Abstract
Environmental stresses adversely affect plant growth and development. A common theme within these adverse conditions is the perturbation of reactive oxygen species (ROS) homeostasis. Here, we demonstrate that the ROS-inducible Arabidopsis thaliana WRKY15 transcription factor (AtWRKY15) modulates plant growth and salt/osmotic stress responses. By transcriptome profiling, a divergent stress response was identified in transgenic WRKY15-overexpressing plants that linked a stimulated endoplasmic reticulum-to-nucleus communication to a disrupted mitochondrial stress response under salt-stress conditions. We show that mitochondrial calcium-flux sensing might be important for regulating an active mitochondrial retrograde signaling and launching an appropriate defense response to confer salt-stress tolerance.
Keywords: abiotic stress signaling, cell expansion, microarray, hydrogen peroxide
Abiotic stresses limit plant performance and productivity. Although signaling mechanisms and metabolic responses may differ, most types of abiotic stresses affect the cellular redox homeostasis and result in an enhanced accumulation of reactive oxygen species (ROS) (1). ROS accumulation had long been considered a toxic event. Stress-induced ROS accumulation is, however, not necessarily a symptom of cellular dysfunction but also is a signal to adjust cellular machineries to changing environmental and developmental conditions (2–6). Plants constantly adjust ROS levels by a diversified network of production and scavenging mechanisms (7, 8). Gene-expression studies of mutants lacking ROS-scavenging enzymes have demonstrated that ROS signals and abiotic stresses share substantial similarities in gene regulation (9, 10). In support, several regulatory proteins of ROS-mediated signaling are also central regulators of abiotic stress responses involved in temperature, salinity, and osmotic stresses (2, 3, 11).
In addition to their effects on the cellular redox state, abiotic stresses can also perturb the functioning of organelles, such as mitochondria and chloroplasts that, in turn, activate feedback mechanisms by which the nuclear gene expression is modified to sustain and/or restore the organellar functions. These organelle-to-nucleus signaling events are often termed retrograde regulation (12–14). Whereas significant progress has been made toward the understanding of chloroplast retrograde signaling (15, 16), less is known about mitochondria-to-nucleus signaling in plants (12, 14). Mitochondrial retrograde regulation (MRR) in plants has been studied primarily in mitochondrial mutants, such as the cytoplasmic male sterility II in tobacco and prohibitin3 in Arabidopsis, and as a response to disruption of the mitochondrial function by chemical inhibitors, demonstrating a role for mitochondria in sensing stress and directing the cellular response into recovery or cell death (14, 17–20). Although evidence is emerging for the convergence of mitochondrial retrograde signals and abiotic stress pathways (20–23), the molecular crosstalk mechanisms and upstream regulators of the MRR signaling cascade in plants remain largely unknown. Recently, a role was proposed for ABSCISIC ACID INSENSITIVE4 (ABI4), an APETALA2-type transcription factor, in mediating mitochondrial signals to regulate the expression of ALTERNATIVE OXIDASE1a (AOX1a) (24). Because ABI4 is also a regulator of plastid retrograde signaling to repress photosynthetic gene expression (13, 16) and of ABSCISIC ACID (ABA) signaling (25), it might act as a molecular interface between retrograde and anterograde regulatory signals.
Here, we show that the hydrogen peroxide (H2O2)-responsive transcription factor WRKY15 functions as a negative regulator of salt- and osmotic-stress tolerance in Arabidopsis thaliana. Molecular phenotyping of WRKY15-overexpressing plants under salt stress revealed a pivotal role for MRR in mediating salt-stress tolerance. Furthermore, our results indicate that mitochondrial calcium-flux sensing is important for the activation of the mitochondrial stress response.
Results and Discussion
WRKY15 Is Induced by Oxidative and Salt Stresses.
The group IId WRKY15 transcription factor is an early H2O2-responsive gene (At2g23320) (9, 26, 27). Gene-expression analysis using Genevestigator revealed that, besides H2O2 treatments, low CO2 availability, and pathogen infections, WRKY15 expression was also induced by salt stress (28). Salt- and oxidative-stress responsiveness of the WRKY15 transcript was confirmed by quantitative RT-PCR analysis (Fig. S1 A and B). The WRKY15 protein is predominantly located in the nucleus (29). To determine spatial and developmental expression patterns of WRKY15 in Arabidopsis, the WRKY15 promoter:β-glucuronidase (GUS) reporter gene fusion constructs were examined. In 3-d-old seedlings, GUS staining was primarily strong in the hypocotyl-to-root transition zone and in the root tip (Fig. S1C). In 9-d-old and mature seedlings, WRKY15 was expressed in the shoot apical meristem, trichomes, and trichome socket cells of young leaves (Fig. S1 D–G). In mature seedlings, GUS staining was also observed in the outer epidermal cell layer of leaf petioles (Fig. S1E). In roots, the WRKY15 promoter activity was detected mainly in the vascular cylinder, at lateral root initials, and in the root tip (Fig. S1 H–M). Thus, the WRKY15 gene is expressed mainly in young, growing, and vascular tissues.
WRKY15 Overexpression Promotes Leaf Growth and Plant-Biomass Production Through a Stimulated Cell-Expansion Rate.
