Abstract
Plant steroid hormones, brassinosteroids (BRs), play important roles in plants. BRs regulate the expression of several thousand genes, half of which are induced and the other half repressed by the hormone. BRs signal through plasma membrane-localized receptor kinase brassinosteroid-insensitive 1 (BRI1), BRI1-associated receptor kinase (BAK1), and several intermediates to regulate the protein levels, cellular localizations, and/or DNA binding of BRI1–EMS suppressor 1 (BES1)/brassinazole-resistant 1 (BZR1) family transcription factors. Although BES1 is known to interact with other transcription factors, histone-modifying enzymes, and transcription elongation factors to activate BR-induced genes, how BES1 mediates the BR-repressed gene expression is not known. Here, we show that BES1 interacts with myeloblastosis family transcription factor-like 2 (MYBL2), a transcription repressor, to down-regulate BR-repressed gene expression. The loss-of-function mybl2 mutant enhances the phenotype of a weak allele of bri1 and suppresses the constitutive BR-response phenotype of bes1-D. The results suggest that suppression of BR-repressed gene expression is required for optimal BR response. Moreover, MYBL2 is a substrate of glycogen synthase kinase 3 (GSK3)-like kinase brassinosteroid-insensitive 2 (BIN2), which has been well established as a negative regulator in the BR pathway by phosphorylating and inhibiting the functions of BES1/BZR1. Unlike BIN2 phosphorylation of BES1/BZR1 leading to protein degradation, BIN2 phosphorylation stabilizes MYBL2. Such dual role of phosphorylation has also been reported in WNT signaling pathway in which GSK3 phosphorylation destabilizes β-catenin and stabilizes Axin, a scaffolding protein facilitating the phosphorylation of β-catenin by GSK3. Our results thus establish the mechanisms for BR-repressed gene expression and the integration of BR signaling and BR transcriptional network.
Keywords: transcription repression, protein stability, plant growth, hormone signaling
A group of plant steroid hormones, named brassinosteroids (BRs), regulate many processes in plant growth, development, and responses to biotic and abiotic stresses. Loss-of-function mutants display dwarf phenotypes with reduced cell elongation, dark-green and epinastic leaves, reduced apical dominance, altered vascular patterning, delayed senescence, male sterility, and late flowering (1–3). By contrast, gain-of-function mutants such as bes1-D have long hypocotyls, leaf petioles, curly leaves, and early leaf senescence (4).
Genetic and molecular studies in Arabidopsis have greatly advanced our understanding of the BR signaling pathway (5–8). BRs are perceived by a plasma membrane-bound receptor brassinosteroid-insensitive 1 (BRI1) (9–11). BR signaling leads to the dephosphorylation of a family of plant-specific transcription factors, defined by their founding members BRI1–EMS suppressor 1 (BES1) and brassinazole-resistant 1 (BZR1) with an atypical basic helix–loop–helix (bHLH) DNA binding domain (12, 13). In the absence of BR, the negative regulator BRI1 kinase inhibitor 1 (BKI1) binds to BRI1 and inhibits its function (14, 15). BR binding to BRI1 leads to the release of BKI1, which in turn sequesters 14-3-3 proteins that inhibit BES1/BZR1 function (16). At the same time, BR also promotes the association of BRI1 with coreceptor BAK1 and a series of phosphorylation events (17–20). Activated BRI1 likely signals through BSK1 and CDG1 kinases as well as BSU1 phosphatase to inhibit brassinosteroid-insensitive 2 (BIN2) kinase (21–24). The inhibition of BIN2 and dephosphorylation by protein phosphatase 2A (PP2A) phosphatase allow accumulation of BES1/BZR1 in the nucleus (25). LCMT, a leucine C-terminal methyltransferase, activates PP2A, which dephosphorylates BRI1 and appears to turn off the BR signaling (26). BRI1 autophosphorylation can also lead to self deactivation (27).
In the absence of BRs, BIN2 phosphorylates BES1/BZR1 and their homologs to inhibit their function (28, 29). In the presence of BRs, BIN2’s kinase activity is inhibited, leading to the accumulation of dephosphorylated BES1 and BZR1 in the nucleus and subsequent regulation of gene expression.
Several genome-wide microarray experiments in Arabidopsis have demonstrated that BRs regulate thousands of target genes, activating and repressing about equal numbers of them (4, 30–35). BES1 and BZR1 target genes have been identified using chromatin immunoprecipitation (ChIP)-chip (ChIP followed by genomic tiling arrays) methods (36, 37). The genome-wide analysis suggests that both BR responsive element (BRRE) and E-box sequences are enriched in BES1 and BZR1 targets with BRRE site preferred in BR-repressed genes and E-boxes more predominant in BR-induced genes. Both BES1 and BZR1 target many transcription factors, some of which have been functionally characterized (38–43).
