Abstract
Selection of aptamers from nucleic acid libraries by in vitro evolution represents a powerful method of identifying high-affinity ligands for a broad range of molecular targets. Nevertheless, a sizeable fraction of proteins remain difficult targets due to inherently limited chemical diversity of nucleic acids. We have exploited synthetic nucleotide modifications that confer protein-like diversity on a nucleic acid scaffold, resulting in a new generation of binding reagents called SOMAmers (Slow Off-rate Modified Aptamers). Here we report a unique crystal structure of a SOMAmer bound to its target, platelet-derived growth factor B (PDGF-BB). The SOMAmer folds into a compact structure and exhibits a hydrophobic binding surface that mimics the interface between PDGF-BB and its receptor, contrasting sharply with mainly polar interactions seen in traditional protein-binding aptamers. The modified nucleotides circumvent the intrinsic diversity constraints of natural nucleic acids, thereby greatly expanding the structural vocabulary of nucleic acid ligands and considerably broadening the range of accessible protein targets.
Keywords: SELEX, pseudoknot, modified nucleotide
Since the advent of SELEX (Systematic Evolution of Ligands by EXponential enrichment) 22 years ago (1, 2), aptamers have been described that bind specifically and with high affinity to many different types of targets, including proteins, peptides, and small molecules (3). Binding interactions between aptamers and their targets are characterized by shape complementarity, polar contacts, hydrogen bonding interactions, and charge–charge interactions (3–5). Other than base stacking interactions, hydrophobic contacts, which are known to make key contributions to protein–protein interactions (6–8), have been notably limited, reflecting the lack of such moieties in nucleic acid libraries typically used in SELEX.
We have recently shown that augmenting the diversity of randomized libraries with functional groups absent in natural nucleic acids can dramatically improve the success rate of SELEX, especially against difficult protein targets (9, 10). We have named this unique class of binding reagents SOMAmers (Slow Off-rate Modified Aptamers), to account for their distinct composition and binding properties. Among the different types of modifications we have tested, functional groups with hydrophobic character have typically yielded SOMAmers with the highest binding affinity. Although the contribution of such functional groups to the outcome of SELEX experiments has been quite apparent (9), the structural basis for the effect of these “side chains” on folding and binding has been unclear. Here, we report two cocrystal structures of related SOMAmers bound to a protein target, platelet-derived growth factor B (PDGF-BB), solved at a resolution of 2.2 Å and 2.3 Å. The structures elucidate the striking impact of the hydrophobic aromatic functional groups in creating novel intramolecular motifs and their extensive participation in shaping the contact surface with the native protein. By combining nucleic acid secondary structure elements (hairpins, pseudoknots) with protein-like features (hydrophobic patches), SOMAmers enhance the vocabulary of structural biology and suggest the next frontier in the area of directed evolution to create unique high-affinity reagents.
Results
Selection and Optimization of PDGF-BB SOMAmers.
Platelet-derived growth factors (PDGF-A, -B, -C, and -D) are ubiquitous mitogens and chemotactic factors for many connective tissue cells (11). PDGFs occur as disulfide-linked dimers that exert their biological effects by dimerizing their cognate tyrosine kinase receptors, leading to autophosphorylation and signal transduction (12). SOMAmers were selected to bind PDGF-BB using SELEX conditions that favor slow dissociation rates, starting from a DNA library composed of 40 randomized positions containing benzyl-dU (Bn-dU), -dA, -dC, and -dG. Following eight rounds of selection, the affinity-enriched pool contained multiple copies of a high-affinity clone and its variants. Systematic truncation led to a 29-nucleotide core sequence (SL1) that retained full binding activity (Fig. 1A). SL1 exhibited high-affinity binding to PDGF-BB (Kd = 20 pM) and PDGF-AB (Kd = 60–80 pM) but ∼100-fold weaker binding to PDGF-AA (Fig. 1A and Fig. S1 A and D). Binding to the more distantly related PDGF-CC and PDGF-DD isoforms was essentially undetectable (Fig. S1D). Reducing the disulfide bonds of PDGF-BB with DTT diminished binding affinity by >100-fold, showing that SL1 selectively recognizes the native conformation of its target. Overall, SL1 binds to PDGF-BB with affinity that compares favorably to previously reported PDGF aptamers (13, 14). SL1 potently inhibits PDGF-BB–induced phosphorylation of PDGF β-receptors (PDGFRβ) in cultured human fibroblasts (Fig. 1A and Fig. S1B).
