involved in synthesis of secondary metabolites, proteins exposed on the cell surface, and defense mechanisms, as well as transport of exogenous compounds (2–4). This suggests that the high turnover of genes in bacteria could result from adaptations to rapidly changing biotic interactions. This idea supports the emerging view that microbial diversity is not solely explained by adaptation to abiotic conditions (5, 6) and indicates that interactions within populations and communities play a large role in creating genomic diversity.
Some of the better-understood types of interactions in bacteria are those mediated by siderophores, small molecules that strongly bind poorly soluble iron. Under iron-limiting conditions, siderophores are excreted outside the cell, where they form complexes with ferric-iron (7). The ferric-iron–siderophore complex is then recognized by specific outer-membrane receptors that transport it back inside the cell. Because any cell that carries specific outer-membrane transporters can take them up, siderophores can be considered a prime example of a public good. They can be exploited by “cheaters,” which do not bear the cost of production but nonetheless reap the benefits brought by the iron–siderophore complex. These systems were among the first to be studied as public goods in in vitro experiments (8, 9). They have also been widely studied for their role in pathogenicity (7, 10) and in natural environments such as the ocean, where they may play an important role in iron acquisition under limiting conditions (11–13). Because of the dilute nature of the aquatic environment, it has been suggested that production of public goods such as siderophores is an efficient strategy only in environments where local cell densities are high (14). In line with this idea, siderophore production has been described in heterotrophic marine bacteria such as Vibrio, Alteromonas, and Marinobacter (13), which normally exploit nutrient patches with high local cell densities and potentially high siderophore reabsorption rates.
Although the dynamic emerging from the interaction between cheaters and producers has been extensively studied using mathematical models and synthetic microbial systems (8, 15), it has not been conclusively demonstrated that complex bacterial populations in the environment engage in the stable or recurrent interactions that are assumed in these models. For example, in environments such as the ocean, where sympatric microbial populations and species are diverse and dispersal rates are high, it is unclear whether closely related genotypes assemble reproducibly into nonclonal, locally interacting and coevolving populations, as opposed to forming random assemblages. Therefore, validating the role of social interactions such as public good games in natural populations has potential to provide support for the existence of stable social structure and for the hypothesis that pervasive genomic microdiversity in the environment in part reflects differences in social roles.
The present study seeks to unravel the evolutionary and ecological dynamics of siderophore production and cheating in natural microbial populations, using a collection of >1,700 marine isolates of Vibrionaceae. These isolates are organized in phylogenetic clusters of highly related but nonclonal genotypes. Clusters of isolates are differentiated by their propensity to associate with different classes of resources, operationally defined by separation into four size classes in the planktonic environment. The smallest class encompasses dissolved organic and inorganic nutrients, primarily exploited by free-living cells, whereas all other size classes represent collections of different types of particles and organisms, onto which bacteria can attach and form biofilms. Because these genotypic clusters group samples of ecologically and genetically similar organisms, they are hypothesized to represent samples from wild ecological populations, and as such they serve as a platform for studies of evolutionary and ecological dynamics of bacteria in the wild.
To explore the evolutionary and ecological dynamics of siderophores, we performed a large-scale phenotypic screen aimed at discerning the variability of the siderophore production among ecologically distinct Vibrionaceae populations. Next, we leveraged genomic analysis of 61 sequenced isolates from these populations to identify the genomic basis of phenotypic variation. Finally, we integrated these results with the habitat partitioning of the Vibrionaceae in the ocean to provide an environmental perspective on how the interaction between ecological and evolutionary phenomena influences iron-acquisition strategies.
Results and Discussion
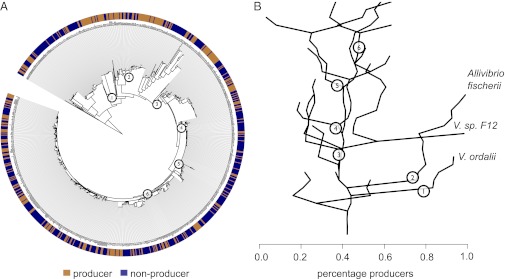
Our high-throughput phenotypic screening (Materials and Methods) indicated that siderophore production is a patchy trait within Vibrio populations: clusters of close relatives showed an average frequency of producers of approximately 40% even in the most shallow branches of the phylogeny where the most recently diverged isolates are found (Fig. 1). Three exceptions with high frequency of producers were observed in clusters of the Vibrio sp. F12 (16, 17), Allivibrio fischerii, and Vibrio ordalii. These latter two are known from the literature to be associated with animal hosts (18, 19), where iron acquisition is likely a critical factor in colonization (20). For all other sampled populations, the production trait is patchily distributed across the tree, implying that it is frequently gained and lost within lineages. This suggests variable selective pressure along each lineage to either produce siderophores or to cheat.
