Abstract
The viral sensor MDA5 distinguishes between cellular and viral dsRNAs by length-dependent recognition in the range of ∼0.5–7 kb. The ability to discriminate dsRNA length at this scale sets MDA5 apart from other dsRNA receptors of the immune system. We have shown previously that MDA5 forms filaments along dsRNA that disassemble upon ATP hydrolysis. Here, we demonstrate that filament formation alone is insufficient to explain its length specificity, because the intrinsic affinity of MDA5 for dsRNA depends only moderately on dsRNA length. Instead, MDA5 uses a combination of end disassembly and slow nucleation kinetics to “discard” short dsRNA rapidly and to suppress rebinding. In contrast, filaments on long dsRNA cycle between partial end disassembly and elongation, bypassing nucleation steps. MDA5 further uses this repetitive cycle of assembly and disassembly processes to repair filament discontinuities, which often are present because of multiple, internal nucleation events, and to generate longer, continuous filaments that more accurately reflect the length of the underlying dsRNA scaffold. Because the length of the continuous filament determines the stability of the MDA5–dsRNA interaction, the mechanism proposed here provides an explanation for how MDA5 uses filament assembly and disassembly dynamics to discriminate between self vs. nonself dsRNA.
Keywords: antiviral, innate immunity, filament dynamics, helicase, viral RNA
Effective antiviral immunity depends on accurate recognition of viral RNAs by pattern-recognition receptors in the innate immune system. Double-stranded RNA (dsRNA) often accumulates in virally infected cells in the form of the viral genome or its replication intermediates (1). A duplex RNA structure initially was considered a unique viral signature that was sufficient to initiate an antiviral response through dsRNA receptors, such as Toll-like receptor 3 (TLR3), protein kinase R (PKR), Melanoma Differentiation-Associated protein 5 (MDA5), and RIG-I. However, dsRNA also is present in many cellular RNAs, including microRNA precursors and transcripts from Alu retrotransposons (2, 3). Accumulating evidence suggests that several dsRNA receptors discriminate between viral and cellular RNAs by recognizing chemical modifications (e.g., pseudouridylation) and end structures (e.g., 5′-triphosphate, 5′ppp) (4–6). Sensitivity to these additional RNA features allows robust and accurate viral RNA detection and host discrimination. Interestingly, immune signaling mediated by MDA5 was shown to be independent of the 5′ppp group and the 7-methyl guanosine cap on dsRNA but instead correlates with dsRNA length over a range of ∼0.5–7 kb, sizes which exceed the typical lengths of cellular dsRNAs (7). This sensitivity to length is in contrast to that of its paralog, RIG-I, which binds to dsRNA ends containing 5′ppp (8, 9) and displays a strong preference for shorter (<1–2 kb) dsRNA (7, 10). Other dsRNA receptors, such as PKR and TLR3, require only 20–80 bp for optimal binding and activation of an immune response (11, 12). These findings suggest that MDA5 uses a unique molecular mechanism to sense dsRNA on the basis of length.
MDA5 shares with RIG-I the same domain architecture and signaling adaptor, MAVS (13). MDA5 and RIG-I contain two N-terminal tandem caspase activation recruitment domains (CARD) that mediate binding to MAVS, a central DExD/H motif helicase domain responsible for RNA-dependent ATP hydrolysis, and a C-terminal domain (CTD) that in RIG-I is involved in RNA binding (14, 15). Domain truncation studies and recent crystal structures suggest that RIG-I is autorepressed in the absence of viral RNAs (13, 16-19). Binding of viral RNA triggers ATP hydrolysis of RIG-I and interaction with MAVS, which subsequently induces oligomerization of MAVS and activation of downstream IRF3/7 and NF-κB signaling pathways (20). Similar structural changes and signal transmission mechanism have been proposed for MDA5 (17), but how MDA5 uses the same structural architecture to achieve vastly different RNA specificity is currently unknown.
We have shown previously that MDA5, but not RIG-I, cooperatively assembles into filaments upon dsRNA binding (21). Using electron microscopy (EM), we found that the MDA5 filament consists of ring-structured elements stacked along the length of the dsRNA, with each ring occupying ∼14 bp. Each ring represents a binding unit and was proposed to be a dimer (21); however, the recent structures of RIG-I, which also displayed a ring shape around dsRNA (16-18), and the close homology between MDA5 and RIG-I suggest that each ring in the MDA5 filament corresponds to a monomer. We further demonstrated that MDA5 filament formation correlates with signaling in the cell. For example, MDA5 binds to various types of nucleic acids with comparable affinities, but only dsRNA, which activates MDA5 signaling in the cell, triggers filament formation in vitro (21). In addition, a partial loss-of-function mutant, I923V, is defective in filament formation and disassembles more rapidly upon ATP hydrolysis, further indicating the importance of filament formation in MDA5 function (21). Binding to dsRNA induces MDA5 to hydrolyze ATP, which in turn triggers rapid filament disassembly (21, 22). However, it still is unclear precisely how MDA5 filaments assemble and disassemble during ATP hydrolysis and how the dynamic interplay between the filament assembly and disassembly processes contributes to MDA5’s ability to discriminate among dsRNAs on the basis of their length. In this paper, we report bulk kinetics assays, single-molecule fluorescence experiments, and time-dependent EM, and reveal unexpected kinetic mechanisms that enable MDA5 to function as a “molecular ruler” for viral RNA recognition.
Results
MDA5 Filament Is a Cooperative Entity Distinct from Beads-on-a-String Configuration.
We examined whether the MDA5 filament represents a cooperative oligomer formed via specific protein–protein interactions or a mere consequence of saturating RNA with protein. To distinguish between these two possibilities, we incubated MDA5 with increasing amounts of dsRNA and examined the size of the oligomers accumulating on dsRNA by an EMSA and EM. If the filament formation simply reflects RNA-mediated bridging, fewer MDA5 molecules would accumulate on individual RNA molecules with increasing concentrations of dsRNA. We found that, even when dsRNA was in excess over protein, the MDA5:dsRNA complex displayed the same mobility in the native gel, suggesting invariant complex size (Fig. 1A). In contrast to MDA5, the size of the RIG-I:dsRNA complex decreased with increasing concentrations of dsRNA, suggesting that RIG-I represents a beads-on-a-string type of oligomer that is distinct from the cooperative MDA5 filament.
Fig. 1.
ATP hydrolysis increases the binding preference of MDA5 for longer dsRNA. (A) EMSA of MDA5 and RIG-I with increasing amounts of 112-bp dsRNA. Protein (0.3 μM) was incubated with 112-bp dsRNA [30 nM (lane 1)–150 nM (lane 5)]. 3′-Fluorescein-labeled dsRNA was maintained at 10 nM, and the ratio of protein to binding sites (BS) was calculated assuming that each monomer occupied 14 bp. (B) Representative electron micrographs of MDA5 in complex with 2,012-bp dsRNA at a protein-to-binding site ratio of 1.8 (Left) or 0.31 (Right) in the presence of 0.5 mM ADP⋅AlF4. (C) EMSA and representative class averages of filaments formed by the CARD deletion mutant MDA5h on 112-bp dsRNA in the presence of 0.5 mM ADP⋅AlF4. EMSA was performed as in A. (D) Competition binding assay (mean ± SD, n = 2–3). Fluorescein-labeled reporter dsRNA (112 bp, 0.18 μg/mL) and unlabeled competitor dsRNA (21–2,012 bp, 0.06–43.74 μg/mL) were premixed and incubated with MDA5 (80 nM). The level of the reporter complex was monitored by EMSA in the presence of increasing amounts of competitor dsRNA (Fig. S2) and was plotted with fitted competition binding curves, which yielded the IC50. (E) Competition EMSA of MDA5 with dsRNAs of various lengths with ADPCP or ATP (2 mM). Experiments were performed as in A with a fixed concentration of competitor dsRNAs (0.54 μg/mL). The relative level of labeled complex with respect to 62-bp competitor dsRNA was plotted (mean ± SD, n = 3). (F) Competition EMSA of RIG-I as in B (mean ± SD, n = 3). The two bands corresponding to RIG-I:RNA complexes with ADPCP suggest binding to each end of dsRNA. The presence of multiple ill-defined bands with ATP is consistent with the translocation of RIG-I on dsRNA (37).