Fig. 1.
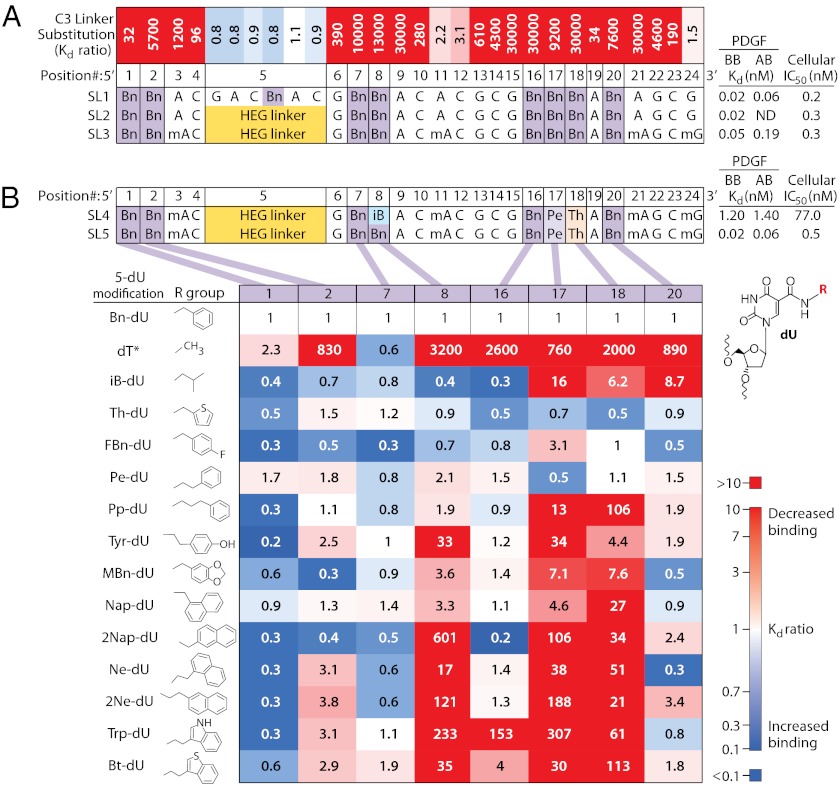
Optimization of PDGF-BB SOMAmer. (A) C3-linker substitution scanning is expressed as the ratio of Kd values (substituted/unsubstituted; top row) with each single C3 substitution compared with SL1. Kd ratios of >1 (red) and <1 (blue) denote a decrease or increase in binding affinity, respectively (see color scale bar in B). Sequences for several variants are shown, along with Kd values to PDGF-BB and PDGF-AB and the IC50 value in a cellular phosphorylation assay (Materials and Methods and Fig. S1). Structures of 5-modified dU residues are shown in B. (B) The eight Bn-dU residues in SL3 were systematically substituted with alternate 5-position modifications of dU, and Kd ratios (compared with SL3) for each substitution are expressed as described in A. Asterisk on dT denotes a direct link of the methyl group to the 5-position. A, dA; Bn, benzyl-dU; C, dC; G, dG; HEG, hexaethylene glycol; iB, isobutyl-dU; mA, 2’-O-methylA; mG, 2’-O-methylG; Pe, phenethyl-dU; Th, thiophene-dU.
To assess the importance of individual nucleotides in SL1, we systematically replaced each nucleotide with a flexible linker (C3 spacer) that lacks both the sugar ring and the base but maintains the same number of carbons between phosphates (Fig. 1A). At most positions, even a single substitution was extremely deleterious to PDGF-BB binding. This linker-scan also revealed several nonessential positions, including the 3′ terminal dG residue, a six-nucleotide stretch d(GAC[Bn-U]AC) near the 5′ end, and a d(AC) dinucleotide in the center. This internal hexanucleotide could be replaced with a single hexaethylene glycol (HEG) linker (SL2, Fig. 1A). We also identified four positions that could accommodate 2′-O-methyl substitutions, resulting in the considerably shorter and more nuclease-resistant variant SL3 (Fig. 1A).
Modified Nucleotide Structure Activity Relationship and Affinity Maturation.