Fig. 1.
Siderophore production is an intermediate frequency trait in natural Vibrio populations. (A) The distribution of producers and nonproducers mapped onto the isolate phylogeny (Materials and Methods). The tree is based on the genetic marker hsp60 and comprises different genotypic clusters previously found to have cohesive ecology and hypothesized to represent samples from natural ecological populations. (B) Representation of the data shown in A in terms of percentage of producers descending from each internal node of the phylogeny. Among the populations with a high incidence of producers are animal host-associated V. ordalii and A. fischerii. The clade descending from node 3 corresponds to close relatives of V. splendidus (referred to as the V. splendidus-like group). Nodes 4, 5, and 6 correspond to the most recent common ancestors of V. crassostreae, V. cyclotrophicus, and V. splendidus. Within this large group, the patchy distribution of producers suggests that siderophore biosynthesis genes are rapidly gained and lost, possibly by recombination.
Using 61 sequenced genomes from the isolate collection (Table S1), we established the genetic basis of siderophore production variability in the natural Vibrio populations. We found a strong match between measured production and the presence of siderophore biosynthesis gene clusters in the genome annotation. Of the 61 isolates tested (25 siderophore positives, 36 negatives) there were only three discrepancies (false negatives) between the functional assay and the genome annotation data, indicating that the patchiness of the trait is primarily the result of gene loss and/or independent acquisitions, as opposed to differential gene regulation or errors in the phenotypic screening assay.
Because siderophore biosynthesis genes are often coded in large gene clusters linked with their specific transporters (21), we expected that nonproducers would lack both biosynthesis genes and the cognate high-affinity receptors. Further inspection of the genomes, however, showed that nonproducers have evolved by selective loss of biosynthesis genes but retention of the specific receptors, consistent with the canonical model of a cheater. For this analysis, we focused on the Vibrio splendidus-like clade, consisting of distinct subpopulations of species V. splendidus, Vibrio crassostreae, Vibrio tasmaniensis, and Vibrio cyclotrophicus, thought to specialize in exploiting different marine particulate nutrient patches (16). In these populations, transitions between production and nonproduction seem to have occurred multiple times because of the extreme patchiness of the production trait. The most common siderophores in this clade, vibrioferrin and aerobactin, are encoded by superoperons in which biosynthesis genes are adjacent to the specific siderophore receptors (22, 23). Contrary to our expectation, whereas biosynthesis genes had been excised or replaced, the adjacent receptors in the nonproducers were kept intact (Figs. 2, S2, and S3). Although putative aerobactin cheaters were distributed on different branches within the V. splendidus-like clade, the genomic region harboring the siderophore receptors was identical in gene composition and order in these strains (Fig. S1). This observation indicates that nonbiosynthetic gene clusters had a common evolutionary origin and that these clusters are alternative operon variants that are maintained in the population.
Fig. 2.
Evolution of “cheating” explains patchiness of siderophore production trait in the V. splendidus-like group. (A) Phylogenetic relationship, siderophore production, and siderophore synthesis and transport genes for sequenced strains in the V. splendidus-like group. The tree is based on 66 single-copy genes present in all of the sequenced isolates. The “sid-prod” column refers to the outcome of the phenotypic screen; “aer-syn” stands for aerobactin biosynthesis genes, “aer-txpt” for aerobactin-specific transport genes, “vf-syn” for vibrioferrin biosynthesis genes, and “vf-txpt” for vibrioferrin-specific transport genes. (B and C) Examples of configurations of siderophore synthesis and transporter gene clusters in producer and nonproducer strains. The figures show that nonproducer phenotype evolved from excision of the biosynthesis genes from the complete synthesis-transport cluster. Black and red stars in A indicate the presence of similar “cheater” gene cluster configurations as those shown in B and C (Figs. S1 and S2).