We next used EM to examine filament formation on longer dsRNA (2,012 bp), which is difficult to examine with EMSA because of the limited resolving power. When an excess amount of MDA5 was incubated with 2,012-bp dsRNA, MDA5 filaments were ∼700–800 nm long. When dsRNA was present in excess over protein, MDA5 filaments were shorter but still were ∼250 nm long (corresponding to a coverage of ∼600 bp) (Fig. 1B), suggesting that MDA5 preferentially binds adjacent to another MDA5 molecule rather than at random positions. The incomplete coverage of 2,012-bp dsRNA probably reflects multiple nucleation events that are more likely to occur with longer dsRNA than with shorter ones such as 112-bp dsRNA (as discussed in more detail in Fig. 5). The CARD-deletion mutant (MDA5h) also exhibited similar cooperativity in dsRNA binding and filament morphology consisting of stacked ring structures, similar to that of the full-length MDA5 filament (Fig. 1C) (21). The similarity between MDA5 and MDA5h filaments suggests that CARD is not involved in the core filament formation and that the ring–ring contact involves the helicase domain and CTD. Consistent with the EMSA result in Fig. 1A, no filaments were observed with RIG-I (Fig. S1B).
Fig. 5.
MDA5 binds to dsRNA via multiple, rate-limiting nucleation steps to assemble discontinuous filaments on a single dsRNA. (A) Schematic of the single-molecule fluorescence assay. The filament assembly reaction was monitored using TIRF single-molecule microscopy upon the addition of fluorescently labeled MDA5 or MDA5h to surface-immobilized dsRNA (t = 0) (Methods and Materials). (B) Representative traces of filament assembly reactions using Alexa 647-labeled MDA5hSNAP (50 nM) and 2,012-bp dsRNA. The immediate increase in fluorescence upon MDA5hSNAP injection (t = 0) indicates the background noise, which also was observed without dsRNA. (C) A model of stepwise assembly reaction. The alternation between elongation and pause in assembly traces indicates the involvement of multiple nucleation events. Elongation ends abruptly upon encounter of dsRNA ends or neighboring filament termini. The insufficiency of a single nucleus to propagate and saturate the entire length of dsRNA suggests unidirectional elongation of MDA5 filaments. The direction of elongation likely is determined by asymmetric binding of a nucleus to symmetric dsRNA with an equal probability of facing either direction. (D) A representative electron micrograph of full-length MDA5 (0.3 μM) in complex with 2,012-bp dsRNA (1.2 μg/mL; i.e., 0.34 μM MDA5 binding site). Gaps (arrows) indicate the presence of multiple filaments propagated from independent nuclei on a single dsRNA. (E) Histogram of the elongation rates of MDA5hSNAP filaments calculated from the slope of the first linear elongation phase of ∼350 assembly traces. Fluorescence intensity was converted to base pair numbers by assuming 100% coverage of 512 bp at saturation. (F) Averaged traces of filament propagation of MDA5, MDA5h, and MDA5hSNAP synchronized by the initial nucleation event (n = 331, 257, and 542, respectively). (G) Histogram of the nucleation times of MDA5hSNAP (50 nM) on 512-bp dsRNA and the single-exponential fit. (H) Dependence of the nucleation time (mean value) on dsRNA length (n = 250–500).
Intrinsic Affinity of MDA5 for dsRNA Depends Moderately on dsRNA Length.
We next asked whether MDA5 filament formation is sufficient to explain the length-dependent signaling activity of MDA5. To address this question, we compared the affinity of MDA5 for dsRNA of various lengths. We used a competition binding assay in which formation of a complex of MDA5 and a 3′-fluorescein–labeled 112-bp dsRNA was measured in the presence of increasing concentrations of unlabeled competitor dsRNAs of 21–2,012 bp (Fig. S2). We included nonhydrolyzable ATP analog, β,γ-methyleneadenosine 5′-triphosphate (ADPCP), in the reaction to mimic the ATP-bound state expected under cellular conditions. Competitor dsRNAs were compared at the same mass concentration, instead of the same molar concentration, because individual MDA5 binding units (ring in Fig. 1C) occupy ∼14 bp independent of the length of dsRNA (21, 22). Comparison of the competitor concentrations required for 50% inhibition of the labeled complex formation (IC50) revealed that MDA5 has higher affinity for longer dsRNAs (Fig. 1D). The most abrupt transition in affinity was observed between 21 and 28 bp, which can accommodate one and two binding units, respectively, suggesting that contacts between the adjacent binding units are important for the stability of the MDA5:dsRNA complex. However, the impact of the length beyond ∼100 bp is moderate, with a twofold reduction in IC50 from 112- to 2,012-bp dsRNA in comparison with a 13-fold reduction in IC50 from 21- to 112-bp dsRNA (Fig. 1D).
ATP Hydrolysis Increases the Length Dependence of the Apparent Affinity of MDA5 for dsRNA.
A potential role for ATP hydrolysis in length discrimination was queried by performing the competition assay in the presence of either ATP or ADPCP. Compared with ADPCP, significantly enhanced length bias was observed in the presence of ATP; the relative preference of 2,012-bp over 62-bp dsRNAs increases from 15% with ADPCP to 70% with ATP (Fig. 1E). The observed affinity of MDA5 to dsRNA in the presence of ATP hydrolysis is referred to henceforth as “apparent” affinity to distinguish it from “intrinsic” affinity in the absence of ATP hydrolysis. In contrast to MDA5, RIG-I displays an opposite length bias with a preference for shorter dsRNAs in the presence of both ATP and ADPCP (Fig. 1F). This result is consistent with RIG-I recognition of dsRNA ends (4), which decrease in concentration with increasing length of RNA at equivalent mass amounts. The similarity between the ATP and ADPCP reactions for RIG-I suggests that the observed length bias in the ATP-dependent apparent affinity is a distinct characteristic of MDA5.
ATP Hydrolysis Rate Reflects the Length-Dependent Apparent Affinity of MDA5.
Because ATP hydrolysis occurs only when MDA5 is bound to dsRNA (21), we asked whether the length-dependent apparent affinity is reflected in the bulk ATP hydrolysis rate. We compared the ATP hydrolysis rate of MDA5 using both in vitro transcribed model dsRNAs of 62–2,012 bp (Table S1) and purified genomic rotavirus dsRNAs of ∼1,080, 1,611, 2,550, and 3,300 bp. To ensure an equivalent number of MDA5 binding sites for each reaction, equal masses of dsRNA were used for all RNAs. Comparable stimulation of ATP hydrolysis was observed in viral and model dsRNAs of similar lengths (Fig. 2A), suggesting that sequence is not important. Instead, the ATP hydrolysis rate was strongly dependent on the length of the dsRNA (Fig. 2A), and this dependence is more pronounced at low concentrations of MDA5 and dsRNA (Fig. 2B). The relative rate of ATP hydrolysis with 1,080-bp dsRNA with respect to 3,300-bp dsRNA increased from 9 to 97% as MDA5 and RNA concentrations increased from 0.06 μM and 0.06 μg/mL to 0.3 μM and 4.8 μg/mL, respectively.
Fig. 2.