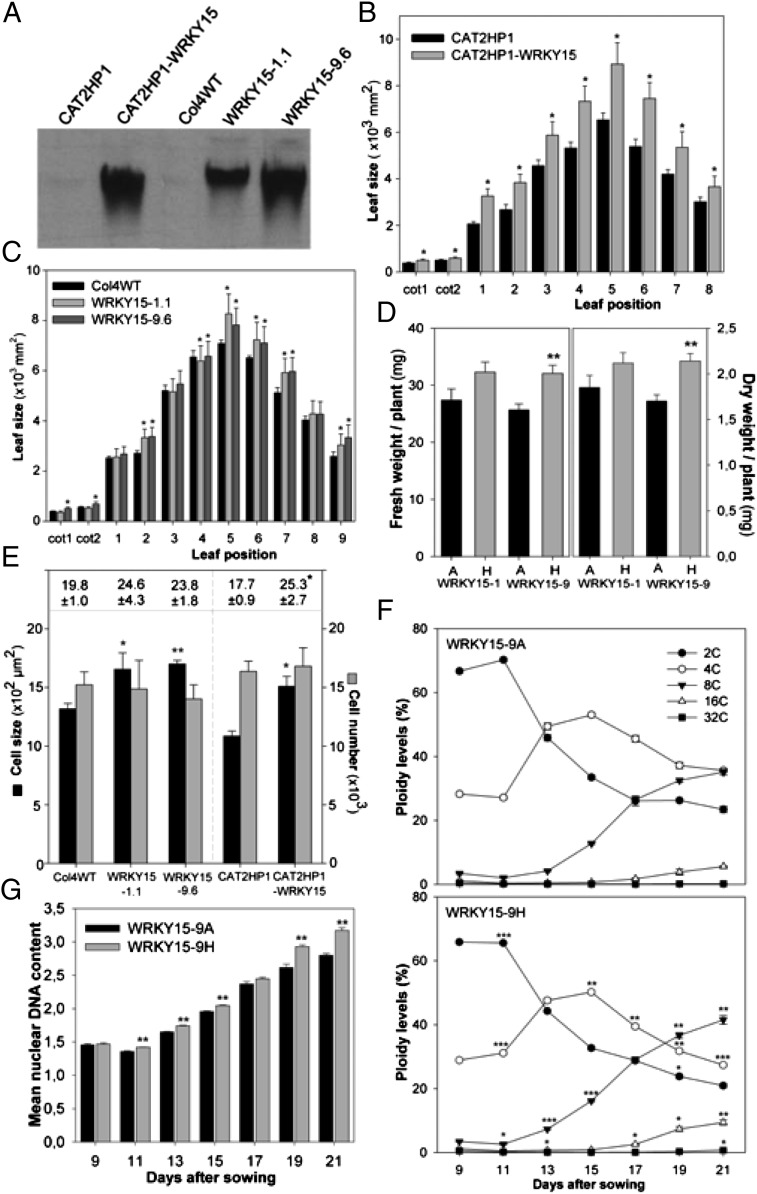
We generated transgenic plants that constitutively and ectopically expressed WRKY15 under control of the cauliflower mosaic virus 35S promoter in both a Columbia-4 wild-type (Col4WT) and a catalase-deficient background (CAT2HP1) in which WRKY15 was originally identified as an H2O2-responsive gene (9, 26, 30). Three independent WRKY15 overexpression (designated WRKY15OE) lines with high transgene expression were selected for further analysis (Fig. 1A). Although similar in germination rates and early development as control plants, fully grown WRKY15OE plants exhibited an increased leaf area (Fig. 1 B and C), resulting in a 15–25% increased plant biomass (Fig. 1D).
Fig. 1.
Enhanced leaf growth and biomass production in WRKY15OE plants. (A) WRKY15 transcript abundance in CAT2HP1, Col4WT, and WRKY15OE plants by RNA gel-blot analysis. (B and C) Leaf area of all individual leaves of 3-wk-old control and WRKY15OE lines in CAT2HP1 (B) and Col4WT background (C). Error bars show SEM (n = 12). Cot, cotyledon. (D) Total fresh and dry weight of 3-wk-old azygous (A) and homozygous (H) WRKY15OE plants (Col4WT background). Error bars show SEM (n = 10). (E) Cell size and number in first leaves of 3-wk-old seedlings. Leaf area is shown at the top of the frame. Data represent averages ± SEM (n = 10). (F) DNA ploidy levels during development of the first leaf pair in azygous (WRKY15-9A) and transgenic (WRKY15-9H) plants. Leaves were harvested at indicated time points. Error bars show SEM (n = 6). (G) Changes in endoreduplication index (2C × 1 + 4C × 2 + 8C × 4 + 16C × 8) calculated from data in F. Error bars show SEM (n = 6). *P < 0.05; **P < 0.005; ***P < 0.0001 (Student t test).
Because the final leaf size is determined by cell-division and cell-expansion rates, we assessed the cell numbers and cell size of abaxial epidermis cells in 21-d-old plants. In WRKY15OE plants, cell numbers did not change, but the average cell size increased (Fig. 1E), indicative of increased cell expansion. A common mechanism by which plants control cell size is the repeated replication of their DNA, resulting in cellular polyploidy, a process termed endo-reduplication (31, 32). DNA ploidy-level analysis revealed that the number of cells with an 8C and 16C content was significantly higher in WRKY15OE plants than that of control plants (Fig. 1F). Moreover, from day 11 on, WRKY15OE plants displayed an increased endoreduplication index, which correlates with the mean nuclear DNA content per cell (33), indicating that endoreduplication was stimulated (Fig. 1G). Taken together, in WRKY15OE and WT plants, the transition from the mitotic cycle to the endocycle is equivalent, as evidenced by a similar cell number, but in WRKY15OE plants, endoreduplication is intensified, correlating with the increased cell size.