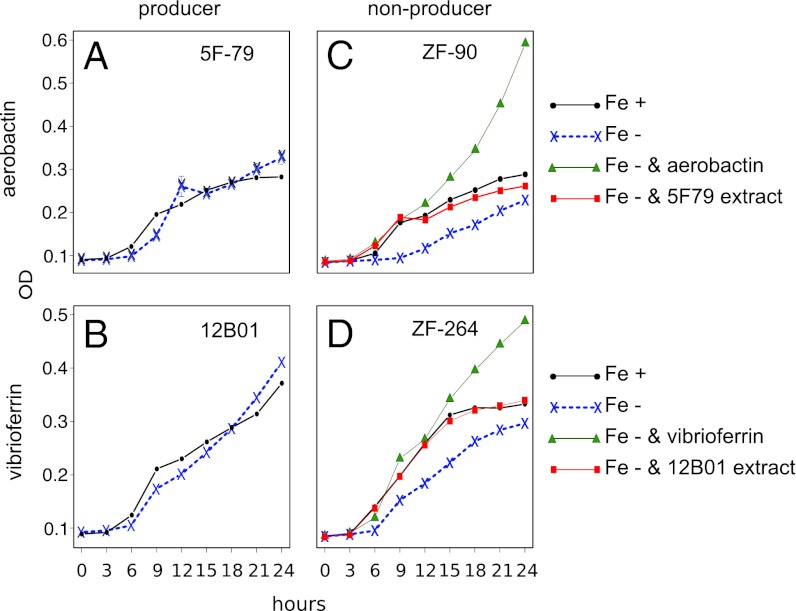
Consistent with their putative cheater role, individuals that have lost the ability to produce siderophores suffered a clear decrease in growth efficiency when grown in iron-poor media (Materials and Methods). By contrast, the growth of producers was similar in both iron-rich and iron-poor media (Fig. 3 A and C). Moreover, when supplementing the iron-poor media with pure siderophores, putative cheaters were able to enhance their growth, showing that they could effectively take up the specific siderophores under those conditions (Fig. 3). Insensitivity to iron-limited conditions was also observed in nonproducers when the media was supplemented with the <3 kDa fraction (small molecules) of the cell-free supernatant from siderophore producers (Materials and Methods), showing that the cross-feeding interaction is possible in nature. Based on a combination of genomic comparison and biological assays, our analysis suggests that within the studied Vibrio populations there is fine-grained differentiation in social roles, whereby some individuals have evolved to exploit exogenous pools of siderophores, whereas others remain self-sufficient.
Fig. 3.
Growth assays confirm bioinformatic predictions of siderophore biosynthesis and utilization capabilities. (A and B) Growth of producer strains 5F-79 and 12B01 in M9-based media (Fe+) and in media supplemented with 100 µM EDDA (Fe−), an iron-specific chelator. The results show that both siderophore producer strains 5F-79 (aerobactin) and 12B01 (vibrioferrin) are insensitive to EDDA addition. By contrast, nonproducers ZF-90 and ZF-264 (C and D), which have specific receptors for aerobactin (aer) and vibrioferrin (vf), respectively, suffered an appreciable effect when grown in Fe− media (100 µM EDDA for ZF-90, 50 uM EDDA for ZF-264). This effect was counteracted by addition of the corresponding siderophore aerobactin (80 µM) or vibrioferrin (70 µM) and by cell-free extract from corresponding producers 5F-79 and 12B01.
Having established the variability in siderophore production and its genetic basis, we asked what habitats and population dynamics favor cheaters or producers in the marine environment. Focusing on the V. splendidus-like group containing small subpopulations with recent and frequent transitions between particle attached and free-living life-styles (Fig. S3), we measured the correlation between the siderophore production trait and the size fraction associated to each isolate. To that end, we used an adaptation of Felsenstein’s independent contrast method to control for the fact that traits measured for each strain (e.g., siderophore production) are not independent observations but related through the phylogeny (Materials and Methods) (24). Doing this we could control for spurious correlations that may appear as a result of the differential associations between clades and traits and not because of a real dependency between the traits. In addition, we accounted for the effect of phylogenetic uncertainty by calculating the distribution of correlation values on a set of 100 bootstrap trees.