To examine the contribution of each of the eight benzyl side chains to binding, we performed another series of systematic point substitutions by chemically synthesizing 5-position variants with a custom-made library of modified dU phosphoramidites. For this purpose, we designed a library to allow us to probe the microenvironment of each of the positions by varying the size, polarity, disposition of H-bond donors and acceptors, linker length, and orientation of the 5-position substituents. In choosing the functional groups for this analysis, we aimed to include variations on a theme of the original modification (in this case, the benzyl group), amino acid side chains overrepresented in complementarity determining regions (CDRs) of antibodies (such as tryptophan and tyrosine) (15, 16), and “privileged” fragments of small-molecule drugs (17). In a sense, we endeavored to combine elements of affinity maturation in antibodies and structure–activity relationship optimization in medicinal chemistry. Although we used a single modified nucleotide during SELEX, post-SELEX optimization is constrained only by the synthetic accessibility of the modified monomers and compatibility with solid-phase synthesis.
The effect of individual substitutions of the benzyl group with 14 alternative moieties at the 5-position is summarized in Fig. 1B, with relative affinities expressed as dissociation constant ratios. Substitution with dT, which only has a methyl group at the 5-position, represents the most drastic change, and in that sense is comparable to alanine scanning mutagenesis in proteins (18). Not surprisingly, this was the least tolerated substitution at six of the eight modified nucleotide positions. The exceptions were nucleotides 1 and 7, where this substitution was well tolerated. These two positions also tolerated many other substitutions, with some replacements yielding up to fivefold improvement in binding affinity (Fig. 1B). In contrast, nucleotides 8, 17, and 18 exhibited the highest sensitivity to changes. The best single substitutions were then combined, yielding additional variants including SL4 and SL5 (Fig. 1B). SL5, which combined phenethyl-dU (Pe-dU) at nucleotide 17 and thiophene-dU (Th-dU) at nucleotide 18, showed excellent binding to both PDGF-BB and PDGF-AB (Fig. 1B). It is worth noting that the affinity of the originally selected SOMAmer was already so high (Kd = 20 pM) that it approached the detection limit of the binding assay, so it is possible that the degree of affinity improvement is underestimated. We have applied similar post-SELEX optimization strategies to other SOMAmers with weaker initial binding (e.g., Kd values ranging from 100 pM to >10 nM) and have observed affinity improvements of up to 100-fold.
Three-Dimensional Structure of the SOMAmer:PDGF-BB Complex.
Aside from affinity optimization, post-SELEX synthesis of variants of the original parent sequence generates a rich pool of ligands for high-throughput crystal hit identification. After initial crystallization and small-scale (10–15 nmol) optimization trials, SL4 and SL5 produced crystals with a rodlike crystal habit that resulted in synchrotron X-ray diffraction data sets at 2.3 Å (for SL4) and 2.2 Å (for SL5). The structure of the SL5:PDGF-BB complex was solved by molecular replacement, using truncated coordinates of PDGF-BB as the search model (Materials and Methods and Table S1 and Fig. S2).