BES1 interacts with many other transcription regulators to activate BR target gene expression (12, 34, 43, 44). However, how BES1/BZR1 act to repress gene expression remains largely unknown. In this paper, we demonstrate that myeloblastosis family transcription factor like-2 (MYBL2), previously shown to be involved in anthocyanin biosynthesis gene expression (45, 46), is a substrate of BIN2 and is stabilized by BIN2 phosphorylation. We show that MYBL2 interacts with BES1 to repress BR-repressed genes. Our study thus identified a previously unknown substrate of BIN2 in the BR pathway and established a mechanism by which BES1 represses target genes expression.
Results
MYBL2 Is a Direct Target of BES1 and Its Expression Is Repressed by BR and BES1.
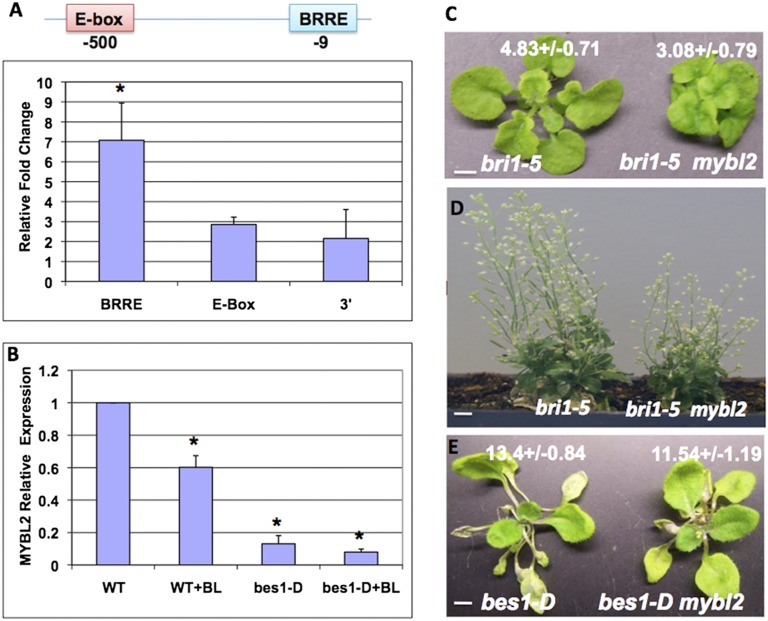
Recent ChIP-chip and gene expression studies indicated that MYBL2 was a direct target of BES1 and BZR1 (36, 37). We performed an independent ChIP PCR experiment to confirm the results (Fig. 1A). Anti-BES1 antibody and a control antibody were used to perform the ChIP assay. There are two putative BES1 binding sites in MYBL2 promoter: an BRRE at −9 bp relative to transcription start site and an E-box at −500 bp. ChIP-qPCR results indicated that BES1 was enriched significantly at BRRE site, but not at the E-box or at 3′-untranslated region (3′-UTR). The preference of BES1 binding to BRRE on MYBL2 promoter suggests that BES1 represses MYBL2 expression. To test the hypothesis, MYBL2 expression was examined in wild-type (WT) and bes1-D seedling plants with or without brassinolide (BL) (the most active BR) treatment (Fig. 1B). The expression of MYBL2 is reduced by BL to about 60% in WT and to less than 20% in bes1-D mutant. These results confirm that MYBL2 is a direct target of BES1 and is repressed by BR through BES1.
Fig. 1.
MYBL2 is repressed by BES1 and acts as a positive regulator in the BR pathway. (A) BES1 targets the BRRE site on MYBL2 promoter. ChIP was performed with anti-BES1 antibody in WT seedlings. The bindings of BES1 at BRRE site (−9 bp), E-box (−500 bp), and 3′ region of the MYBL2 gene were examined by qPCR. The 5s rRNA was used as internal control. (B) The expression of MYBL2 was examined by quantitative RT-PCR in 2-wk-old WT and bes1-D seedlings with or without 1,000 nM BL treatment for 2.5 h. (C and D) The phenotypes of bril-5 mutants and bril-5 mybl2 double mutants at different growth stages. (Scale bars: C, 2.5 cm; D, 1 cm.) (E) The phenotype of 4-wk-old bes1-D mutant and bes1-D mybl2 double mutant. (Scale bar, 5 cm.) The average petiole lengths of the longest (fifth) leaf for each genotype are indicated (C and E). The average and SDs were from 10 plants. The difference was significant as analyzed by Student’s t test (*<0.05).
MYBL2 Is a Positive Regulator in the BR Pathway.