Because no true loss-of-function T-DNA insertion mutants are available, transgenic plants containing artificial microRNA (amiR) constructs targeting WRKY15 were generated (34). These WRKY15-amiR plants showed a strong reduction in WRKY15 levels (below 20% of WT levels), were smaller than WT plants, and displayed a decreased average leaf cell area (Fig. S2 A and B). High WRKY15 transcript abundance in young and growing tissues (Fig. S1), together with the altered cell expansion upon WRKY15 perturbation (see Fig. 1 E–G for WRKY15OE plants and Fig. S2 A and B for WRKY15-amiR plants), support its involvement in plant growth and possibly endoreduplication, either directly or indirectly as a result of enhanced growth processes of which the relative contribution is not known.
Elevated WRKY15 Expression Increases Sensitivity to Osmotic and Oxidative Stresses.
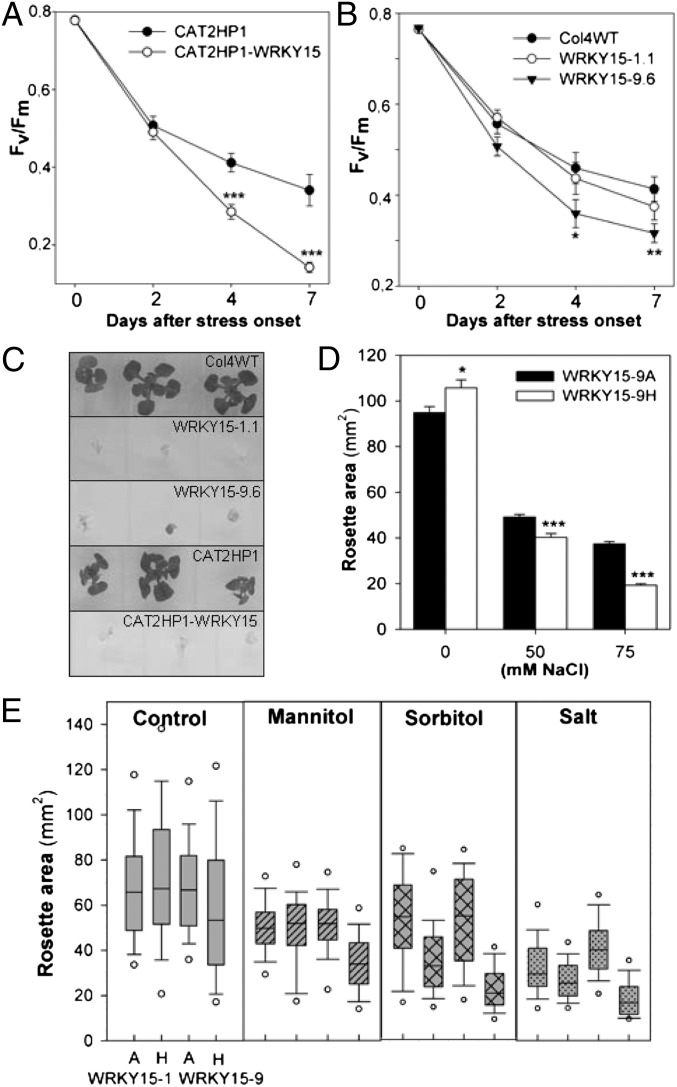
Transgenic Arabidopsis plants with perturbed WRKY15 expression were assessed for altered phenotypes when exposed to abiotic stress conditions. For oxidative stress, we used a bioassay in which photorespiration is induced by restricting gas exchange within Petri plates. Chlorophyll fluorescence was measured and the maximum quantum efficiency of photosystem (PS)II (Fv/Fm) was determined, which is an effective measure of plant stress (35). In CAT2HP1-WRKY15 plants, the decrease in Fv/Fm was stronger than in control CAT2HP1 plants, hinting at an increased sensitivity to oxidative stress (Fig. 2A). In the Col4WT background, the catalase activity inhibitor 3-aminotriazole (3-AT) was used to impose H2O2 stress and mimic catalase deficiency. Again, WRKY15OE plants were more susceptible to oxidative stress (Fig. 2B). WRKY15-amiR plants did not perform differently from WT plants when subjected to oxidative stress (Fig. S2C).
Fig. 2.
Stress sensitivity of WRKY15OE plants. (A and B) Maximum quantum efficiency of PSII (Fv/Fm) in leaves of CAT2HP1 and CAT2HP1-WRKY15 plants after exposure to photorespiration-promoting conditions (A) and Col4WT and WRKY15OE plants after exposure to photorespiration-promoting conditions in the presence of the catalase inhibitor 3-AT (3 µM) (B). Error bars show SEM (n = 18 plants). (C) Four-wk-old control and WRKY15OE plants germinated and grown on 100 mM NaCl. (D) Rosette area of 3-wk-old azygous (A) and transgenic (H) WRKY15OE plants grown on increasing salt concentrations. Error bars show SEM (n = 5 plates with 15–30 plants). (E) Rosette area of 3-wk-old azygous (A) and transgenic (H) WRKY15OE plants grown under control conditions (control), 25 mM mannitol, 100 mM sorbitol, and 75 mM NaCl (salt) stress. For each experiment, 80 plants per line and condition were used. The line represents the median; the lower and upper boundaries, the 25th and 75th percentiles; and the whiskers, the 10th and 90th percentiles. The outlying points are shown to the 5th/95th percentiles. *P < 0.05; **P < 0.01; ***P < 0.0005 (Student t test).