Structure Overview.
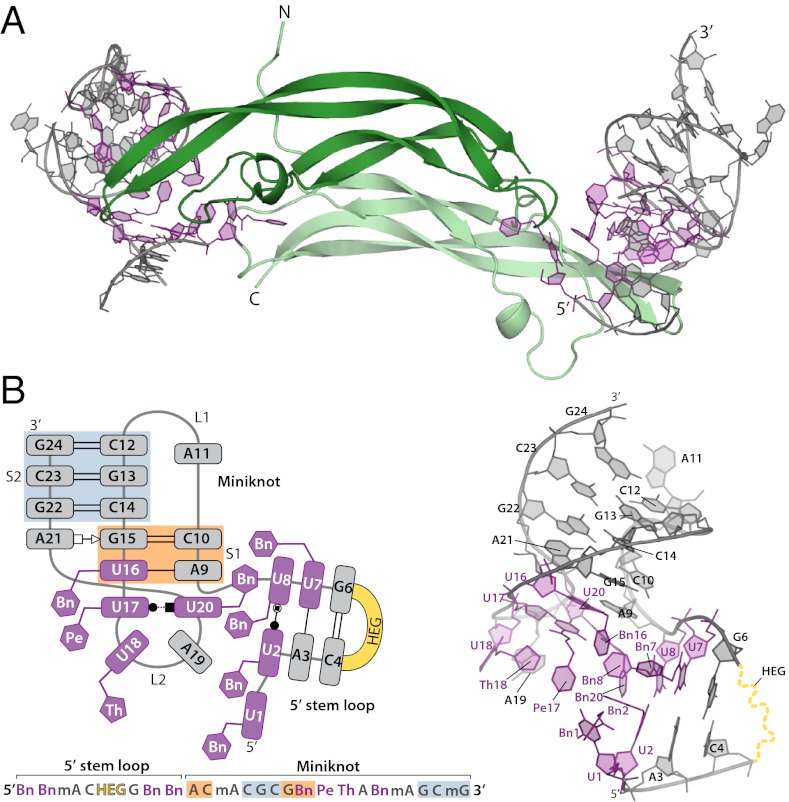
The monomeric subunits of PDGF-BB form twisted β-sheets that dimerize in an antiparallel orientation characteristic of the cystine knot family of proteins (19). SL5 binds two homologous sites at either end of the long axis, crossing the homodimer interface and contacting each of the three PDGF loops (Fig. 2A). The SOMAmer is composed of two domains connected by a network of hydrophobic aromatic interactions (Fig. 2B). At the 5′ end, a short stem is capped with a HEG loop (disordered in the crystal structure), whereas the remainder of the molecule folds into an extraordinarily small H-type pseudoknot (20), with modified nucleotides clustering at the stem loop/pseudoknot junction. Remarkably, all eight modified nucleotides are in contact with PDGF. Seven modified nucleotides cluster together along a hydrophobic groove on the protein, whereas Bn-dU1 adopts an extended conformation, following a channel at the PDGF homodimer interface. Two natural nucleotides also contact PDGF, and the remaining natural nucleotides contribute to internal structure (Fig. 2 and Fig. S3).The secondary structure elements of SL5, a stem loop and a pseudoknot, are well-known nucleic acid structural motifs. However, replacement of certain conventional bases with modified nucleotides offers novel functional groups for alternative interactions. This distinguishing feature of SL5 results in an extensive hydrophobic surface for protein binding as well as unique intramolecular contacts between canonical and modified nucleotides.
Fig. 2.
SOMAmer structure overview. (A) PDGF-BB homodimer bound to SOMAmer SL5. Dark and light green are PDGF-B chains 1 and 2, respectively. SOMAmers are shown in gray, with modified nucleotides highlighted in purple. (B) Schematic representation of SL5 showing backbone trace and base pairing patterns seen in the cocrystal structure (Left; color scheme as in A; miniknot stem 1, orange box; miniknot stem 2, blue box; see Fig. 1B for structures). Base pairs are coded according to the nomenclature of Leontis and Westhof (48). Stick/cartoon view of SOMAmer SL5 with colors and approximate orientation to match the schematic on the left (Right).
Miniknot: Smallest Known H-Type Pseudoknot.
Although the 3′ end of SL5 exhibits hallmark characteristics of an H-type pseudoknot (21), this categorization understates the unconventional nature of this signature “miniknot” motif. Compared with the structure of the smallest reported H-type pseudoknot, which requires 21 nucleotides (22), the SL5 miniknot consists of a mere 16 nucleotides (Fig. 2B). Furthermore, deletion of the terminal mG24:dC12 base pair of stem 2 (S2) results in undiminished binding affinity (Fig. S4C), demonstrating the functional integrity of a 14-nucleotide miniknot. With unprecedented backbone twists and stacking interactions, the miniknot represents a distinct pseudoknot variant in which unusually small size is attained via stabilization contributed by packing of the hydrophobic moieties of the modified nucleotides.