To determine the function of MYBL2 in BR responses, we obtained a T-DNA insertion line (SALK_126807) at 3′-UTR of MYBL2 (Fig. S1). RT-PCR analysis indicated that there is no detectable transcript in the mutant (Fig. S1A), as reported previously (45). We created double mutants of mybl2 with bri1-5, a weak loss-of-function allele of BR receptor BRI1 (47), and bes1-D, a gain-of-function mutant in the BR pathway (4). Although the single mutant mybl2 did not display obvious growth phenotype in either seedling or adult stage, double-mutant analysis indicated that mybl2 enhanced bri1-5 phenotype and suppressed bes1-D phenotype (Fig. 1 C and D and Fig. S1B). The bri1-5 mybl2 double mutant showed more severe reduced growth or dwarf phenotype than bri1-5 either in vegetative or inflorescent stage (Fig. 1 C and D). The bes1-D mybl2 double mutant had shorter petioles in adult plants and shorter hypocotyls at seedling stage compared with bes1-D (Fig. 1E and Fig. S1C). These genetic studies demonstrate that MYBL2 plays a positive role in the BR pathway.
MYBL2 Interacts with BES1 in Vitro and in Vivo.
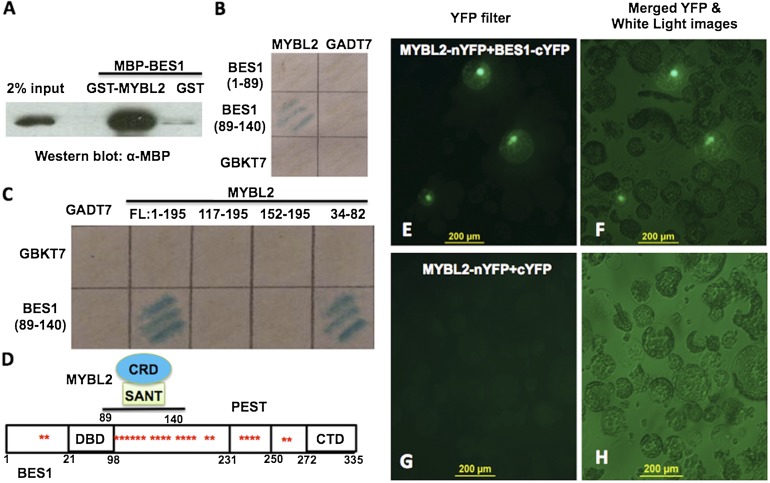
We have previously showed that BES1-induced MYB30 interacts with BES1 to activate BR-induced genes and thus amplify the BR signaling (43). We hypothesize that BES1-repressed MYBL2 may interact with BES1 in the down-regulation of BR-repressed genes. Yeast two-hybrid experiments indicated an interaction between full-length BES1 and MYBL2 (Fig. S2A). GST pull-down experiment confirmed this interaction (Fig. 2A). GST-MYBL2, but not GST alone, pulled down a significant amount of MBP-BES1 protein, demonstrating a direct interaction between MYBL2 and BES1.
Fig. 2.
BES1 interacts with MYBL2 in vitro and in vivo. (A) GST pull-down using GST, GST-MYBL2, and MBP-BES1. BES1 was detected by Western blotting with anti-MBP antibody. (B) MYBL2 interacts with BES1 (amino acids 89–140) in yeast as detected by β-galactosidase (LacZ) activity. (C) MYBL2 interacts with the phosphorylation domain of BES1 (amino acids 89–140) through its SANT domain (amino acids 34–82). (D) A model shows BES1 protein structure and MYBL2’s corresponding domain involved in the interaction. CRD, MYBL2 C-terminal repression domain; CTD, BES1 C-terminal domain; DBD, BES1 DNA binding domain. The asterisk (*) indicate BIN2 phosphorylation sites. (E–H) MYBL2 interacts with BES1 in vivo by BiFC. Cotransformation of MYBL2-nYFP and BES1-cYFP led to the reconstitution of YFP activity in Arabidopsis protoplasts [(E) under YFP filter and (F) merged microscopic images from YFP and white light], whereas coexpression of MYBL2-nYFP and cYFP did not produce any positive YFP signal (G and H).
Several truncated GST-BES1 were used to map the domain in BES1 required for the interaction with MYBL2. Although BES1 with deletion to amino acid 89 still interacts with MYBL2, BES1 with deletion to amino acid 140 lost the interaction, suggesting that amino acids 89–140 in BES1 are important for the interaction (Fig. S2B). Indeed, yeast two-hybrid experiment confirms that amino acids 89–140 in BES1 were sufficient for the interaction with MYBL2 (Fig. 2B). To map the specific domain in MYBL2 for the interaction, a series of truncated-MYBL2 constructs were generated for yeast two-hybrid experiments. Fig. 2C showed that the SANT domain of MYBL2, amino acids 34–82, is both necessary and sufficient to interact with BES1. These results indicate that part of the BIN2-phosphorylation domain of BES1 (amino acids 89–140) and the SANT domain (amino acids 34–82) of MYBL2 are required for the interaction between these two proteins (Fig. 2D).