To examine salt-stress responses, control, WRKY15OE, and WRKY15-amiR plants were germinated and grown on medium containing increased salt concentrations. On 100 mM NaCl, control plants could still grow and remained green, whereas the growth of WRKY15OE plants was inhibited and chlorosis was initiated (Fig. 2C). The hypersensitivity of WRKY15OE plants toward salt stress was similar in both Col4WT and CAT2HP1 backgrounds (Fig. 2C) and was salt dose–dependent (Fig. 2D). The rosette area of WRKY15-amiR plants grown on 100 mM NaCl was 10–15% reduced compared with WT plants (Fig. S2 D and E), but this growth reduction was of similar magnitude as observed in the absence of stress, indicating that there was no additive effect of the salt treatment.
To assess whether overexpression of WRKY15 also altered the responsiveness to osmotic stresses, plants were germinated and grown in the presence of 25 mM mannitol, 100 mM d-sorbitol, or 75 mM NaCl. On salt or sorbitol, again the rosette area of WRKY15OE plants was significantly reduced, whereas mannitol stress only affected growth in the strongest overexpression line (WRKY15-9H) (Fig. 2E).
To determine the effect of salt-stress sensitivity at the cellular level, we examined the first leaf pairs of 2-wk-old WRKY15OE plants grown under control and salt-stress conditions (50 mM NaCl) by transmission-electron microscopy. This mild salt stress already initiated cellular degeneration in WKRY15OE leaves, as evidenced by the presence of deteriorated cells and large intercellular spaces (Fig. S3 A–D). This cellular phenotype was not observed under nonstressed conditions (Fig. S3 E–H).
Transcript Profiling Reveals Induced Unfolded Protein Response and Impaired Mitochondrial Stress Response in WRKY15OE Plants.
RNA of three independent replicates of WT and WRKY15OE seedlings grown in the absence and presence of 50 mM NaCl was hybridized to Affymetrix GeneChip Arabidopsis Tiling 1.0R arrays. A two-factor ANOVA revealed 598 up-regulated and 750 down-regulated transcripts in WRKY15OE plants, independently from salt-stress treatment (Table S1). Among the up-regulated transcripts, genes involved in the endoplasmic reticulum (ER) stress response were significantly enriched (36–38) (Fig. S4A and Tables S1 and S2). This response, also known as the unfolded protein response (UPR), is an evolutionarily conserved transcriptional response that is triggered by the accumulation of unfolded or misfolded proteins in the ER lumen and is essential to maintain ER homeostasis (37). The UPR triggers (i) enhancement of protein- folding activities by induction of ER-resident molecular chaperones, foldases, and high-capacity Ca2+-binding proteins; (ii) increase in protein degradation capacity to remove improperly folded proteins; and (iii) in mammals, also attenuation of translation to limit the entry of nascent polypeptides when conditions are unsuitable for proper folding (36, 39, 40). Besides the induction of core UPR genes (37) (Fig. S4B), WRKY15 overexpression also significantly repressed transcript levels of proteins involved in protein synthesis (Fig. S4A and Tables S1 and S2), indicative of a complete ER stress response in WRKY15OE plants. Growth on more severe salt-stress conditions intensified the induction of core UPR genes in WRKY15OE plants (Fig. S3 C and D), which correlated with their increased salt-stress–sensitivity phenotype (Fig. 2D). Integration of salt-adaptation responses and ER stress signaling was observed before in mutants defective in protein N-glycosylation (41, 42). Dissimilar to WRKY15OE plants, salt-/osmotic-stress sensitivity in these mutants was associated with root-tip swelling and enhanced lateral-root development that resulted from a constitutive UPR activation caused by the reduced protein N-glycosylation levels (41, 42). Affinodetection of high-mannose-type N-glucan–containing glycoproteins in WRKY15OE plants identified an altered protein N-glycosylation pattern only in salt-stressed WRKY15OE plants (Fig. S4E), whereas they constitutively induced the UPR already without stress. These observations, together with the distinct shoot-sensitivity phenotype of WRKY15OE plants compared with N-glycosylation mutants, suggest that divergent processes might operate in salt-stress adaptation. To assess the impact of drug-induced ER stress, plants were grown on tunicamycin, an ER-stress–inducing drug that inhibits protein N-glycosylation and disulfide bonding. In WRKY15OE plants, the rosette area was 20–40% reduced compared with WT plants (Fig. S4F), indicating an increased sensitivity toward drug-imposed ER stress.
For 497 transcripts, the expression was significantly affected by the combination of salt stress and WRKY15 overexpression, among which, 363 transcripts were induced by salt stress in WT plants but impaired in WRKY15OE plants (Table S3). More than half of these 363 genes had a predicted mitochondrial localization (43) and included genes of the so-called mitochondrial dysfunction regulon (MDR) (18/29; P = 5.81 × e−28; Fisher’s exact test) that were differentially regulated in response to mitochondrial dysfunction and environmental stress (20, 22). MDR genes are highly coregulated with AOX1a and might be part of the mitochondria-to-nucleus retrograde signaling pathway or MRR (14, 20) (Fig. S5A).