The ATP hydrolysis rate reflects the length-dependent RNA binding by MDA5. (A) ATP hydrolysis rates of MDA5 (0.3 μM) and RIG-I (30 nM) with model dsRNAs prepared by in vitro transcription (▲) and rotavirus genomic dsRNAs (●) (4.8 μg/mL) (mean ± SD, n = 3). (B) Relative ATP hydrolysis rate of MDA5 with rotavirus dsRNAs at different concentrations of RNA and MDA5 (mean ± SD, n = 4). Rates were normalized against the rate measured with 3.3 kb dsRNA. (C) Relative ATP hydrolysis rates of MDA5 (0.3 μM) with 112- or 1,012-bp dsRNA (4.8 μg/mL) containing variant 5′ functional groups, different sequences (Table S1), and different internal structures (a mismatch, bulge, or gaps) (mean ± SD, n = 3–4). Rates were normalized against the rate measured with intact 112-bp or 1,012-bp dsRNAs.
In contrast to MDA5, ATP hydrolysis by RIG-I is stimulated more efficiently by shorter dsRNAs (Fig. 2A), consistent with a higher affinity of RIG-I for short dsRNA (Fig. 1F). These results suggest that the opposite dsRNA length bias observed in the apparent affinity of MDA5 and RIG-I can be recapitulated by bulk ATP hydrolysis rate.
Continuous dsRNA Structure Is Important for Efficient ATP Hydrolysis.
To test the importance of internal and end dsRNA structures, we measured ATP hydrolysis rates using 112-bp dsRNAs with variant 5′ functional groups (5′ppp or 5′ hydroxyl), different sequences (Table S1), and with a 4-nt mismatch, a bulge, or a gap at the center of the duplex (Fig. 2C). The 5′ functional group, dsRNA sequence, and internal mismatch had only a small (<10%) effect on the ATP hydrolysis rate. A central gap or bulge within 112-bp dsRNA lowered the rate by 25–30%, a level similar to that observed with 62-bp dsRNA at equivalent mass amount (Fig. 2C). These observations suggest that MDA5 recognizes a gap or bulge as a dsRNA break. To support this interpretation further, we also tested a 1,012-bp dsRNA with four 243-bp duplex regions separated by three 10-nt gaps (Fig. 2C). The hydrolysis rate of MDA5 with the gapped 1,012-bp dsRNA was ∼50% lower than with intact 1,012-bp dsRNA and was between the rates seen with 512-bp and 112-bp dsRNAs (Fig. 2C). Because the gaps account for less than 3% of the total base pairs, the reduction of ∼50% reduction is unlikely to be caused by reduced binding sites but instead suggests the importance of a continuous, intact duplex structure for recognition by MDA5.
MDA5 Dissociates More Rapidly from Shorter dsRNA During ATP Hydrolysis.
The enhanced length dependence of MDA5 measured by apparent affinity and ATP hydrolysis rate prompted us to examine the role of ATP in filament disassembly kinetics and to compare the disassembly kinetics of MDA5 and RIG-I. To accurately measure RNA bound protein, we developed a pull-down kinetic assay to directly monitor the level of MDA5 or RIG-I bound to dsRNA during disassembly (Fig. 3A). We preformed the MDA5:dsRNA or RIG-I:dsRNA complex, initiated dissociation by adding a mixture of ATP and heparin, and then quenched the reaction at discrete time points with ADP⋅AlF4, an ATP analog that blocks ATP hydrolysis by MDA5 (21). Heparin traps MDA5 and RIG-I and sequesters unbound proteins during dissociation (at 100-fold excess of dsRNA) (Fig. S3A). Upon quenching, dsRNA-bound MDA5 was isolated rapidly using streptavidin magnetic beads and was measured by quantitative SDS/PAGE. For quantitation, we labeled MDA5 with fluorescein using sortase (23), which has little effect on the RNA binding and ATP hydrolysis activity of MDA5 (21). Comparison of the RNA-bound fraction at 0 and 2 min suggests that within 2 min 40% and 92% of MDA5 dissociates from 512-bp dsRNA with ADPCP and ATP, respectively (Fig. 3B). In contrast to MDA5, less than 10% of RIG-I dissociated within 2 min of incubation with either ADPCP or ATP (Fig. 3B). This result suggests that ATP hydrolysis promotes dissociation of MDA5, but not of RIG-I, from dsRNA.
Fig. 3.
ATP hydrolysis promotes dissociation of MDA5, but not RIG-I, from dsRNA at a rate inversely proportional to dsRNA length. (A) Schematic of pull-down dissociation kinetic assay. The level of MDA5 bound to biotinylated dsRNA was monitored at discrete time points during dissociation by using streptavidin magnetic bead pull-down. MDA5 was fluorescently labeled for quantitation on SDS/PAGE. See Fig. S3 for details. (B) A representative SDS/PAGE image of MDA5 (Left) or RIG-I (Right) from dissociation pull-down assays at t = 0 or 2 min (2 m) using 512-bp dsRNA with either ATP or ADPCP (2 mM). Nonspecific binding (BG) was measured by using nonbiotinylated dsRNA. (C) Analysis of dissociation kinetics of MDA5 with ATP or ADPCP. Biotin pull-down was performed as in B using dsRNAs of 112–2,012 bp (mean ± SD, n = 3).
We next examined the length-dependence of the dissociation kinetics of MDA5 on using dsRNAs of 112–2,012 bp. Comparison of the declining levels of the RNA-bound fraction of MDA5 indicates that MDA5 dissociates from dsRNA in a length-dependent manner (Fig. 3C). We observed that 50% of MDA5 dissociates within ∼7 s, 15 s, ∼30 s, and 1 min from 112-, 512-, 1,012-, and 2,012-bp dsRNA, respectively. This result demonstrates an inverse correlation between RNA length and the dissociation rate of MDA5 during ATP hydrolysis.
MDA5 Binds to dsRNA with Slow Kinetics, Amplifying the Impact of Length-Dependent Dissociation.
We next asked whether the length-dependent dissociation is sufficient to explain dsRNA length-dependent apparent affinity of MDA5 during ATP hydrolysis. If dsRNA binding were significantly more rapid than dissociation, the impact of differential dissociation rates would be diminished. To measure the RNA-binding kinetics of MDA5, we used a modified pull-down kinetic assay in which the coincubation time of MDA5 and dsRNA was varied and heparin was added at discrete time points to quench the binding reaction before biotin pull-down (Fig. 4A). Time evolution of the dsRNA-bound fraction indicates that RNA binding progresses over ∼10 min, with an apparent rate (kobs) of 0.29 × 10−2 s−1 with 512-bp dsRNA (Fig. 4A). A comparable binding rate was observed using time-dependent EMSA (Fig. S4A) and with 112-, 1,012-, and 2,012-bp dsRNAs (Fig. 4A). First-order approximation of kobss measured at four concentrations of MDA5 yielded an association rate constant (kon) of ∼0.5 × 104 M−1⋅s−1 (Fig. 4B), a value significantly lower than the binding rates of other well-characterized nucleic acid-binding proteins (108–10 M−1⋅s−1) (24).
Fig. 4.
Slow dsRNA binding amplifies the length dependence of MDA5. (A) Analysis of the RNA-binding kinetics of MDA5 using biotin pull-down. As with the dissociation assays in Fig. 3, the level of MDA5 bound to dsRNA was monitored using streptavidin magnetic beads and was quantitated by using the fluorescein tag on MDA5 (Methods and Materials). On the right is the time course of the bound fractions and fitted single exponential curves (mean ± SD, n = 3). (B) The binding rate constant (kon) was obtained from the first-order approximation of the apparent binding rate (kobs, obtained from Fig. S4B) against MDA5 concentration. (C) Time evolution of the ATP hydrolysis reaction initiated by the addition of ATP to the preformed complex of MDA5 (0.3 μM) and 112-bp dsRNA (0.6 μg/mL) (dsRNA→ATP) or by the addition of dsRNA to the preformed complex of MDA5 and ATP (ATP→dsRNA) (mean ± SD, n = 3). (D) Initial lag period of the ATP hydrolysis reactions of MDA5 (0.3 μM) using different concentrations of 112-bp dsRNA at 100 or 150 mM NaCl (mean ± SD, n = 3). The lag period was estimated from the linear extrapolation of the reaction time course as in C. (E) Relative ATP hydrolysis rates of MDA5 (0.3 μM) bound to model dsRNAs of 62- to 2,012 bp (4.8 μg/mL) at 100 or 150 mM NaCl (mean ± SD, n = 3). Rates are normalized against the rate measured with 2,012-bp dsRNA.