We further tested in vivo interaction between MYBL2 and BES1 by bimolecular fluorescence complementation (BiFC) experiment with MYBL2 fused to N-terminal YFP (MYBL2-nYFP) and BES1 fused to C-terminal YFP (BES1-cYFP). When MYBL2-nYFP and BES1-cYFP were cotransformed into Arabidopsis protoplasts, strong fluorescence signal was observed in the nucleus (Fig. 2 E and F). In contrast, there was no fluorescence observed when MYBL2-nYFP and cYFP were cotransfected (Fig. 2 G and H). Taken together, these results demonstrate that MYBL2 and BES1 interact with each other in vitro and in vivo.
MYBL2 Facilitates BES1 in Down-Regulating BR-Repressed Gene Expression.
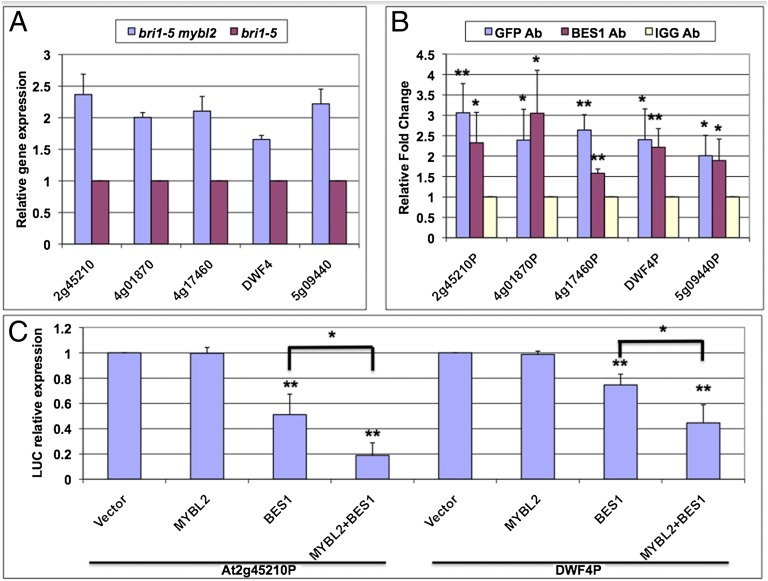
It has been shown previously that MYBL2 represses gene expression by interacting with bHLH transcription factor TT8 (45). Based on the interaction between BES1 and MYBL2, we hypothesize that MYBL2 is recruited by BES1 to down-regulate BR-repressed genes. The expression of seven BR-repressed genes, which are direct targets of BES1, were examined in bri1-5 mybl2 and bri1-5 mutants. The expression of five of the seven tested genes increased significantly in bri1-5 mybl2 compared with bri1-5 (Fig. 3A), suggesting that MYBL2 is responsible for the repression of a portion of the BR-repressed genes. To confirm that MYBL2 indeed are targeted to these genes, we performed ChIP PCR assays with MYBL2-GFP transgenic plants using anti-GFP, anti-BES1 antibodies, and normal IgG as control. Both MYBL2 and BES1 binding were enriched on all five target gene promoters (Fig. 3B and Fig. S3). To further confirm that BES1 and MYBL2 cooperate in the down-regulation of the BR-repressed genes, we constructed promoter-luciferase (LUC) reporter constructs with At2g45210 and DWF4 genes and assayed their regulation by BES1 and MYBL2 in tobacco leaves by a transient expression experiment (Fig. 3C). Although BES1 led to reduced gene expression on both reporter genes, MYBL2 alone showed no effect. However, when MYBL2 and BES1 were coexpressed, the expression of the reporter genes was further reduced compared with BES1. We also tested the repression effect of BES1 with truncated MYBL2 protein that lacked interaction domain (SANT domain); there was no synergistic repression effect on these two reporter constructs (Fig. S4). All of the results demonstrated that MYBL2 forms a complex with BES1 to inhibit BR-repressed genes expression.
Fig. 3.
MYBL2 cooperates with BES1 to inhibit BR-repressed gene expression. (A) BR-repressed genes were increased in bri1-5 mybl2 double mutant compared with bri1-5. Quantitative RT-PCR was performed with RNA from 4-wk-old plants. (B) MYBL2 is targeted to BR-repressed genes in vivo. ChIP using MYBL2-GFP transgenic plants with anti-GFP antibody, anti-BES1 antibody, and normal IgG control. The bindings of MYBL2 and BES1 to indicated gene promoters are detected by qRT-PCR flanking the BRRE sites. The 5s rRNA was used as internal control. (C) Transient gene expression assays were performed in tobacco leaves with At2g45210-LUC and DWF4-LUC reporter genes cotransfected with BES1 and/or MYBL2 via Agrobacterium. The relative expression levels were normalized with total protein. The average and SDs were from three biological repeats. The significant difference was analyzed by Student’s t test (*<0.05, **<0.01).