Integration of Calcium-Mediated Signaling, MDR Expression, and Stress Tolerance.
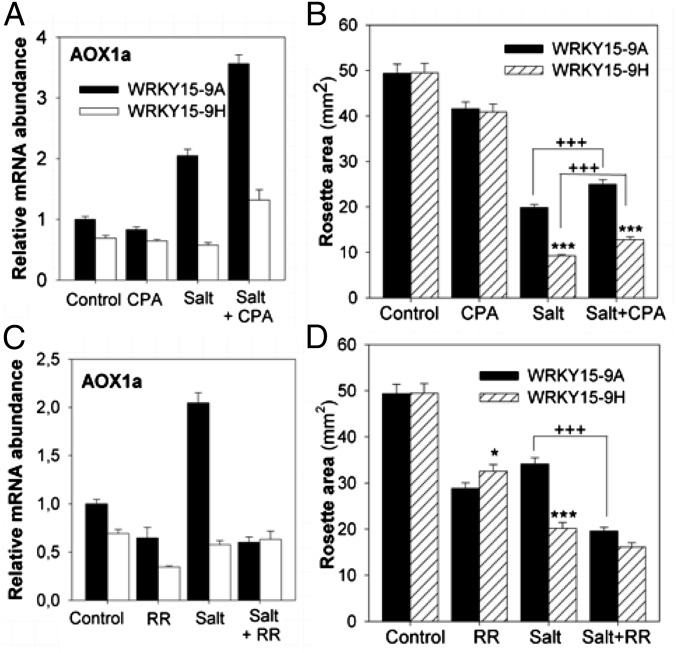
Because the salt-stress–sensitive phenotype of WRKY15OE plants coincided with UPR activation and an impaired mitochondrial stress response, the question arises of how these two distinct organelle-to-nucleus signaling pathways are interconnected with the salt-stress sensitivity. Besides N-glycosylation, disulfide bonding, and protein assembly, the ER also contributes to the intracellular Ca2+ homeostasis. Both the regulated release and leakage of Ca2+ from the ER Ca2+ stores are controlled by the sarco-/endoplasmic reticulum Ca2+ ATPase (SERCA) that maintains the internal Ca2+ storage (44). In mammalian cells, a tight communication exists between the ER and mitochondria with Ca2+ as the potential mediator signal (45, 46). By analogy, a significant enrichment for Ca2+-induced genes was found among the WRKY15-regulated genes (47) (Fig. S5B and Tables S1 and S3). To assess the involvement of Ca2+-mediated interorganellar signaling in WRKY15OE plants, we evaluated the effect of cyclopiazonic acid (CPA) (which is a specific SERCA inhibitor and, thereby, increases the cytosolic Ca2+ concentration) (48) on MDR gene expression. During salt stress, CPA clearly intensified the MDR gene expression in WT plants (see Fig. 3A for AOX1a and Fig. S5C for three additional MDR genes), indicating that increased cytosolic Ca2+ concentrations promoted MDR. Furthermore, CPA alleviated significantly the salt-stress–induced growth reduction (Fig. 3B), indicative for a potential link between Ca2+-mediated MDR activation and salt-stress tolerance. In contrast, in WRKY15OE plants, CPA only mildly enhanced MDR gene expression and growth performance under salt stress, suggesting that the potential increase in cytosolic Ca2+ provoked by CPA addition was not sufficient to overcome the WRKY15-dependent inhibitory effect on MDR gene expression (Fig. 3 A and B and Fig. S5C). To assess whether mitochondrial Ca2+-flux sensing was deregulated in WRKY15OE plants, we determined the effect of ruthenium red (RR) on the salt-stress–induced MDR gene expression. RR abolishes the uptake of cytosolic Ca2+ by mitochondria through inhibition of the Ca2+ uniporter channel located in the inner mitochondrial membrane (49). During salt stress, RR reduced MDR gene expression in WT plants to levels similar to those in salt-treated WRKY15OE plants (see Fig. 3C for AOX1a and Fig. S5D for three additional MDR genes) and mimicked the stress-sensitivity phenotype of WRKY15OE plants (Fig. 3D). These results indicate that mitochondrial Ca2+-flux sensing might be necessary to launch a salt-stress response, involving MDR gene expression. Whether this cascade of events is solely necessary for a proficient defense response enabling plants to withstand moderate salt-stress conditions remains to be elucidated.
Fig. 3.
Interconnection of salt-stress tolerance and MDR induction involving Ca2+ fluxes in the ER–mitochondria axis. (A) Transcript abundance of AOX1a in azygous control (WRKY15-9A) and transgenic WRKY15OE (WRKY15-9H) plants grown without (control) or with 5 μM CPA, 50 mM NaCl (salt), or both (salt+CPA). (B) Rosette area of control and WRKY15OE plants grown with CPA and/or salt (75 mM NaCl). Data represent average ± SEM (n = 3 plates with 15–30 plants). (C) Transcript abundance of AOX1a in control and WRKY15OE plants grown without (control) or with 10 μM RR, 50 mM NaCl (salt), or both (salt+RR). (D) Rosette area of control and WRKY15OE plants grown with RR and/or salt (50 mM NaCl). Error bars in B and D show SEM (n = 3 plates containing 15–30 plants). *P < 0.05 and ***P < 0.0001 (Student t test, WRKY15OE vs. WT); +++P < 0.0001 (Student t test, salt+CPA/RR vs. salt).