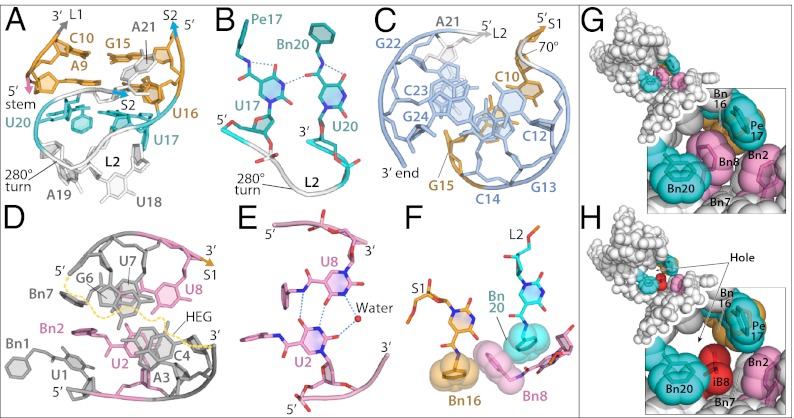
Miniknot stem 1 (S1) formally consists of just two Watson–Crick base pairs (Fig. S5A), whereas loop 2 (L2) is composed nominally of five bases: Pe-dU17, Th-dU18, dA19, Bn-dU20, and mA21. Although interactions between L2 and S1 are a defining feature of pseudoknots, they are typically limited to H-bonding. In contrast, the SOMAmer miniknot makes atypical loop-to-stem stacking interactions, supported by unconventional base pairing. In particular, S1 is stabilized by stacking with a noncanonical Bn-dU17:Bn-dU20 base pair derived from L2 (Fig. 3A and Fig. S5B), effectively creating a three-base-pair S1 with a novel backbone discontinuity. In contrast to previously described U:U imino carbonyl base pairs, the Pe-dU17:Bn-dU20 base pair uses a single H-bond between N3 of Bn-dU17 and the carbonyl oxygen in the amide linker of Bn-dU20 (Fig. 3B). The syn conformation about the glycosyl bond of Bn-dU20 impedes H-bonding with Bn-dU17 but allows Bn20 to stack with Bn-dU8 base without steric clashing with the sugar of Bn-dU8. The unconventional Pe-dU17:Bn-dU20 base pair is made possible by a 280° turn in the backbone between nucleotides 18 and 20 (Fig. 3A). This dramatic strand reversal allows the Bn-dU20 base to stack with the sugar of dA9 and form a hydrogen bond with Pe-dU17. Importantly, the Pe-dU17:Bn-dU20 base pair derives additional stabilization through hydrophobic interactions conferred by the modified nucleotides; the ethylene (linker) portion of the Pe-dU17 side chain is directed toward Bn16 (CH::π), whereas its benzyl group is stacked in π-π edge-to-face interactions with Bn2 and Th18 (Fig. S5C). One additional interaction between L2 and S1 is a base triple (mA21:dG15:dC10; Fig. S5E), a recurrent motif in pseudoknots (23). This is the only long-range tertiary interaction in SL5 that does not involve the modified nucleotides.
Fig. 3.
Modified nucleotides facilitate unique structural features of the PDGF SOMAmer. (A) S1 and L2 side view. Color scheme for bases and backbone is as follows. Miniknot stem 1: Watson–Crick base pairs, orange; noncannonical base pair, cyan; loop 2, white; stem 2, blue; 5′ stem loop: Watson–Crick base pairs, gray; noncannonical base pair, pink. (B) Detail of noncanonical base pair between Pe-dU17 and Bn-dU20. (C) Axial view of S2. (D) Axial view of the 5′ stem domain highlights deviation from B-form DNA (Table S2). (E) Noncanonical base pair between Bn-dU2 and Bn-dU8. (F) Bn8 makes edge-to-face π-π interactions with Bn16 and Bn20 that define the topology of the interdomain junction. The effect of a single modified nucleotide substitution at the junction is evident in the space-filling images of SOMAmer SL5 (G) versus SL4, where iB8 group is shown in red (H).
Loop 1 (L1) consists of a single extruded nucleotide, mA11, that allows the backbone to make a tight 94° turn, with the intrastrand phosphate distance between mA11 and dC12 compressing to just 5.9 Å (Fig. S5F). H-type pseudoknots often have one or two nucleotides in L1, which typically form hydrogen bonds with S2 and stack into the helical junction (22, 24). The extruded L1 nucleotide is necessary to keep the structure condensed so that the 5′ stem domain can interface with the miniknot through the hydrophobic moieties of the modified nucleotides. As expected, the extruded base is not conserved (Fig. S4) and can be replaced with a single C3 spacer (Fig. 1A); however, its deletion abrogates binding, presumably due to interference with the miniknot formation.