MYBL2 Is a Substrate of BIN2 and Is Stabilized by BIN2 Phosphorylation.
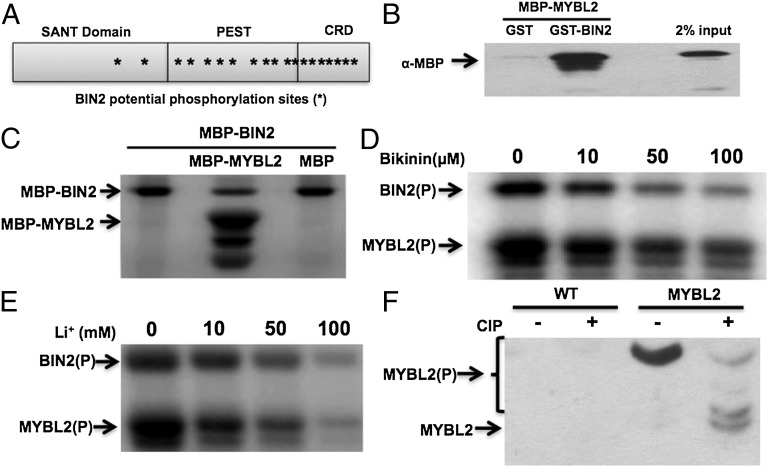
BIN2 is a negative regulator in the BR pathway, which phosphorylates and inhibits BES1 and BZR1 (4, 48–50). BIN2 belongs to glycogen synthase kinase 3 (GSK3) family, whose substrates contain repeats of a short consensus sequence S/TxxxS/T (x corresponds to any amino acid residues). There are 19 potential phosphorylation sites for BIN2 in the predicted MYBL2 protein (Fig. 4A), which prompted us to test whether MYBL2 was a substrate of BIN2. We first tested the interaction between BIN2 and MYBL2 by GST pull-down experiment. Fig. 4B showed that there was a direct interaction between MYBL2 and BIN2 in vitro. We then performed an in vitro kinase assay with MBP-BIN2 and MBP-MYBL2. BIN2 indeed phosphorylated MYBL2, but not MBP (Fig. 4C). Bikinin, a small molecule, and lithium chloride (LiCl) have been found to inhibit the kinase activities of BIN2 and its close homologs (48, 51). Both Bikinin and Li+ can inhibit BIN2 phosphorylation of MYBL2 as well as BIN2 autophosphorylation (Fig. 4 D and E).
Fig. 4.
MYBL2 is a substrate of BIN2 kinase in BR signaling. (A) The structure of MYBL2. The asterisk (*) indicates potential BIN2 phosphorylation sites. The SANT domain, putative PEST domain, and C-terminal repression domain (CRD) in MYBL2 are indicated. (B) GST pull-down experiments using GST, GST-BIN2, and MBP-MYBL2. MYBL2 was detected by Western blotting with anti-MBP antibody. (C) BIN2 phosphorylates MBP-MYBL2 but not MBP in the in vitro kinase assay. The arrows indicate phosphorylated-MYBL2 or autophosphorylated-BIN2. (D) BIN2 phosphorylation of MYBL2 is inhibited by Bikinin in kinase assay in vitro. (E) The phosphorylation of MYBL2 by BIN2 was inhibited by LiCl in kinase assay in vitro. (F) MYBL2 is phosphorylated in vivo. MYBL2 immunoprecipitated from transgenic plants was treated with calf-intestinal alkaline phosphatase (CIP) and separated on SDS/PAGE gel containing Phos-tag reagent (NARD Institute). CIP treatment produced several fast-migrating bands. The arrows indicate the phosphorylated or unphosphorylated MYBL2.
To test whether MYBL2 exists as phosphorylated form in plants, MYBL2-GFP protein was immunoprecipitated from MYBL2-GFP transgenic plants and subjected to phosphatase [calf-intestinal alkaline phosphatase (CIP)] treatment (Fig. 4F). CIP treatment leads to several fast-migrating bands on a SDS/PAGE gel containing Phos-tag reagent that binds to phosphorylation groups and reduce protein mobility (Materials and Methods), indicating that the MYBL2-GFP exists as phosphorylated form in plants.