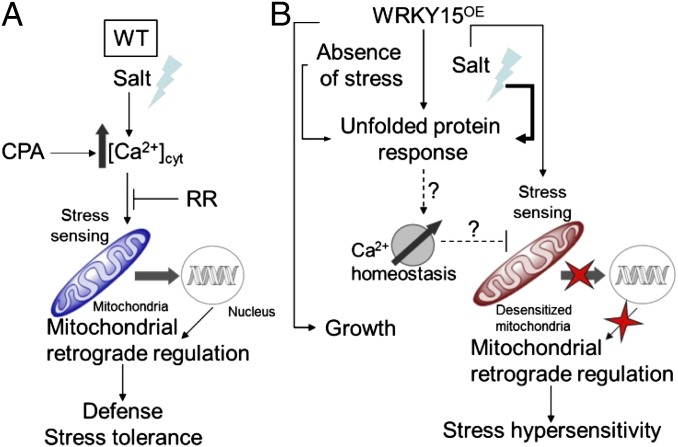
Because the ER acts as a dynamic Ca2+ reservoir and UPR proteins are important in regulating the activity of the SERCA pump (44, 50), constitutive UPR activation in WRKY15OE plants might possibly disturb the cellular Ca2+ homeostasis. During intracellular Ca2+ signaling, mitochondria rapidly take up Ca2+ from the cytosol via the Ca2+-selective uniporter channel located at the inner membrane (49). Mitochondrial Ca2+ uptake has a biphasic dependence on cytosolic Ca2+ because it is facilitated by Ca2+/calmodulin (CaM) (Ca2+-activated CaM) and inactivated by sustained cytosolic Ca2+ levels (51). Therefore, uncontrolled endoplasmic Ca2+ release in WRKY15OE plants might facilitate an unceasing mitochondrial Ca2+ uptake, potentially causing mitochondrial desensitization. Hence, during salt stress, signal-induced Ca2+ fluxes might not be sensed and no MDR gene expression is activated in WRKY15OE plants, possibly causing salt-stress sensitivity (Fig. 4).
Fig. 4.
Hypothetical model for failure to induce MRR causing stress hypersensitivity in WRKY15OE plants. (A) Under salt stress, increase in cytosolic Ca2+ concentrations and possible sensing by the mitochondria that subsequently activate a retrograde signaling cascade to launch a defense response, rendering the plants tolerant to salt stress. Application of CPA, which increases cytosolic Ca2+ levels, promotes this mitochondrial response, whereas addition of RR, which blocks mitochondrial Ca2+ uptake, impairs mitochondrial retrograde signaling and stress tolerance. (B) In the absence of stress, WRKY15 overexpression promotes growth, but also the transcriptional activation of an UPR that is intensified under salt stress. Constitutive induction of the UPR might possibly disturb the cellular Ca2+ homeostasis, presumably leading to mitochondrial desensitization. Upon salt stress, signal-induced Ca2+ fluxes might not be sensed and no retrograde signaling cascade might be activated to trigger the appropriate defense response causing the salt-stress hypersensitivity. Whether this cascade of events is solely responsible for a proficient defense response enabling plants to withstand moderate salt-stress conditions remains to be elucidated.
Is WRKY15 a Transcriptional Regulator of MRR?
The failure to activate MDR gene expression during salt stress in WRKY15OE plants suggests that WRKY15 might function as a repressor of MRR. Substantial evidence indicates that many genes are repressed by WRKY transcription factors bound to their promoters (52), and particular insights into repressor functions of WRKY proteins were obtained with PcWRKY1, OsWRKY71, and HvWRKY1 and HvWRKY2 (53, 54). In addition, chromatin immunoprecipitation studies revealed that W-box sequences in the promoters of pathogen-defense genes are constitutively occupied by WRKY transcription factors, even in the absence of pathogen infection or elicitor treatment (55). Upon a specific stimulus, allosteric interactions might cause the release of WRKY factors from their cognate W-box elements and its possible replacement by other WRKY proteins. Therefore, WRKY proteins are thought to act in a network of mutually competing participants with temporal displacement by other WRKY family members in a stimulus-dependent manner (53, 55). Interestingly, active repression of basal expression has already been reported for AOX1a, which is widely used as a model to study MRR (14, 19, 56). In a deletion study of the AOX1a promoter, a strong repressor element had been identified that relieves the repression of AOX1a upon stress application (57). Furthermore, three W-box motifs were found within the AOX1a promoter region, of which one is located within the 93-bp MRR region that is important for full AOX1a induction upon treatment with antimycin A (AA) or monofluoroacetate (MFA) that chemically perturb mitochondrial function (18). A mutant lacking this W-box motif (mut1) had a strongly reduced response to AA and MFA, suggesting a potential role for WRKY proteins in the regulation of AOX1a gene expression (18). Besides in AOX1a, W-box motifs were significantly overrepresented in the promoters of WRKY15-repressed genes (Tables S1 and S3). Therefore, it is possible that, in the absence of stress, MDR gene expression is repressed by WRKY15, which is either (i) inactive and constitutively occupies the W boxes or (ii) actively represses the basal gene expression. Upon salt stress, the WRKY15 activity might be modified, thereby derepressing or activating MDR gene expression.