The slow RNA-binding kinetics of MDA5 manifests as a lag phase in the ATP hydrolysis time course (Fig. 4C). When the reaction was initiated by the addition of dsRNA to the preformed MDA5:ATP complex (ATP→dsRNA), the ATP hydrolysis reaction progressed with an initial lag of 1.2 min, but no lag was detectable in the reaction initiated by the addition of ATP to the preformed MDA5:dsRNA complex (dsRNA→ATP) (Fig. 4C). Consistent with the notion that the lag is caused by slow RNA binding, the duration of the lag phase was dependent on the concentration of dsRNA but not on its length (Fig. 4D and Fig. S5A). In contrast, RIG-I exhibited no significant kinetic delay in either the ATP→dsRNA or dsRNA→ATP reaction at any of the RNA concentrations tested (Fig. S5 B and C). This result suggests that RNA binding is the rate-limiting step in the ATP hydrolysis reaction of MDA5 but not of RIG-I.
To examine whether the dsRNA binding kinetics of MDA5 is dependent on the salt concentration in the reaction condition, we performed pull-down kinetics and ATP hydrolysis (ATP→dsRNA) assays in the buffer containing 100 mM NaCl (as opposed to 150 mM NaCl in reactions described above). We found that reduced ionic strength accelerated RNA binding and decreased the initial lag phase and binding affinity (Fig. 4D and Fig. S6 A and B). We further examined the initial lag phase as a function of the salt concentration between 100–200 mM for both NaCl and KCl. The lag phase increased sharply between ∼125 mM and ∼175 mM NaCl or KCl (Fig. S6C). At salt concentrations above 175 mM, ATP hydrolysis was barely detectable, and the initial lag phase could not be determined reliably. This strong dependence on the salt concentration suggests that the MDA5–dsRNA interaction is primarily electrostatic and that the slow binding kinetics observed with 140–160 mM NaCl/KCl likely recapitulates the binding kinetics of MDA5 in the cell, which contains 139 mM K+ and 12 mM Na+ (25). To be consistent with the known intracellular ion concentration, we used 150 mM NaCl in all of our experiments except when examining the effect of salt concentration.
To examine the importance of slow RNA binding for the length dependence of MDA5, we measured the ATP hydrolysis rate using dsRNAs of various lengths (62–2,012 bp) in a buffer containing 100 mM NaCl in which RNA binding is accelerated. The length dependence of the ATP hydrolysis rate was diminished in 100 mM NaCl; the relative rate of 112 bp vs. 2,012 bp increased from 26% in 150 mM to 61% in 100 mM NaCl (Fig. 4E). Thus, accelerated RNA binding leads to reduced dependence on length, underscoring the importance of slow RNA-binding kinetics in the ATP-dependent discrimination of length by MDA5.
Slow dsRNA Binding Is Caused by a Kinetic Barrier to MDA5 Filament Nucleation.
To understand the slow binding kinetics of MDA5 in the context of filament assembly, we used single-molecule fluorescence microscopy. We immobilized 3′-biotinylated dsRNA on the streptavidin-coated surface of a flow cell, introduced fluorescently labeled MDA5, and monitored the increase in fluorescence using total internal reflection fluorescence (TIRF) microscopy (Fig. 5A). Because of the limited labeling efficiency of MDA5 using sortase (∼15%), we also labeled MDA5 by fusing it to a SNAP tag, a variant of the O6-alkylguanine-DNA alkyltransferase (26), which could be labeled with Alexa 647 at ∼50–70% efficiency (as measured by fluorophore optical densities relative to protein concentration). We found that SNAP-tagged MDA5 (MDA5SNAP) was insoluble, but the SNAP-tagged CARD-deletion mutant (MDA5hSNAP) retained the solubility (Fig. S1) and dsRNA-dependent ATP hydrolysis activity of the untagged MDA5h (Fig. S7A). Because MDA5h also forms filaments (Fig. 1C and Fig. S7B) and binds to dsRNA with similarly slow kinetics as MDA5 (Fig. S7C), we used MDA5hSNAP along with sortase-labeled MDA5 and MDA5h in our single-molecule analyses.
To examine the level of nonspecific binding of MDA5 to the flow cell surface, we injected fluorescently labeled MDA5hSNAP into a flow cell containing immobilized 112-bp dsRNA or an empty flow cell. Unbound MDA5hSNAP was washed out, and a fluorescence image was recorded. Comparison of the MDA5 fluorescence signal showed that both the frequency and the fluorescence intensity of surface-bound MDA5hSNAP were approximately five to seven times higher in the 112-bp dsRNA-coated flow cell than in the empty flow cell (Fig. S8). This result suggests that nonspecific binding of MDA5 is minimal and can be distinguished unambiguously from filaments formed on dsRNA.
Examination of the single-filament assembly traces revealed an initial lag phase of ∼1–7 min and two to five steps of linear elongation alternated by pauses (Fig. 5B). The linear growth curve is consistent with a steady filament elongation, whereas sharp transitions from elongation to pause suggest stalling of filament elongation upon encountering dsRNA ends or neighboring filaments on the same dsRNA molecule (Fig. 5C). Alternations between elongation and pause have been observed for other filamentous proteins, such as RecA and Rad51 (27, 28), and have been interpreted as a consequence of multiple independent nucleation events on the same nucleic acid molecule. To examine the possibility of multiple nucleation of MDA5, we visualized MDA5 filaments by negative-stain EM (Fig. 5D). We used MDA5 in a slightly substoichiometric amount relative to dsRNA to allow visual identification of filament boundaries. We observed occasional gaps in MDA5 filaments that likely represent regions of dsRNA devoid of MDA5 between distinct filaments propagated from independent nuclei (Fig. 5D). This result indicates that the stepwise assembly observed in the single-molecule traces (Fig. 5B) is caused by multiple nucleation events. The insufficiency of a single nucleus to saturate the entire length of dsRNA further suggests unidirectional elongation of MDA5 filaments.
To measure the relative contributions of elongation vs. nucleation kinetics to overall filament assembly, we analyzed the histogram of the elongation rates. To ensure measurement of a single-filament elongation rate, the rates during the first linear growth phase were analyzed from 542 individual traces. The later stages of growth were excluded from the analysis because of a higher probability of concurrent elongation of multiple independent filaments. An average rate of ∼10 arbitrary units (a.u.)⋅s−1 was obtained for 512-bp dsRNA, which corresponds to 5 bp⋅s−1 or 0.35 binding unit⋅s−1 (0.7 s−1) (Fig. 5E), assuming 100% coverage at saturation. A similar rate was observed for 2,012-bp dsRNA, suggesting that the elongation rate is independent of dsRNA length (Fig. 5E). To examine how the rate of MDA5 incorporation changes during the course of assembly, we averaged 542 individual assembly traces synchronized by the initial nucleation point. The average growth curve is hyperbolic with a slower rate of assembly in the later stage of the reaction (Fig. 5F). This slower rate is unlikely to be caused by a decrease in the intrinsic elongation rate. Instead, it likely reflects a shorter duration of each elongation phase because of the higher probability of encountering other filaments on the same dsRNA molecule and a greater requirement for de novo nucleation steps to fill the remaining short, disconnected gaps. Similar averaged traces were obtained for MDA5 and MDA5h labeled by sortase (Fig. 5F).