The Watson–Crick base pairs of miniknot S1 (dA9:Bn-dU16, dC10:dG15) assemble by the favored H-type pseudoknot arrangement in which strand one of S2 leads directly into strand two of S1, providing efficient stacking of the stems (25). The three base pairs of S2 are composed entirely of Watson–Crick interactions and form a slightly undertwisted B-form helix (Fig. 3Cand Table S2). This understwisting results in helical parameters that more closely resemble A-form helices, as expected for pseudoknot topology; however, the relevance of these calculations is equivocal, given the short length of the helices in this structure. S2 does not form a conventional coaxial stack with S1 due to severe helical overwinding at the junction (twist angle of 70°) formed by dC10:dG15 of S1 and dC14:dG22 of S2. Continuous stacking of the stems is nevertheless maintained as dC14 stacks with dG15, and dG22 stacks with mA21 from the base triple (Fig. 3C). The extensive helical twist at this junction is necessary to allow mA21 to bridge the major groove of S2 while broadening the minor groove for base triple formation. This configuration is typical in pseudoknots with one or two nucleotides in L1 (22, 24).
Modified Nucleotides Stabilize 5′ Stem and Hinge Between Domains.
The SL5 5′ stem is composed of two Watson–Crick base pairs (mA3:Bn-dU7 and dC4:dG6) and a noncanonical Bn-dU2:Bn-dU8 base pair at the base of the stem (Fig. 3D). The Bn-dU2:Bn-dU8 base pair contains two hydrogen bonds, a typical 4-carbonyl-N3 and a unique 4-carbonyl from Bn-dU2 base to amide linker of Bn-dU8 bond (Fig. 3E). Analysis of related sequences in the affinity-enriched pool shows that the length and base composition of the 5′ stem can change, with the notable exception of the invariant Bn-dU:Bn-dU brace at the base of the stem (Fig. S4 A and B), highlighting the importance of this noncanonical base pair in the overall structure and function of SL5. Stability of the 5′ stem helix is further bolstered by stacking of dU8, Bn20, and Pro82 of PDGF (Fig. S6H). The 5′ stem-loop and miniknot domains of SL5 converge where the backbone makes a sharp 111° bend. Significant twist angles and radial displacement of the base pairs in the 5′ stem (Table S2) result in bases 2–4 and 6–7 having greater stacking overlap (because of helix undertwisting) than in conventional B-form helices, whereas Bn-dU8 base is shifted out and Bn-dU7 base stacks with the amide linker of Bn-dU8 (Fig. 3D). This atypical helix facilitates critical interactions with the rest of SL5 and with PDGF; Bn-dU8 base stacks with Bn20, whereas Bn8 lies perpendicularly between the rings of Bn16 and Bn20 in consecutive π-π edge-to-face interactions. These long-range tertiary interactions define a precise hinge between the miniknot and the stem-loop domains (Fig. 3F and Fig. S5D). The lack of curvature between the first two nucleotides prevents clashing of Bn-dU1 base with Bn2, augmenting stacking of the rings (Fig. 3D). Bn2 sits in the middle of a hydrophobic cluster created by Bn7 and Bn8 (from the 5′ stem) and Bn16, Pe17, and Bn20 (from the miniknot) (Figs. 2B and 3D and Fig. S5D). This hydrophobic cluster contributes to stabilization of the SOMAmer, supported by the observation that SL5 exhibits a melting temperature (Tm) of 64 °C, which is >30 °C higher than its analog that lacks the modified nucleotides (Fig. S7).
Bn-dU8 Is Critical for Junction Topology and Functional Activity.
In addition to SL5, we also solved the structure of SL4, which is identical to SL5 except for the replacement of Bn-dU8 with isobutyl-dU (iB-dU). When iB-dU8 was combined with Pe-dU17 and Th-dU18 in variant SL4, the SOMAmer showed substantially weaker binding (∼20- to 50-fold vs. SL5) and a 75-fold lower in vitro inhibitory activity (Fig. 1A and Fig. S1B). The smaller nonaromatic isobutyl side chain cannot form the energetically favorable π-π edge-to-face stacking seen with the benzyl side chains of Bn-dU20, Bn-dU8, and Bn-dU16 in SL5 (Fig. 3 G and H). This creates a hole in the center of the hydrophobic cluster at the protein interface, effectively unlocking the hinge between the 5′ stem and the miniknot domains. The structural effect of this substitution is directly analogous to a Phe to Leu mutation in the hydrophobic core of a protein. Such protein mutations are well described (26–28) and usually have a significant destabilizing effect. Junctions between secondary structure motifs are known to play a critical role in determining nucleic acid tertiary structure (29).