A possible mechanism by which WRKY15 might regulate gene expression is through an interaction with CaM. Ca2+/CaM-mediated transcriptional regulation has been reported for several transcription factors, modulating both their DNA-binding ability and transcriptional activity (58). WRKY15 contains a conserved Ca2+-dependent CaM-binding domain (CaMBD) and has CaM-binding ability (59). To evaluate the involvement of Ca2+/CaM-mediated regulation, we generated transgenic plants that overexpressed a mutant form of WRKY15 with amino-acid substitutions at two of the six conserved hydrophobic CaMBD residues (WRKY15-F79RL86ROE). Similar amino-acid substitutions in the CaMBD of WRKY7, also a member of the WRKYIId subfamily, completely abolished CaM binding (59). WRKY15-F79RL86ROE plants with similar transgene expression levels as WRKY15OE plants did not respond differently from WRKY15OE plants (Fig. S5 E–G). This observation might indicate that the transcriptional control is either (i) independent from Ca2+/CaM-mediated signals and regulated by a different mechanism [such as Ca2+-dependent (de)phosphorylation], or (ii) dependent on the Ca2+/CaM threshold because the effect of WRKY15 overexpression might be much more profound than any potential effect of WRKY15 on MDR gene expression during normal salt-stress signaling, or (iii) mediated by an upstream (CaM-dependent) regulator.
Materials and Methods
Plant Material and Growth Conditions.
Transgenic WRKY15 plants of A. thaliana (L.) Heynh. were obtained as described in SI Materials and Methods. Plants were grown in vitro on Murashige and Skoog (MS)-containing agar medium at 21 °C and 65–80 μmol⋅m−2⋅s−1 in a 16-h/8-h light/dark regime.
Stress Treatments.
For the photorespiration-promoting conditions, plants were grown on MS agar medium for 2.5 wk. Then, the plates were transferred to a continuous light regime after replacing the surgical tape (Micropore; 3M) that sealed the plates by two layers of Parafilm M (Alcan) to restrict gas exchange. Treatments were done in the presence or absence of the catalase inhibitor 3-AT (3 µM). The maximum efficiency of the PSII photochemistry (Fv/Fm) was determined using a PAM-2000 chlorophyll fluorometer and ImagingWin software application (Walz). For salt stress, plants were germinated and grown on MS agar medium containing 50, 75, or 100 mM NaCl. For mannitol and sorbitol stress, the MS agar medium was supplemented with 25 mM mannitol or 100 mM d-sorbitol, respectively.
Site-Directed Mutagenesis of the WRKY15 CaM-Binding Domain.
The two hydrophobic amino acids, F79 and L86, important in CaM binding of WRKY15 (59) were substituted with Arg (denoted as F79R and L86R) with the GeneTailor Site-Directed Mutagenesis System (Invitrogen). For primer sequences, see Table S4. Generation of transgenic plants overexpressing the WRKY15-F79RL86R ORF was as described for overexpression plants.
Microarray Analysis.
Triplicate batches of shoot material of 20 Col4WT and WRKY15-9.6 plants (growth stage 1.05) (60) germinated and grown on half-strength MS agar medium supplemented with 0 or 50 mM NaCl were harvested for total RNA isolation. Details on RNA preparation, microarray hybridization, data processing, and statistical analysis are provided in SI Materials and Methods. The microarray data are available at the GEO database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20494).
Quantitative RT-PCR Analysis.
RNA isolation, cDNA synthesis, and quantitative RT-PCR analyses were performed as described (61) using SYBR Green (Invitrogen) and gene-specific primers (Table S4). Actin-related protein 7 (deregulated in only 7 of the 1,685 conditions in Genevestigator) (28) was used for the normalization of relative transcript levels.
Microscopy and Flow-Cytometric Analyses.
Size and number of abaxial pavement cells in leaves and the nuclear DNA content distribution were determined as described (26).
Supplementary Material
Acknowledgments
We thank Debbie Rombaut for technical assistance and Dr. Stefanie De Bodt for help with Fisher’s exact tests. This work was supported by Ghent University (Geconcerteerde Onderzoeksacties Grant 12051403 and Multidisciplinary Research Partnership “Ghent BioEconomy” Project 01MRB510W), the Agency for Innovation by Science and Technology (Grant 040134/Bayer), the Research Foundation-Flanders (Grant G.0350.04N), and the Scientific Exchange Program Flanders-France (Tournesol T.2008.21). S.V. and A.I. were, respectively, postdoctoral and predoctoral fellows of the Research Foundation-Flanders. P.M. was the recipient of a Marie Curie Intra-European Fellowship for Career Development (PIEF-GA-2009-235827).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20494).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217516109/-/DCSupplemental.