To examine the nucleation kinetics, we analyzed the histogram of the duration of the initial lag phase in the single-molecule assembly traces. The nucleation time displays a quasi-exponential distribution with a mean value of 223 s (Fig. 5G), corresponding to the nucleation rate of ∼0.004 s−1 at 50 nM MDA5hSNAP. The nucleation time decreases with RNA length (Fig. 5H), indicating nucleation at random positions along dsRNA, consistent with the lack of dsRNA sequence specificity (Fig. 2 A and C). The nucleation time also was strongly dependent on the salt concentration (125–150 mM) in the reaction buffer (Fig. S9), and no filaments were observed at salt concentrations above 175 mM. The observed sensitivity of the nucleation time to the salt concentration is consistent with the bulk kinetic assay (Fig. S6) and again supports the electrostatic nature of the MDA5–dsRNA interaction. Comparison of the nucleation kinetics of MDA5hSNAP and sortase-labeled MDA5h showed similar nucleation rates despite the approximately threefold difference in labeling efficiency (Fig. 5H). This result suggests that rapid filament elongation immediately following nucleation likely compensates for the low labeling level of MDA5h and that estimation of the nucleation rate is insensitive to labeling efficiency. Nucleation of full-length MDA5 is ∼40% faster than that of MDA5h (Fig. 5H) but still is >100-fold slower than elongation (for 512-bp dsRNA), suggesting that nucleation is the rate-limiting step for both MDA5 and MDA5h filament assembly.
MDA5 Dissociates from the Ends of Individual Filaments Grown from Independent Nuclei.
To understand the mechanism for length-dependent dissociation, we monitored the disassembly process using a similar single-molecule approach. We immobilized 3′-biotinylated dsRNA in complex with MDA5hSNAP, washed out unbound MDA5 and RNA, injected ATP to trigger MDA5hSNAP dissociation, and monitored the decrease in fluorescence intensity over time (Fig. 6A). The fluorescence signal decayed rapidly with the addition of ATP, but not with ADPCP or with ATP and EDTA, (Fig. 6A), suggesting that the rapid fluorescence decay is caused by ATP-driven filament disassembly. The ATP-driven dissociation rate was largely independent of the laser power, suggesting that the contribution from photobleaching is minimal (Fig. S10A). Comparison of the dissociation kinetics with 512-, 1,012-, and 2,012-bp dsRNAs showed that the dissociation rate is inversely proportional to the length of dsRNA (Fig. 6B), consistent with the results from the pull-down kinetic assays (Fig. 3C). Furthermore, an internal gap, bulge, or mismatch increased the dissociation rate (Fig. S10B), suggesting the importance of dsRNA continuity for filament stability during ATP hydrolysis. Similar dissociation trends were observed for full-length MDA5 (Fig. S10C), but the rates were ∼40% lower than for MDA5hSNAP.
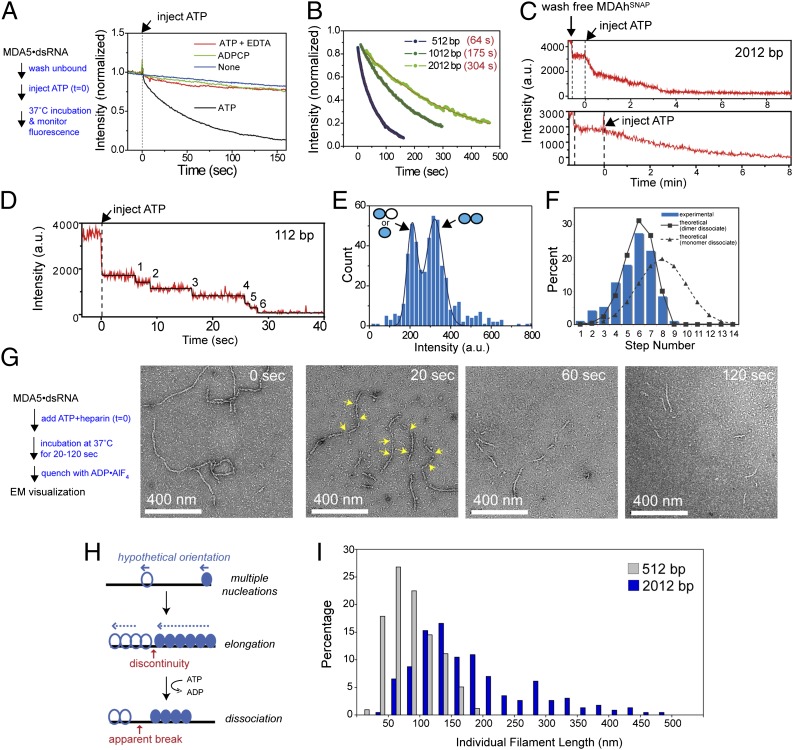
Fig. 6.
MDA5 dissociates from individual filament ends. (A) Averaged traces of filament disassembly reaction. MDA5hSNAP filaments were preformed on 512-bp dsRNA and were immobilized on the flow cell surface, and unbound filaments were washed out. Disassembly reactions were initiated by the addition of 2 mM ATP, ADPCP, or ATP with 0.5 mM EDTA at t = 0. Single-molecule TIRF microscopy was performed as in Fig. 5. n = ∼500–700 for each sample. (B) Averaged traces of MDA5hSNAP dissociating from 512-, 1,012-, and 2,012-bp dsRNAs and their single-exponential fits with time constants (τ) (n = 763, 570, and 258, respectively). (C) Representative traces of Alexa 647-labeled MDA5hSNAP dissociating from surface-immobilized 2,012-bp dsRNA. (D) A representative trace of MDA5hSNAP dissociating from 112-bp dsRNA fitted with a step function (black). (E) Histogram of step intensities during disassembly of 112-bp filaments from 95 independent traces and fitted double Gaussian function (black line). Filled and blank circles indicate labeled and unlabeled MDA5 monomers, respectively. (F) Histogram of the number of step decrements during disassembly of 112-bp filaments from 95 independent traces. Predicted distribution curves (black lines) are derived from a probability theory (SI Materials and Methods). (G) Representative electron micrographs of MDA5 2,012-bp dsRNA filaments during disassembly. Disassembly was initiated by the addition of ATP and heparin and was quenched with ADP⋅AlF4 at the indicated time points before EM (Methods and Materials). Arrows at t = 20 s indicate filament gaps. (H) A model of MDA5 filament disassembly. We propose that individual filaments disassemble independently from their ends, generating apparent internal breaks from regions containing filament discontinuities. (I) Histogram of individual filament lengths at t = 20 s after addition of ATP (n = 415 and 230 for 512-bp and 2,012-bp dsRNAs, respectively). The reaction was performed as in F.
Examination of disassembly traces of individual filaments showed approximately one to five steps of linear decay occasionally alternated with pauses of ∼10–30 s (Fig. 6C). The linear decay indicates sequential end disassembly. Reasons for the occasional short pauses are unclear, but the pauses could be caused by stochastic delays in ATP hydrolysis, as has been seen for RecA and Rad51 filaments (28, 29). To determine the unit of dissociation, we used 112-bp dsRNA because it accommodates approximately seven or eight MDA5 binding units, allowing the counting of individual dissociation events through fitting the trace with step functions (Fig. 6D). Histogram analysis of the step decrement in intensity reveals a bimodal distribution (Fig. 6E), which is expected for dissociation of dimers that are singly or doubly labeled but does not exclude the possible dissociation of individual monomers. The ratio of the median values of the two peaks (210 vs. 321 a.u.) is less than 2, likely reflecting self-quenching of adjacent fluorophores within a filament (30). In addition, the histogram of the number of steps involved in 112-bp filament disassembly showed that disassembly evolves in approximately five to seven stepwise decrements (Fig. 6F), as is consistent with the notion that filament disassembles via dissociation of individual binding units.