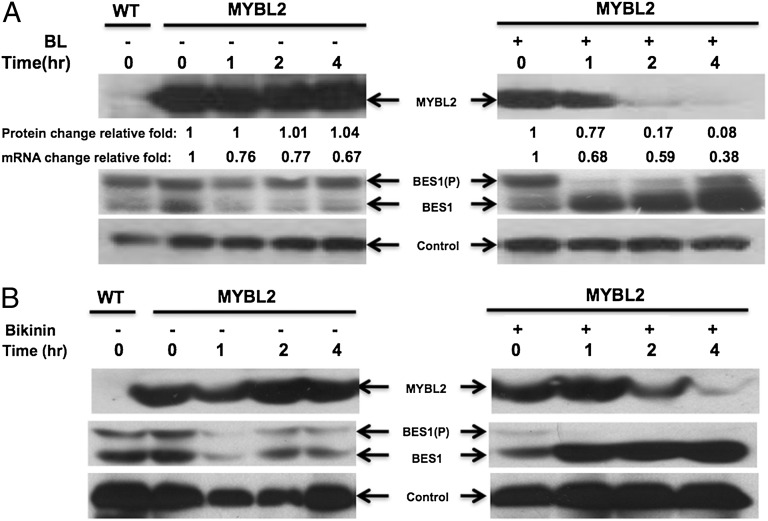
To further investigate the function of the BIN2 phosphorylation on MYBL2, MYBL2-GFP protein level in transgenic plants was examined by Western blotting. Fig. 5A showed that, in the absence of BL, MYBL2-GFP protein level remains mostly constant. However, in the presence of BL, MYBL2-GFP protein decreased more than 10-fold after 4 h of treatment, whereas its mRNA level only decreased by about 40%. As previously demonstrated, BL treatment leads to the accumulation of unphosphorylated BES1 (Fig. 5A, Middle). Because BL treatment is well established to reduce BIN2 activity, we conclude that BIN2-phosphorylated MYBL2 is stable in the absence of BL and is unstable as unphosphorylated form in the presence of BL.
Fig. 5.
BL and BIN2 inhibitors destabilize MYBL2 protein. (A) BL treatment destabilizes MYBL2 protein. Four-week-old MYBL2-GFP transgenic plants were treated with or without 1 μM BL for indicated periods of time and used to prepare protein to detect MYBL2 (Top), BES1 (Middle), and a control protein (Bottom). MYBL2 protein levels were quantified using Alphalmager 3400, and MYBL2 mRNA levels were quantified by qRT-PCR. (B) Bikinin treatment induces the degradation of MYBL2 protein. MYBL2-GFP transgenic plants were treated with or without 100 μM Bikinin for indicated periods of time and used to prepare protein to detect MYBL2 (Top), BES1 (Middle), and a control protein (Bottom).
To test the hypothesis, BIN2 inhibitor Bikinin was also applied to plants to test the effect of BIN2 phosphorylation on MYBL2. MYBL2 protein decreased significantly after 2 h with Bikinin treatment (Fig. 5B, Top), whereas the dephosphorylated form BES1 accumulated at the same time (Fig. 5B, Middle). We further examined MYBL2 protein level in bri1-5, in which BR signaling is blocked and BIN2 is thus constitutively active. The protein level of MYBL2 significantly accumulated compared with WT (3.9 times; Fig. S5), which is more than the MYBL2-GFP transcript accumulation (2.3 times) in the bri1-5.
Proteasome inhibitor MG132 was used to test whether BL-induced decrease of MYBL2 protein was due to proteasome-mediated degradation. When the plant samples were treated by BL together with MG132, the MYBL2 protein is significantly accumulated (Fig. S6). This result indicated that MYBL2 degradation by BL is dependent on the proteasome-mediated pathway. Taken together, all of the results demonstrate that MYBL2 is regulated by BIN2.
Discussion
Recent studies indicated that BRs activate and repress about equal number of genes, and BES1/BZR1 transcription factors play an essential role in mediating BR-regulated gene expression (36, 37). BES1/BZR1 interacts with other transcription factors (BIM1, MYB30, and PIF4), chromatin-modifying enzymes (REF6/ELF6 histone demethylase), and transcription elongation factor (IWS1) to regulate BR-induced gene expression. However, how BES1 and its homologs repress gene expression is not well established. In this study, we found that BES1 interacts with one of its targeted transcription factors, MYBL2, to repress the BR-repressed gene expression. This MYBL2-mediated repression is required for BR-regulated plant growth, as mybl2 knockout mutant enhances the weak loss-of-function BR mutant (bri1-5) and suppresses gain-of-function BR mutant (bes1-D). We further found that MYBL2 is a substrate of BIN2 kinase, and BIN2 phosphorylation of MYBL2 stabilizes the protein. The regulation of MYBL2 by BIN2 is opposite of BIN2 phosphorylation of BES1, which destabilizes BES1. The study thus provides insights into the regulation of BR transcriptional network by BR signaling.
MYBL2 is a small MYB protein that has only one R3-MYB (SANT) domain. MYBL2 interacts with TT8, a bHLH protein, to repress anthocyanin biosynthesis gene expression (45, 46). MYBL2 has a repression motif (TLLLFR) at its carboxyl terminus (45), which might interact with TOPLESS family corepressors (52). Several lines of evidence support the role of MYBL2 as a transcription corepressor for BES1. First, BES1 interacts with MYBL2 both in vitro and in vivo, through the SANT domain of MYBL2 and a specific region of BES1 (amino acids 89–140). It was proposed that the SANT domain of MYBL2 does not interact with DNA directly due to the replacement of several critical residues required for DNA binding (45). Our findings that the SANT domain of MYBL2 does not bind to DNA and is involved in interaction with BES1 corroborate this conclusion. Second, MYBL2 and BES1 act cooperatively to repress BR-repressed gene expression in a transient expression assay. Last, several BR-repressed genes are up-regulated in mybl2 mutant in bri1-5 background.