The results of the correlation analysis (Fig. 4) show that association with large particle sizes in the ocean is correlated with a higher frequency of cheaters, whereas the free-living stage (<1 µm) is positively correlated with siderophore production. We find that these correlations are significant, with P values <0.01, and not driven by a few transitions on a few clades (Fig. S4). The correlation between siderophore production and the free-living lifestyle is the strongest among the four size fractions. For the other size fractions, the distribution of correlation values shifts progressively toward negative values, with the lowest correlation values observed for the largest size fraction. This result shows that the frequency of producers changes in a predictable manner, based on the size of the particle to which the population is specialized.
Fig. 4.
Cheaters within the V. splendidus-like clade are more successful in larger particles than in smaller ones. (A) Spearman correlations calculated between the phylogenetic contrasts of the “siderophore production” trait and the different sampling size fractions (SOM). The distributions cover values calculated for 100 bootstraps of the hsp60 tree for the 1,013 vibrio isolates. (B) Cartoon representation of the impact of particle size on frequency of siderophore producers. Our results suggests that social interactions are more relevant on large particles, possibly owing to the higher cell densities and long periods of attachment facilitating the accumulation and exploitation of public goods.
These results suggest that the opportunity for cheating is highest on large particles. One hypothesis for the observed trend is that higher local iron concentrations on large particles preclude the need for siderophore production. However, the observation that nonproducers maintain receptors while selectively losing biosynthesis genes (Fig. 2) indicates that social cheating—and not only environmental selection—drives changes in siderophores producer abundance. We hypothesize that the observed correlation is explained by differences in the structure of social interactions between particle-attached vs. nonattached bacteria. Vibrios are copiotrophs, which can rapidly expand their populations under favorable conditions, in the extreme leading to blooms that can dominate a bacterioplankton community (25). Between blooms, some vibrios have a tendency to attach to particles, whereas others remain planktonic (16). Marine particles can harbor dense communities of attached bacteria with high metabolic activity (26), and therefore vibrios attached to particles experience relatively constant high cell densities in their local environment. On the other hand, those that remain free-living or attached to smaller aggregates fluctuate from high cell densities during blooms to low densities during their planktonic phase. This type of bottleneck population dynamics experienced by free-living vibrios has been shown to favor the abundance of cooperators in public good games (27, 28). On the other hand, the constant high cell densities in the local environment of particle-attached bacteria could favor the establishment of cheaters that “scrounge” public goods from producers (29). This proposed model, based on the specific population dynamics associated to different microenvironments, provides an explanation for the variation in frequency of producers observed between particle attached and free-living Vibrio.
Conclusion
Our results suggest that public good interactions—in particular those mediated by siderophores—could play an important role in structuring natural bacterial populations in the ocean. By studying the patterns of social interactions imprinted on bacterial genomes, we have documented the genomic events that led to transitions between siderophore production and cheating. The evidence suggests that these trends are mediated by dynamic differential gain and loss of siderophores gene cluster variants. Phylogenetic analysis shows that although populations retain an intermediate frequency of producers, transitions occur frequently along individual lineages, so that producers and cheaters coexist in a dynamic equilibrium whereby public good output is stable at the population level but not at the individual level. These results contrast with the idea that functional characterizations of genotypes and populations are interchangeable, as is frequently assumed. Our results further provide evidence that the incentive to cheat in public good games can drive genomic diversity, as recently suggested by the Black Queen Hypothesis (30), and that this force is relevant at the scale of closely related but nonclonal genotypes in natural populations.
Finally, the data indicate that the marine microenvironment influences public good interactions in predictable ways, in this case with particle size influencing the frequency of cheaters via its effect on population dynamics. Marine particles are known to harbor dense microbial communities with high metabolic activity and potential for interactions (26, 31, 32), and they are important features of the marine ecosystem (26). Here, we propose that public good interactions impact the evolution and dynamics of the microbial communities that colonize and grow on marine particles and that public good game theory could be applied to understand microbial diversity in these important microenvironments.
Because of the dilute nature of the marine environment, the role of siderophores as a successful iron acquisition strategy in the ocean is often questioned (14). It has been suggested that siderophore production may only be beneficial in high cell density environments where high local siderophore concentrations can counteract the effect of diffusion kinetics (14). On the surface, this idea would seem to contradict our result showing a negative correlation between the frequency of producers and particle size. However, as mentioned above, the Vibrio strains isolated from the free-living fraction most likely experience episodic population dynamics reaching high cell densities during punctual bursts of growth. We hypothesize that the difference in population dynamics between small size fraction and large particle populations could explain the correlations reported in Fig. 4. Therefore, our study suggests that not only diffusion kinetics, but also the dynamics of social interactions need to be considered to explain the distribution of siderophore producers in the marine environment.