To understand the mechanism of the filament disassembly process, and in particular the mechanism by which dissociation kinetics is affected by neighboring molecules and the length of dsRNA, we used EM to visualize the disassembly intermediates. As with the dissociation pull-down assay, preformed filament was incubated with ATP in the presence of heparin trap, and ATP hydrolysis was quenched at discrete time points with ADP⋅AlF4 (Fig. 6G). To capture early disassembly intermediates, we reduced the ATP hydrolysis rate by using a low concentration of ATP (0.25 mM vs. 2 mM) (Fig. S10D). Within 20 s, one to five internal breaks appeared along 2012-bp filaments (Fig. 6G). As the reaction proceeded, fewer segments of filaments were visible, and these filaments had larger gaps or ambiguous connectivity, as is consistent with end disassembly of individual filaments on a single dsRNA (Fig. 6H).
To examine whether end disassembly of individual filaments can explain the length-dependent dissociation rate shown in Fig. 6C, we compared the distribution of individual filament lengths on 512-bp and 2,012-bp dsRNA 20 s after the addition of ATP (Fig. 6I). Both 512- and 2,012-bp filaments showed a distribution of lengths significantly shorter than expected for the respective full-length filaments (∼200 and ∼800 nm, which were the filament lengths observed at saturation), with average lengths of 115 and 193 nm, respectively (Fig. 6I). With 2,012-bp filaments, however, ∼30% of the population showed filament lengths longer than 200 nm, the theoretical limit of a 512-bp filament (Fig. 6I). The presence of longer filaments on longer dsRNAs provides at least a partial explanation for dsRNA length-dependent dissociation kinetics.
ATP Hydrolysis Promotes Formation of Long, Continuous Filaments and Delays Disassembly.
The dissociation experiments discussed above were performed with filaments assembled without ATP. We next asked how ATP hydrolysis during filament assembly, as would occur in the cell, would affect length-dependent dissociation kinetics. Previous studies on RecA revealed that ATP-driven dissociation induces the rearrangement of RecA molecules from multiple independent filaments to form a single, continuous filament (31). Such rearrangement could occur via faster dissociation of shorter filaments, which would allow continued elongation of longer filaments on the same nucleic acid lattice (Fig. 7A). Repair of filament discontinuities would remove small gaps generated between independent filaments and increase the filament-packing density, which can be measured by the fluorescence intensity of filaments at saturation (31). We first asked whether such similar ATP-driven repair also can be observed with MDA5 filaments. We analyzed filament-assembly reactions in the presence of ATP and ADPCP using single-molecule fluorescence assays and compared the averaged traces (Fig. 7B). Net filament assembly occurs more slowly with ATP than with ADPCP because of the contribution of ATP-triggered dissociation during the assembly reaction. However, at completion of assembly, filaments are saturated to a 20% higher level with ATP than with ADPCP (Fig. 7B), suggesting that more continuous filaments form when ATP hydrolysis is allowed during assembly. A similar increase in packing density was observed when ADPCP was replaced by ATP during filament assembly (Fig. S11A). Consistent with the role of ATP in repairing filament discontinuities, filaments formed with ATP dissociated ∼20% more slowly than those formed without ATP (Fig. 7C). To validate further the role of ATP in repairing filament discontinuities, we visualized filament disassembly intermediates using EM. The disassembly intermediates of filaments assembled with ATP contained fewer internal breaks than those assembled without ATP (Fig. 7D). Filaments formed with ATP show an even distribution of length from 100–800 nm, in contrast to the narrow distribution of filaments formed without ATP (Fig. 7E). To examine filament shapes and lengths during ATP hydrolysis steady state, instead of during disassembly reaction, we also visualized filaments in the absence of any quenching reagents (i.e., ADP⋅AlF4) or protein trap (heparin). Similarly continuous filaments with a broad distribution of lengths were observed during steady state (Fig. S11 B and C). These results suggest that, although filament length is affected by both dsRNA length and multiple nucleation events, ATP-mediated repair of filament discontinuities skews the filament length distribution to more accurately reflect the length of the underlying dsRNA scaffold.
Fig. 7.
ATP hydrolysis repairs discontinuities in MDA5 filaments and promotes the formation of longer, continuous filaments. (A) Proposed mechanism by which ATP hydrolysis repairs discontinuities in MDA5 filaments. Multiple filaments propagated from independent nuclei individually undergo cycles of ATP-driven disassembly and elongation, which allow the rearrangement of MDA5 molecules to generate more continuous filaments. (B) Averaged assembly traces of MDA5hSNAP filament on 2,012-bp dsRNA in the presence of 2 mM ADPCP or 0.2 mM ATP using 100 nM MDA5hSNAP. Individual traces were synchronized by the nucleation time (n = 386 and 495 for ADPCP and ATP, respectively). (C) Averaged disassembly traces of MDA5hSNAP filaments initially assembled on 2,012-bp dsRNA with or without ATP (n = 375 and 223, respectively). Disassembly reactions were performed as in Fig. 6C. (D) A representative electron micrograph of disassembly intermediates of MDA5 filaments initially formed with ATP. MDA5 filaments were formed on 2,012-bp dsRNA in the presence of 0.25 mM ATP. Dissociation was initiated at t = 0 by the addition of ATP and heparin and was quenched with ADP⋅AlF4 at t = 20 s. Fewer gaps (arrows) were observed than when filaments were formed without ATP (Fig. 6G). (E) Histogram of individual filament lengths at t = 20 s after the addition of ATP. Filaments were formed initially with ATP (light blue bars, n = 260) as in D or without ATP (dark blue bars) (from Fig. 6I).
Role of CARD in Filament Formation and Dynamics.
The slower disassembly and faster nucleation kinetics of MDA5 filaments in comparison with MDA5h filaments (Fig. 5H and Fig. S10C) prompted the question whether the signaling domain, CARD, contributes to filament formation and dynamics. To address this question, we first examined whether growing filaments can discriminate between MDA5 and MDA5h. We formed filaments using a 1:1 mixture of MDA5 (labeled with Hylite 647) and MDA5hSNAP (labeled with Alexa 546) on 1,012-bp dsRNA and identified a total of 696 filaments displaying fluorescence above the background level (Fig. 8A). Interestingly, only 128 filaments (18%) displayed fluorescence signals from both MDA5 and MDA5hSNAP; the other filaments showed fluorescence signal from either MDA5 (34%) or MDA5hSNAP (48%) (Fig. 8B). The higher number of MDA5hSNAP filaments may reflect the higher labeling efficiency of MDA5hSNAP. The theoretical probability of having a filament formed entirely of either MDA5 or MDA5hSNAP by random binding is nearly zero (2 × 10−22), and thus the above result suggests that MDA5 and MDA5h prefer to recruit molecules of the same kind during filament extension. For the 128 filaments containing both MDA5 and MDA5hSNAP, no relationship was detected between the two fluorescence signals (Fig. 8B), possibly reflecting independent nucleation of MDA5 and MDA5hSNAP filaments on a single dsRNA molecule or the infrequent incorporation of molecules of the other kind during filament extension.
Fig. 8.