In addition to the regulation by BES1 at transcription level, MYBL2 is regulated by BR signaling through BIN2 kinase. BIN2 phosphorylates and inhibits BES1 and its homolog, BZR1, through several mechanisms, including targeted protein degradation, cytoplasmic retension by 14-3-3 proteins, and reduced DNA binding (6, 29, 53). Recent studies suggest that BIN2 can also phosphorylate and regulate other factors involved in BR-regulated processes. For example, it was recently reported that BIN2 phosphorylates and inhibits MAP kinase kinase YODA and transcription factor SPCH to regulate stomatal development (54, 55). BIN2 was also reported to phosphorylate and inhibit ARF2 DNA binding (56). AIF, a BES1/BZR1 target bHLH protein involved in BR responses, and CESTA, implicated in BR biosynthesis, are both reported to be phosphorylated by BIN2 in vitro (41, 57). Rice transcription factor DLT is a BIN2 kinase substrate and mediates BR signaling in rice (58). BIN2 phosphorylation of MYBL2, like that of DLT, is not as effective as BIN2 phosphorylation of BES1/BZR1, which may explain the higher concentrations of bikinin and LiCl required to observe their effects on MYBL2 phosphorylation and protein accumulation.
In contrast to BES1/BZR1 and SPCH, which are destabilized by BIN2 phosphorylation, BIN2 phosphorylation of MYBL2 stabilizes the protein. Our results thus suggest that BIN2 phosphorylation can have different functional consequences. The dual role of GSK-like kinase phosphorylation has been observed in WNT pathway. In the WNT signaling pathway, GSK3 kinase phosphorylates positive regulator β-catenin in a protein complex including scaffolding protein Axin, leading to β-catenin degradation (59). Interestingly, Axin is also phosphorylated by GSK3 kinase and such phosphorylation stabilizes Axin (60), much like the effect of BIN2 phosphorylation on MYBL2 (Fig. S7).
Although MYBL2 acts as a transcriptional repressor, it functions as a positive regulator in the BR pathway, as mybl2 mutant enhances and suppresses the phenotype of bri1-5 and bes1-D, respectively. Why is the positive regulator MYBL2 repressed by BRs and in bes1-D? One possibility is that this regulation represents a feedback regulation mechanism in which excessive BRs or BR signaling down-regulates the pathway through MYBL2. We therefore propose that MYBL2 functions as a “buffer” to fine-tune BR responses (Fig. S8). Global gene expression studies with mybl2 mutants and functional studies of BIN2 regulation of MYBL2 phosphorylation are needed to further test the model.
In conclusion, our studies showed that MYBL2 formed a complex with BES1 to down-regulate BR-repressed genes and BIN2 phosphorylates and stabilizes MYBL2 protein. Like BES1-mediated gene activation, BES1–MYBL2 complex likely interacts with additional cofactors in the regulation of the BR-repressed genes. Identification of these factors and elucidation of the transcriptional network through which BES1 and its corepressors act to repress gene expression are important steps to understand how BRs regulate plant growth and various responses.
Materials and Methods
Plant Materials and Growth Condition.
T-DNA insertion mutant, mybl2, was obtained from Arabidopsis Biological Resource Center, corresponding to line SALK_126807 (61). All of the plants were grown on 1/2MS plates and/or in soil under long day conditions (16-h light/8-h dark) at 22 °C. All the DNA primers used in the study are listed in Table S1.
Plasmid Constructs.
For GFP-tagged transgenic plants, MYBL2 genomic sequence including its native promoter was cloned from wild type and fused with GFP tag into pZP211 vector (62). For recombinant protein purification and GST pull-down assay, MYBL2 coding region was cloned into pETMALc-H vector, whereas BIN2, BES1, and truncated BES1 fragments were incorporated into pET42a(+) (Novagen), respectively. For yeast two-hybrid assays, BES1, the DNA binding domain of BES1, and phosphorylation domain of BES1 were cloned into pGBKT7, whereas MYBL2 as well as its deletion mutation were clone into pGADT7 (Clontech). For BiFC assay, the constructs of N or C terminus of EYFP used were previously reported (44). The coding region of MYBL2 and BES1 were inserted into YFP-N construct and YFP-C construct, respectively.
Transgenic Plants.