These hypotheses open avenues for further research, which should explore the role of public good games among other particle-attached species. In contrast to classic public good game models, the populations studied here are not clonal, and although we have focused our study on one particular type of public good, because of their genome diversity cheaters with respect to one public good could act as cooperators with respect to another. Further research is needed to investigate these types of synergisms. Overall, our study highlights the importance of describing population-based phenomena, for interpreting the mechanisms that generate and sustain the enormous genetic diversity observed in wild populations bacteria.
Materials and Methods
Siderophore Screen of Wild Isolates.
We screened a total of 1,710 Vibrionaceae isolates from seawater collected at the Plum Island Estuary, MA using the well-established Chrome-Azurol S (CAS) assay (33). Biological replicate measurements for both the liquid and solid versions of the assay were performed in high-throughput using a 96-well format. For the solid and liquid versions of the CAS assay, strains were first grown overnight in duplicates in 2216 marine broth (Difco, Becton Dickinson) at room temperature. Overnight cultures were stamped directly onto CAS agar plates (solid assay) and transferred into iron-poor media to induce siderophore production in liquid (SI Materials and Methods). After 48 h cell-free supernatants were mixed in a 1:1 ratio with liquid CAS dye and 2 μL shuttle solution (33). The mixture was incubated in the dark for 15 min, after which absorbance at 630 nm was measured on a Synergy2 filter-based multimode plate reader (Biotek). Siderophore production was considered positive for all absorbances <0.3. The liquid CAS dye and shuttle solution were prepared as previously described (33).
Bioinformatic Analysis of Siderophore Synthesis and Transport Gene Clusters.
Gene finding and annotations were performed using the SEED subsystems (34) and the RAST server (35) on the 61 genomes. Siderophore biosynthesis clusters were identified using a combination of annotation text searches and the software AntiSMASH developed to identify biosynthetic clusters of secondary metabolites (36). Details of the search criteria used for these programs are presented in SI Materials and Methods.
Phylogenetic Correlation Analysis.
We calculated the Spearman rank correlation between the siderophore production and particle size traits associated with each isolate. To correct for the fact that values measured for each strain are not independent of each other (being related through the phylogeny), we used Felsenstein’s independent contrast method (24). Our data are composed of binary values on the leaves of the tree (e.g., 1 production, 0 nonproduction; 1 large-particle associated, 0 otherwise), and we reconstructed states for internal nodes of the tree by calculating the fraction of nonzero values under each node. We then applied Felsenstein’s independent contrast method (24) to obtain a vector of differences between the fractions of producers under sister nodes of the phylogeny. This vector corresponds to evolutionary transitions increasing or decreasing the frequency of producers or strains associated to a specific size fraction. Using the vectors of transitions for the siderophore production and size fraction variables, Spearman rank correlations were calculated (removing nodes where either transition was 0) between the vectors to measure the association between the traits. To reduce the noise in the estimation of frequencies, we did not consider evolutionary transitions between leaves or between leaves and internal nodes. This process was repeated for each of the 100 maximum likelihood bootstrap trees calculated with the FastTree program (37) using the evolutionarily conserved housekeeping gene hsp60. The distributions in Fig. 4 show the results of this analysis for each size fraction. Distributions were smoothed using kernel density estimation as implemented in the function “density” provided in the statistical computing language R.
Growth Enhancement Experiments.
Isolates were grown overnight at room temperature in 2216 marine broth and pelleted (2 min at 9,300 × g) to wash cells using a modified M9 salts solution. Cells were then inoculated (1:1,000) into modified M9 salts growth media (SI Materials and Methods) with (iron-poor) or without (iron-replete) the iron-specific chelator EDDA (ethylenediamine-N,N′-diacetic acid) (Sigma-Aldrich). During incubation at room temperature absorbance at 600 nm was measured every 3 h for 24 h. Purified ferric-aerobactin (EMC Microcollections) and vibrioferrin (generously provided by Carl Carrano, San Diego State University, San Diego, CA) were added at specified concentrations. Extracts from the aerobactin producer 5F-79 and the vibrioferrin producer 12B01 grown under iron-poor conditions were obtained by filtering cell-free supernatant through a 3-kDa membrane using an Amicon Ultra centrifugal unit (Millipore).