Despite markedly weaker target-binding affinity, SL4 exhibits a thermal melting profile similar to that of SL5 in the absence of ligand (Tm values of 62 °C and 64 °C, respectively; Fig. S7A). This is consistent with the notion that the cavity and altered junction topology created by iB-dU8 substitution in SL4 destabilizes the protein-binding interface, while leaving the intradomain structures of the SOMAmer intact. The conformations of free SOMAmers in solution may well be very different from those in the complex with the protein, which could also diminish the relationship between Tm and binding affinity. In fact, because the energetic cost of solvating a large hydrophobic surface of the SOMAmer is likely to be substantial, we expect the uncomplexed SOMAmer to collapse around the hydrophobic side chains and adopt a conformation in which they are partially protected from the solvent (30).
Extensive Hydrophobic Interactions Define the SOMAmer:Protein Interface.
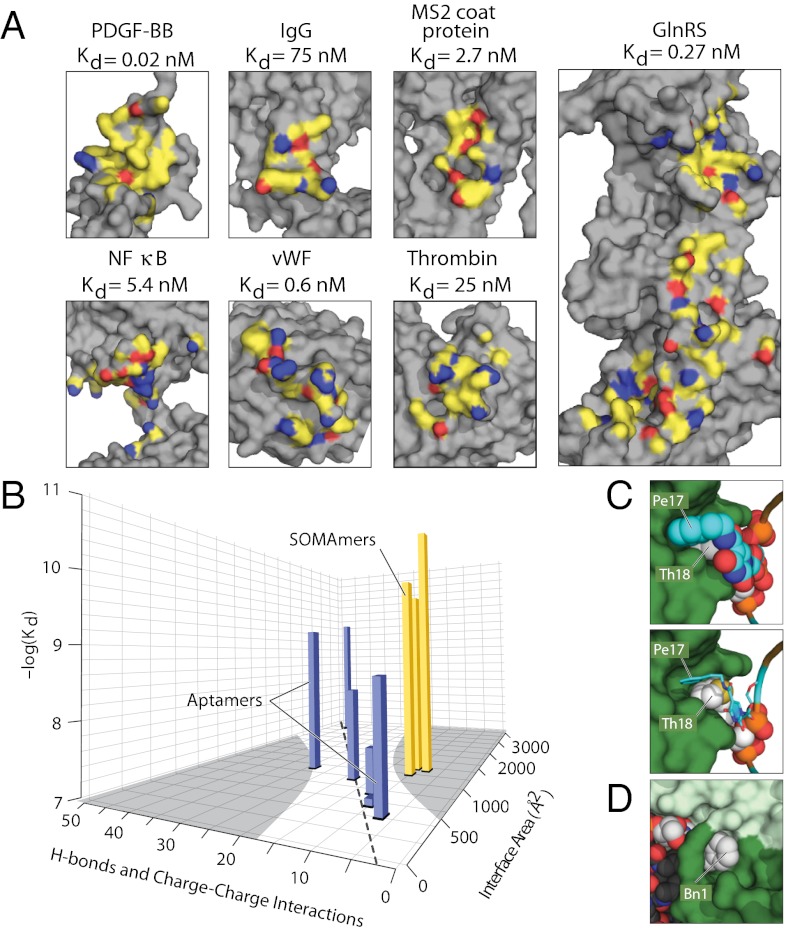
In contrast to previously described protein:aptamer complexes, hydrophobic interactions dominate the interface between SL5 and PDGF (Figs. S3 and S6). Binding to PDGF-BB creates a buried surface area of ∼1,225 Å2 per SOMAmer. The eight modified nucleotides of SL5 create an extensive hydrophobic interface that interacts with 13 nonpolar amino acids of PDGF (Ala35, Phe37, Leu38, Val39, Trp40, Pro42, Cys52, Cys53, Ile75, Ile77, Pro82, Ile83, and Phe84), which account for approximately half of the total nonpolar contacts, with the remainder composed of aliphatic regions of polar or charged amino acids such as Glu24, Arg27, Asn36, Asn54, Asn55, Arg56, Arg73, Lys74, Lys80, Lys85, and Lys86 (Fig. S6). Similar interactions between completely nonpolar residues and nonpolar moeities of charged amino acids are often observed in proteins. Thus, the structural diversity afforded by the modified nucleotides in SOMAmers enables them to mimic the rich repertoire of interactions accessible to proteins. The striking difference in the extent of hydrophobic contacts made by the SOMAmer compared with traditional aptamers is evident when the interface atoms are displayed on the surfaces of the target proteins (Fig. 4A). SL5 exhibits remarkably few polar interactions, having just six H-bonds and one charge–charge interaction with PDGF, despite close proximity to basic amino acids. Relative to the contact surface area, this is significantly lower than what is typical for aptamers. The total number of H-bonds and charge–charge interactions (that is, polar contacts) for six traditional aptamers increases approximately linearly in direct proportion to the interface area (Fig. 4B and Fig. S8A) with a correlation coefficient of 0.91 and an average of 1.9 ± 0.4 polar contacts per 100-Å2 interface area. SL5, as well as two additional SOMAmers in other cocrystal structures, clearly fall outside of the 99% confidence intervals of this trend, with less than half the number of polar contacts per interface area (average of 0.7 ± 0.2 per 100-Å2 interface area), while exhibiting a trend toward higher binding affinities for their targets (Fig. 4B and Fig. S8C). In terms of ligand efficiency (free energy of binding per nonhydrogen contact atom) (31), aptamers and SOMAmers do not appear to be different (Fig. S8C), encompassing a range of values observed with protein-based and small-molecule–based ligands (32). Free energies of binding per interface area are also similar (Fig. S8C). What is different, however, is the value of free energy of binding per polar contact, which is about twice as large for SOMAmers as for aptamers (Fig. S8 B and C), consistent with the notion that SOMAmers derive a larger contribution to binding from hydrophobic interactions.