Discrimination between MDA5 and MDA5h in filament formation. (A) Fluorescence images of filaments formed by a mixture of MDA5hSNAP (labeled with Alexa Flour 546) and MDA5 (labeled with Hylite647) on 1,012-bp dsRNA. A 1:1 mixture of MDA5hSNAP and MDA5 (100 nM each) was incubated with 1,012-bp dsRNA (0.1 nM), and the complex was immobilized on the flow cell surface. Two-color imaging identified 462 filaments with Alexa 546 fluorescence (Left) and 362 filaments with Hylite 647 fluorescence (Right). Among these, 128 filaments showed common, overlapping positions (such as the one in yellow circle), indicating that these 128 filaments contain both MDA5 and MDA5hSNAP. (B) Venn diagram summarizing the results in A and plot of Alexa 546 vs. Hylite 647 fluorescence intensities of the 128 filaments containing both MDA5 and MDA5hSNAP. (C) Representative traces of MDA5 filament dynamics in the mixture of MDA5 and MDA5hSNAP. Preformed filaments of Hylite 647-labeled MDA5 on 512-bp dsRNA were immobilized on the flow cell surface, and unbound filaments were washed out. A 1:1 mixture of Hylite 647-labeled MDA5 and Alexa 546-labeled MDA5hSNAP (100 nM each) was injected into the flow cell with ATP or ADPCP (2 mM). The fluorescence intensities of MDA5 and MDA5hSNAP were normalized against those of the filaments formed by the respective protein alone. (D) Averaged traces of MDA5 filament dynamics in the mixture of MDA5 and MDA5hSNAP. Experiments were performed as in C. n = 325 and 396 for ATP and ADPCP reactions, respectively. (E) Proposed model for length discrimination by MDA5. With ATP, MDA5 filaments continuously switch between assembly and disassembly phases. Because disassembly occurs primarily from filament ends, filaments on short dsRNA more frequently undergo a complete disassembly, which requires slow de novo nucleation for rebinding. In contrast, filaments on long dsRNA alternate between partial disassembly and fast elongation, bypassing nucleation. Thus the slow nucleation kinetics amplifies the time constant (τ) in Fig. 6B.
We next examined whether this preferential selection of MDA5 and MDA5h also occurs during ATP hydrolysis-mediated filament redistribution. We first formed 512-bp filaments using MDA5 (labeled with Hylite 647), washed out the unbound fraction, injected a 1:1 mixture of MDA5 (labeled with Hylite 647) and MDA5hSNAP (labeled with Alexa 546) together with ATP or ADPCP, and monitored the filament redistribution process using two-color real-time imaging. To take into account the differences in the fluorophore and the labeling efficiency of MDA5 and MDA5hSNAP, the fluorescence intensities of Hylite 647 and Alexa 546 were normalized against those of filaments formed by MDA5 and MDA5hSNAP alone, respectively. As expected, ATP, but not ADPCP, triggered filament disassembly and reassembly, as indicated by the large fluctuation in the fluorescence level of MDA5 (Fig. 8C). However, only a small population of MDA5hSNAP was incorporated into the MDA5 filament, resulting in an occupancy of ∼20% on average in the final filament (Fig. 8D). This result suggests that MDA5 dissociated from the filament is more likely to be replaced by another MDA5 molecule than by MDA5hSNAP and further indicates that CARD plays a positive role in the formation of full-length MDA5 filaments.
Discussion
MDA5, a viral RNA sensor, regulates the level of antiviral signaling proportional to dsRNA length, ranging between ∼0.5 and ∼7 kb (7), through a poorly understood mechanism. We have shown previously that MDA5 forms filamentous oligomers along the length of dsRNA (21). The MDA5 filament is distinct from the beads-on-a-string type of oligomers, as evidenced by its preference for binding to dsRNA adjacent to another MDA5 molecule rather than at random positions and the requirement of filament formation for high-affinity interaction with dsRNA (Fig. 1). Filament formation alone, however, does not explain the observed discrimination of length by MDA5 in the cell. MDA5 forms stable filaments on dsRNA as short as ∼50–100 bp, and in the absence of ATP the intrinsic affinity of MDA5 depends only moderately on dsRNA length beyond ∼100 bp (Fig. 1D). During ATP hydrolysis, however, dsRNA binding displays an increased dependence on length recapitulating the length-dependent signaling observed in the cell. To dissect the molecular mechanisms underlying this ATP-driven length discrimination by MDA5, we used a combination of bulk kinetic assays, single-molecule analyses, and EM, which revealed a complex dynamic interplay between assembly and disassembly processes of MDA5 filaments as a key mechanism for dsRNA length discrimination.
MDA5 filaments disassemble upon ATP hydrolysis (21). Our previous ATP hydrolysis assays indirectly suggested that filament disassembly occurs at a rate inversely proportional to dsRNA length. Here we directly measure the length-dependent dissociation kinetics and demonstrate that filaments disassemble from an end. Filament end disassembly, however, does not involve a simple shrinkage of a single filament on a single dsRNA molecule. Instead, filament assembly occurs via multiple nucleation events, which generate multiple independent filaments on a single dsRNA molecule (Fig. 5). ATP hydrolysis triggers independent disassembly of individual filaments from their ends. This disassembly results in an appearance of internal breaks in the filament, which in fact represent gaps between disassembling individual filaments (Fig. 6).
Surprisingly, we identified a seemingly paradoxical role of ATP in promoting the formation of longer and more continuous MDA5 filaments. During dynamic equilibrium between assembly and ATP-driven end disassembly reactions, short filaments disassemble faster than longer ones, allowing continued elongation of longer filaments to a length that more accurately reflects the length of underlying dsRNA scaffolds (Fig. 7A). This positive role of ATP hydrolysis in MDA5 filament assembly, which was unexpected from its previously known role in effecting MDA5 dissociation, underscores the necessity of understanding MDA5 functions in the context of a continuous dynamic cycle of filament assembly and disassembly rather than a linear model of filament nucleation, elongation, and disassembly.
The effect of dsRNA length-dependent filament disassembly is amplified further by the slow nucleation kinetics. In principle, dissociated MDA5 can rebind to dsRNA either by adding onto a growing end of existing filaments or by forming a de novo nucleus. The latter process is significantly (>100 times) slower than the former, probably because of the instability of single monomers or dimers on dsRNA (Fig. 1D). Because short filaments would dissociate more rapidly during end disassembly, de novo nucleation would be a more predominant mechanism for MDA5 to rebind to short dsRNA (Fig. 8E). On the other hand, slower dissociation of MDA5 from longer filaments would allow MDA5 molecules to rebind via a filament elongation mechanism, bypassing the slow nucleation steps (Fig. 8E). Thus, the two distinct turnover cycles of filaments that are differentially used by short and long dsRNA provide an explanation for the preferential accumulation of MDA5 on long dsRNA during ATP hydrolysis. Because the nucleation rate is critically dependent on MDA5 concentration, this model also explains the more pronounced length bias observed at low concentrations of MDA5 as expected in the resting cell state.
The use of slow nucleation and ATP-driven dissociation as a means to discriminate selectively against short dsRNA is reminiscent of the kinetic proofreading mechanisms used by enzymes in DNA replication and protein synthesis pathways (32). These enzymes use multiple irreversible steps to discard incorrect substrates after ligand binding and to enhance the fidelity of the substrate discrimination far beyond the level predicted from differences in intrinsic affinity. Analogously, dynamic instability of MDA5 filaments necessitates continuous cycles of association and dissociation during its interaction with dsRNA, and slow nucleation makes the dissociation of MDA5 essentially irreversible for short dsRNA. This proofreading mechanism prevents the accumulation of MDA5 on relatively short dsRNA, despite its high intrinsic affinity, and thus avoids the inappropriate activation of antiviral signal by cellular dsRNAs.