Supplementary Material
Acknowledgments
We thank Carl J. Carrano for generously providing purified vibrioferrin. Funding for this work was provided by National Science Foundation (NSF) Grant DEB 0821391; the Gordon and Betty Moore Foundation; the Cooperative Agreement between the Masdar Institute of Science and Technology (Masdar Institute), Abu Dhabi, United Arab Emirates and the Massachusetts Institute of Technology (MIT), Cambridge, MA (Reference 02/MI/MI/CP/11/07633/GEN/G/00); Center for Microbial Oceanography: Research and Education NSF Science and Technology Center Award EF0424599; and the Broad Institute’s Scientific Planning and Allocation of Resources Committee program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Whole Genome Shotgun projects have been deposited in the DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank databases (accession nos. AJWN00000000, AHTI00000000, AICZ00000000, AIDA00000000-AIDS00000000, AJYD00000000-AJYZ00000000, and AJZA00000000-AJZQ00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213344109/-/DCSupplemental.
References
- 1.Shapiro BJ, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336(6077):48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36(7):760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 3.Cordero OX, Hogeweg P. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci USA. 2009;106(51):21748–21753. doi: 10.1073/pnas.0907584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher Y, et al. Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. mBio. 2011;2(2):e00335-10. doi: 10.1128/mBio.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ., Jr Genome-genome interactions: Bacterial communities in initial dental plaque. Trends Microbiol. 2005;13(1):11–15. doi: 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence D, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10(5):e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 9.West SA, Buckling A. Cooperation, virulence and siderophore production in bacterial parasites. Proc Biol Sci. 2003;270(1510):37–44. doi: 10.1098/rspb.2002.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: Identification and characterization of genes involved in production of a siderophore. Infect Immun. 2004;72(1):29–37. doi: 10.1128/IAI.72.1.29-37.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mawji E, et al. Hydroxamate siderophores: Occurrence and importance in the Atlantic Ocean. Environ Sci Technol. 2008;42(23):8675–8680. doi: 10.1021/es801884r. [DOI] [PubMed] [Google Scholar]
- 12.Amin SA, et al. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci USA. 2009;106(40):17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandy M, Butler A. Microbial iron acquisition: Marine and terrestrial siderophores. Chem Rev. 2009;109(10):4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Völker C, Wolf-Gladrow DA. Physical limits on iron uptake mediated by siderophores or surface reductases. Mar Chem. 1999;65:227–244. [Google Scholar]
- 15.Damore JA, Gore J. Understanding microbial cooperation. J Theor Biol. 2012;299:31–41. doi: 10.1016/j.jtbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt DE, et al. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320(5879):1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 17.Preheim SP, et al. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol. 2011;13(1):265–275. doi: 10.1111/j.1462-2920.2010.02328.x. [DOI] [PubMed] [Google Scholar]
- 18.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11(2):483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin B. Taxonomy of bacterial fish pathogens. Vet Res. 2011;42(1):20. doi: 10.1186/1297-9716-42-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Ann Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 21.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki K, et al. Identification and transcriptional organization of aerobactin transport and biosynthesis cluster genes of Vibrio hollisae. Res Microbiol. 2006;157(8):730–740. doi: 10.1016/j.resmic.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Tanabe T, et al. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J Bacteriol. 2003;185(23):6938–6949. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 25.Gilbert JA, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6(2):298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon M, Grossart HP, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- 27.Chuang JS, Rivoire O, Leibler S. Simpson’s paradox in a synthetic microbial system. Science. 2009;323(5911):272–275. doi: 10.1126/science.1166739. [DOI] [PubMed] [Google Scholar]
- 28.Cremer J, Melbinger A, Frey E. Growth dynamics and the evolution of cooperation in microbial populations. Sci Rep. 2012;2:281. doi: 10.1038/srep00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 30.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. mBio. 2012;3(2):e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long RA, et al. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl Environ Microbiol. 2005;71(12):8531–8536. doi: 10.1128/AEM.71.12.8531-8536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grossart HP, Kiørboe T, Tang K, Ploug H. Bacterial colonization of particles: Growth and interactions. Appl Environ Microbiol. 2003;69(6):3500–3509. doi: 10.1128/AEM.69.6.3500-3509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 34.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medema MH, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue):W339-46. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.