References
- 1.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7(9):405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 2.Miller G, Shulaev V, Mittler R. Reactive oxygen signaling and abiotic stress. Physiol Plant. 2008;133(3):481–489. doi: 10.1111/j.1399-3054.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- 3.Jaspers P, Kangasjärvi J. Reactive oxygen species in abiotic stress signaling. Physiol Plant. 2010;138(4):405–413. doi: 10.1111/j.1399-3054.2009.01321.x. [DOI] [PubMed] [Google Scholar]
- 4.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17(7):1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11(4):861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 6.Fujita M, et al. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9(4):436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Mittler R, et al. ROS signaling: The new wave? Trends Plant Sci. 2011;16(6):300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Vanderauwera S, et al. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139(2):806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadjev I, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141(2):436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki N, Koussevitzky S, Mittler R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- 12.Nott A, Jung H-S, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 13.Koussevitzky S, et al. Multiple signals from damaged chloroplasts converge on a common pathway to regulate nuclear gene expression. Science. 2007;316(5825):715–719. and erratum (2007) 316(5832):1698. [PubMed] [Google Scholar]
- 14.Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7(3):177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr Opin Plant Biol. 2007;10(6):600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9(5):383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutilleul C, et al. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15(5):1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dojcinovic D, Krosting J, Harris AJ, Wagner DJ, Rhoads DM. Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol Biol. 2005;58(2):159–175. doi: 10.1007/s11103-005-5390-1. [DOI] [PubMed] [Google Scholar]
- 19.Zarkovic J, Anderson SL, Rhoads DM. A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol Biol. 2005;57(6):871–888. doi: 10.1007/s11103-005-3249-0. [DOI] [PubMed] [Google Scholar]
- 20.Van Aken O, et al. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 2007;52(5):850–864. doi: 10.1111/j.1365-313X.2007.03276.x. [DOI] [PubMed] [Google Scholar]
- 21.Smith CA, Melino VJ, Sweetman C, Soole KL. Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol Plant. 2009;137(4):459–472. doi: 10.1111/j.1399-3054.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 22.Skirycz A, et al. Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 2010;152(1):226–244. doi: 10.1104/pp.109.148965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Ries A, Wu K, Yang A, Crawford NM. The Arabidopsis prohibitin gene PHB3 functions in nitric oxide-mediated responses and in hydrogen peroxide-induced nitric oxide accumulation. Plant Cell. 2010;22(1):249–259. doi: 10.1105/tpc.109.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraud E, Van Aken O, Ho LHM, Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009;150(3):1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10(6):1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderauwera S, et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA. 2011;108(4):1711–1716. doi: 10.1073/pnas.1018359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 28.Hruz T, et al. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinform. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inzé A, et al. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012;35(2):308–320. doi: 10.1111/j.1365-3040.2011.02323.x. [DOI] [PubMed] [Google Scholar]
- 30.Vandenabeele S, et al. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004;39(1):45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- 31.Breuer C, Ishida T, Sugimoto K. Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol. 2010;13(6):654–660. doi: 10.1016/j.pbi.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Lee HO, Davidson JM, Duronio RJ. Endoreplication: Polyploidy with purpose. Genes Dev. 2009;23(21):2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horiguchi G, Gonzalez N, Beemster GTS, Inzé D, Tsukaya H. Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J. 2009;60(1):122–133. doi: 10.1111/j.1365-313X.2009.03940.x. [DOI] [PubMed] [Google Scholar]
- 34.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra A, Mishra KB, Höermiller II, Heyer AG, Nedbal L. Chlorophyll fluorescence emission as a reporter on cold tolerance in Arabidopsis thaliana accessions. Plant Signal Behav. 2011;6(2):301–310. doi: 10.4161/psb.6.2.15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15(2):561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urade R. Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 2007;274(5):1152–1171. doi: 10.1111/j.1742-4658.2007.05664.x. [DOI] [PubMed] [Google Scholar]
- 38.Iwata Y, Fedoroff NV, Koizumi N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell. 2008;20(11):3107–3121. doi: 10.1105/tpc.108.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 40.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101(3):249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 41.Koiwa H, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15(10):2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JS, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA. 2008;105(15):5933–5938. doi: 10.1073/pnas.0800237105. and erratum (2008) 105(22):7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35(Database issue):D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int J Biochem Cell Biol. 2009;41(10):1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scorrano L, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: A control point for apoptosis. Science. 2003;300(5616):135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 46.Arduíno DM, et al. ER-mediated stress induces mitochondrial-dependent caspases activation in NT2 neuron-like cells. BMB Rep. 2009;42(11):719–724. doi: 10.5483/bmbrep.2009.42.11.719. [DOI] [PubMed] [Google Scholar]
- 47.Kaplan B, et al. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell. 2006;18(10):2733–2748. doi: 10.1105/tpc.106.042713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361(6410):315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 49.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427(6972):360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 50.John LM, Lechleiter JD, Camacho P. Differential modulation of SERCA2 isoforms by calreticulin. J Cell Biol. 1998;142(4):963–973. doi: 10.1083/jcb.142.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreau B, Nelson C, Parekh AB. Biphasic regulation of mitochondrial Ca2+ uptake by cytosolic Ca2+ concentration. Curr Biol. 2006;16(16):1672–1677. doi: 10.1016/j.cub.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 52.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Ülker B, Somssich IE. WRKY transcription factors: From DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Shen Q-H, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315(5815):1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 55.Turck F, Zhou A, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell. 2004;16(10):2573–2585. doi: 10.1105/tpc.104.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanlerberghe GC, Cvetkovska M, Wang J. Is the maintenance of homeostatic mitochondrial signaling during stress a physiological role for alternative oxidase? Physiol Plant. 2009;137(4):392–406. doi: 10.1111/j.1399-3054.2009.01254.x. [DOI] [PubMed] [Google Scholar]
- 57.Ho LHM, et al. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 2008;147(4):1858–1873. doi: 10.1104/pp.108.121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23(6):2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park CY, et al. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005;579(6):1545–1550. doi: 10.1016/j.febslet.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 60.Boyes DC, et al. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13(7):1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanderauwera S, et al. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA. 2007;104(38):15150–15155. doi: 10.1073/pnas.0706668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.