How then does the MDA5 filament activate antiviral signaling, and how could the filament dynamics explain the length-dependent signaling? The simplest explanation would be that the level of signaling correlates with the level of MDA5 bound to dsRNA. A recent crystal structure of full-length RIG-I revealed that, in the absence of dsRNA, CARD is masked by intramolecular interactions but becomes released upon dsRNA binding (16). A similar conformational change may occur in MDA5, and exposure of CARD upon dsRNA binding may be sufficient to activate signaling. Because the stability of the MDA5–dsRNA interaction is tightly regulated by filament dynamics, which in turn depend on the length of dsRNA, this mechanism provides an explanation for the length-dependent signaling activity of MDA5. Alternatively, dsRNA binding could be insufficient to activate signaling and additionally might require oligomerization of CARD that possibly is promoted by assembly of the MDA5 filament. We found that full-length MDA5 selectively incorporates full-length MDA5 rather than the CARD-deletion mutant MDA5h (Fig. 8 A–D), and the dissociation and nucleation kinetics of full-length MDA5 also suggest a positive role of CARD in filament formation and stability (Fig. 5H and Fig. S10C). It is tempting to speculate that CARD interacts with another CARD within the filament and promotes antiviral signaling. A recent study suggested that both RIG-I and MDA5 require a K63-linked, free-polyubiquitin chain to oligomerize CARD and activate signaling (33), but it also is possible that filament formation plays an additional positive role in CARD oligomerization. Future investigations will focus on a more detailed examination of potential oligomerization or dynamics of CARD within MDA5 filaments and its detailed mechanism for signal activation via MAVS.
MDA5 exhibits a remarkable parallel with distantly related filamentous proteins in the RecA family and shows a marked contrast to the closely related helicase RIG-I. We showed that RIG-I prefers shorter dsRNAs and binds to dsRNA with no apparent kinetic delay (Fig. S5 B and C) and that ATP hydrolysis does not promote its dissociation from dsRNA (Fig. 3B). In contrast, RecA assembles into filaments along DNA, also with slow nucleation kinetics, and disassembles from filament ends upon ATP hydrolysis (34). RecA uses similar filament dynamics to choreograph DNA pairing and strand-exchange reactions during homologous recombination. Our current findings of MDA5 filament dynamics thus provide a framework for understanding MDA5 functions and mechanisms that are distinct from RIG-I and offer an intriguing example of the utilization of the kinetic principles used by the RecA family to function instead as a molecular ruler for antiviral defense.
Materials and Methods
Procedures for material preparation and protein and RNA labeling are described in SI Materials and Methods.
Competition Binding Assay.
EMSA was performed to monitor complex formation of MDA5 with labeled dsRNA in the presence of competitor dsRNAs. A mixture of 3′-fluorescein–labeled 112-bp RNAs (0.18 μg/mL) and unlabeled competitor dsRNAs (0.06–43.74 μg/mL) was incubated with MDA5 (80 nM) in buffer A [20 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2 and 2 mM DTT] with 2 mM ADPCP or ATP at 37 °C for 10 min and analyzed by Bis-Tris native PAGE (Life Technologies). Fluorescence gel images were recorded using a GE FLA9000 scanner.
ATP Hydrolysis Assay.
The ATP hydrolysis assay was performed as previously described (21). Unless otherwise stated, MDA5 (0.3 μM final concentration) was preincubated with 2 mM ATP in buffer A at 37 °C for 3 min, and the reaction was initiated by addition of prewarmed RNA (4.8 μg/mL final concentration), and the reaction was quenched with 50 mM EDTA on ice.
Pull-Down Kinetic Assay.
For dissociation kinetics, fluorescein-labeled MDA5, MDA5h, or RIG-I (0.6 μM) was preincubated with 3′-biotinylated dsRNA (2 μg/mL) in buffer A. Dissociation was initiated by the addition of 200 μg/mL heparin and 2 mM ATP at 37 °C, and the reaction was quenched with 2 mM ADP⋅AlF4 at discrete time points. For binding kinetics, labeled MDA5 (0.3 μM) was incubated with 3′-biotinylated dsRNA (0.6 μg/mL) at 37 °C in buffer A with 2 mM ADPCP for the indicated time periods. The binding reaction was quenched with 60 μg/mL heparin and 2 mM ADP⋅AlF4 and was incubated with streptavidin magnetic beads (New England BioLabs). Beads were washed three times with buffer A and eluted with SDS sample buffer before gel analysis. See SI Materials and Methods for details.
Single-Molecule Fluorescence Assay.
3′-Biotinylated dsRNA (40 pM) was immobilized on the polymer-coated quartz surface of a microfluidic flow cell prepared as previously described (35). For assembly reactions, MDA5, MDA5h, or MDA5hSNAP (50 nM) labeled with Alexa 647 or Hylite 647 in buffer A with 2 mM ADPCP was introduced into the flow cell at t = 0. The assembly reaction was monitored at 37 °C using TIRF microscopy at 0.5–50 Hz with an exposure time of 50 ms. For disassembly reactions, labeled MDA5 or MDA5hSNAP (0.2 μM) in complex with dsRNA (40 pM) was immobilized on the surface of a flow cell. Free protein and RNA were rinsed out using buffer A before injection of ATP (2 mM unless mentioned otherwise) at t = 0. See SI Materials and Methods for details.
Electron Microscopy.
Unless stated otherwise, MDA5 (0.3 μM) and dsRNA (1.2 μg/mL) were preincubated at 37 °C for 8 min to assemble filaments. The filament disassembly reaction was initiated by the addition of a mixture of ATP (0.25 mM) and heparin (120 μg/mL) to preformed filaments at 37 °C and was quenched at discrete time points with 2 mM ADP⋅AlF4 on ice. Immediately after quenching, samples were spotted on carbon-coated grids (Ted Pella) and stained with uranyl formate as described previously (36). Images were collected using a Philips CM10 transmission electron microscope, and filament lengths were measured by using the freehand line option in the program ImageJ (National Institutes of Health).
Supplementary Material
Acknowledgments
We thank S. Harrison for rotavirus particles. A.P. and B.W. are recipients of postdoctoral fellowships from GlaxoSmithKline. T.W. is an Investigator in the Howard Hughes Medical Institute. S. Hohng is the recipient of Grants 2009-0081562 from the Creative Research Initiatives (Physical Genetics Laboratory) and R31-10032 from the World Class University program of the National Research Foundation of Korea. S. Hur is the recipient of a New Investigator Award from Massachusetts Life Sciences Center and a Pew Scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 19884 (volume 109, number 49).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208618109/-/DCSupplemental.
References
- 1.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang HR, et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14(6):1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nallagatla SR, et al. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318(5855):1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 7.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17(7):781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, et al. The structural basis of 5′triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18(8):1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baum A, Sachidanandam R, García-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010;107(37):16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard JN, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci USA. 2008;105(1):258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12(11):5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22(1):41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29(2):169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 15.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147(2):409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 20.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peisley A, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108(52):21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31(7):1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antos JM, et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J Am Chem Soc. 2009;131(31):10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37(Pt 2):343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 25.Lodish H, et al. 2000. Intracellular ion environment and membrane electric potential. Molecular Cell Biology (W. H. Freeman, New York), 4th Ed, Section 15.4.
- 26.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21(1):86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 27.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35(21):7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinstein E, et al. Single-molecule studies of the stringency factors and rates governing the polymerization of RecA on double-stranded DNA. Nucleic Acids Res. 2011;39(9):3781–3791. doi: 10.1093/nar/gkr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Mameren J, et al. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457(7230):745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wendt H, Berger C, Baici A, Thomas RM, Bosshard HR. Kinetics of folding of leucine zipper domains. Biochemistry. 1995;34(12):4097–4107. doi: 10.1021/bi00012a028. [DOI] [PubMed] [Google Scholar]
- 31.van Loenhout MTJ, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37(12):4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36(6):959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha T, Kozlov AG, Lohman TM. Single-molecule views of protein movement on single-stranded DNA. Annu Rev Biophys. 2012;41:295–319. doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]