The construct of MYBL2-GFP driven by its native promoter was transformed into Agrobacterium tumefaciens (strain GV3101), which was used to transform plants by the floral dip method (63). Transgenic lines were selected on 1/2 MS medium plus 50 μg/mL kanamycin. Transgene expression was analyzed by Western blotting.
Gene Expression Analysis.
Total RNA was extracted and purified from 4-wk-old plants of different genotypes using RNeasy Mini Kit (Qiagen). Mx4000 multiplex quantitative PCR system (Stratagene) and SYBR Green PCR Master Mix (Applied Biosystems) were used in quantitative real-time PCR analysis. At2g45210 promoter (1021 bp including 5′-UTR) and DWF4 promoter (972 bp including 5′-UTR) were cloned and used to drive luciferase reporter gene expression. MYBL2, MYBL2△SANT, and BES1 coding region driven by CaMV 35S promoter were cloned into pZP211 vector, respectively. Tobacco leaf transient assay (64) was used to examine the repression effect of MYBL2 on reporter gene expression in the presence or absence of BES1 and/or MYBL2. Equal amount of Agrobacterium cells (measured by OD, adjusted to same with vector-containing strain) was injected into tobacco leaves. The luciferase activities were measured from protein extracts from triplicate samples and measured using Berthold Centro LB960 luminometer with luciferase assay system (Promega). The luciferase levels were normalized by the total protein from each sample.
ChIP.
ChIP was performed as previously described (36). GFP antibody was used to precipitate chromatin from MYBL2-GFP overexpression plants, whereas antibody against BES1 and normal IgG (Sigma) were used as control.
In Vitro Kinase Assay and Detection of in Vivo MYBL2 Phosphorylation.
The in vitro kinase assay was performed as described (4). MYBL2 protein was immunoprecipitated from transgenic plants and treated with or without CIP as described (4). The in vivo phosphorylated MYBL2 was examined by Phos-tag reagent (NARD Institute) with or without CIP treatment as described (65).
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for Arabidopsis T-DNA insertion mutants and Tadao Asami (University of Tokyo) for providing BRZ. The work was supported by National Science Foundation Grants IOS-0546503 and IOS-1122166 (to Y.Y.) and in part by the Plant Science Institute of Iowa State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205232109/-/DCSupplemental.
References
- 1.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111(3):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272(5260):398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 3.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 4.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 5.Gudesblat GE, Russinova E. Plants grow on brassinosteroids. Curr Opin Plant Biol. 2011;14(5):530–537. doi: 10.1016/j.pbi.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23(4):1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye H, Li L, Yin Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol. 2011;53(6):455–468. doi: 10.1111/j.1744-7909.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 9.She J, et al. Structural insight into brassinosteroid perception by BRI1. Nature. 2011;474(7352):472–476. doi: 10.1038/nature10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hothorn M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474(7352):467–471. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120(2):249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 13.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307(5715):1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313(5790):1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 15.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25(3):232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, et al. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell. 2011;21(5):825–834. doi: 10.1016/j.devcel.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Gou X, et al. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8(1):e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh MH, et al. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(2):658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh MH, et al. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc Natl Acad Sci USA. 2010;107(41):17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15(2):220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43(4):561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11(10):1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295(5558):1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 25.Tang W, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13(2):124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, et al. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal. 2011;4(172):ra29. doi: 10.1126/scisignal.2001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh MH, Wang X, Clouse SD, Huber SC. Deactivation of the Arabidopsis BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc Natl Acad Sci USA. 2012;109(1):327–332. doi: 10.1073/pnas.1108321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J. Regulation of the nuclear activities of brassinosteroid signaling. Curr Opin Plant Biol. 2010;13(5):540–547. doi: 10.1016/j.pbi.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vries SC. 14-3-3 proteins in plant brassinosteroid signaling. Dev Cell. 2007;13(2):162–164. doi: 10.1016/j.devcel.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2(9):E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goda H, et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134(4):1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müssig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129(3):1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130(3):1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA. 2010;107(8):3918–3923. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H, et al. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106(18):7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo XM, et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19(6):872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang LY, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21(12):3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, et al. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21(12):3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Z, et al. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22(4):1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58(2):275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, et al. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA. 2008;105(21):7618–7623. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55(6):954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 46.Dubos C, et al. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55(6):940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi T, et al. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121(3):743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150(2):710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(15):10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127(1):14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16(6):594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158(1):423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plant Sci. 2007;12(1):37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482(7385):419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gudesblat GE, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14(5):548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 56.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105(28):9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poppenberger B, et al. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011;30(6):1149–1161. doi: 10.1038/emboj.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tong H, et al. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 2012;24(6):2562–2577. doi: 10.1105/tpc.112.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 60.Yamamoto H, et al. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J Biol Chem. 1999;274(16):10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
- 61.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 62.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25(6):989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 63.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 64.Antony G, et al. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 2010;22(11):3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mao G, et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23(4):1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.