Fig. 4.
Hydrophobic interactions dominate SOMAmer-target binding interface. (A) Interface contact atoms of PDGF in the SOMAmer cocrystal are compared with six currently available cocrystal structures for aptamer:target complexes with Kd values <500 nM (Fig. S8C). Noncontact atoms are colored gray, and atoms that make ≤4 Å contact with the nucleic acid ligand are colored yellow (nonpolar contacts), red (oxygen participating in a H-bond or salt bridge), or blue (nitrogen participating in a H-bond or salt bridge). (B) Plot of the number of polar contacts (hydrogen bonds plus charge–charge interactions) vs. contact surface area vs. reported binding affinities (also Fig. S8) for the six aptamer-target complexes (blue bars) and for three SOMAmers (yellow bars) including the PDGF:SL5 complex and two other SOMAmer:target structures. The area outside the 99% confidence interval for the polar contacts vs. contact surface area trendline defined by the six aptamers is indicated by gray shading on the floor of the figure. (C) Detailed view of shape complementarity of the PDGF:SOMAmer complex as exhibited by Pe-dU17 and Th-dU18 residues in space-filling representations (Left) or Pe-dU17 in stick representation revealing the contours of the thiophene ring of Th-dU18 (Right). PDGF chains 1 and 2 are shown as dark and light green surfaces, respectively. (D) Detail of Bn-dU1 interaction with PDGF.
Charge–charge interactions often contribute less than 0.2 kcal/mol to the stability of a folded protein (33). In contrast, burying just a single methylene group is estimated to contribute ∼1–1.5 kcal/mol to globular protein stability and/or binding interactions (6, 7). SOMAmer structures reveal a strong reliance on hydrophobic interactions, and in this sense their binding to proteins more closely resembles typical protein–protein interactions. Consistent with this observation, the affinity of SL5 for PDGF shows virtually no decrease across a broad range of salt concentrations (0.1–1.0 M NaCl) or pH values (5.0–8.8), in contrast to the effects seen with traditional aptamers (14, 34).
Post-SELEX optimization facilitates fine tuning of shape complementary and hydrophobic packing interactions. For example, the exceptional shape complementary of Pe-dU17 and Th-dU18 at the protein interface (Fig. 4C) is corroborated with the structure–activity relationships (Fig. 1B). Bn-dU1 also forms a unique interaction with PDGF-BB, with the benzyl ring sitting in a tunnel formed by the Cys43–Cys52 disulfide bond and a salt bridge between Glu24 of PDGF chain1 and Arg56 of chain 2 (Fig. 4D). The crystal structure suggests that the binding pocket can accommodate a variety of side chains, including larger bicyclic substituents, thereby enhancing this point of contact with the protein. Indeed, we have identified several modified nucleotide substitutions at this position that confer 5- to 10-fold enhancement in binding affinity (Fig. 1B).
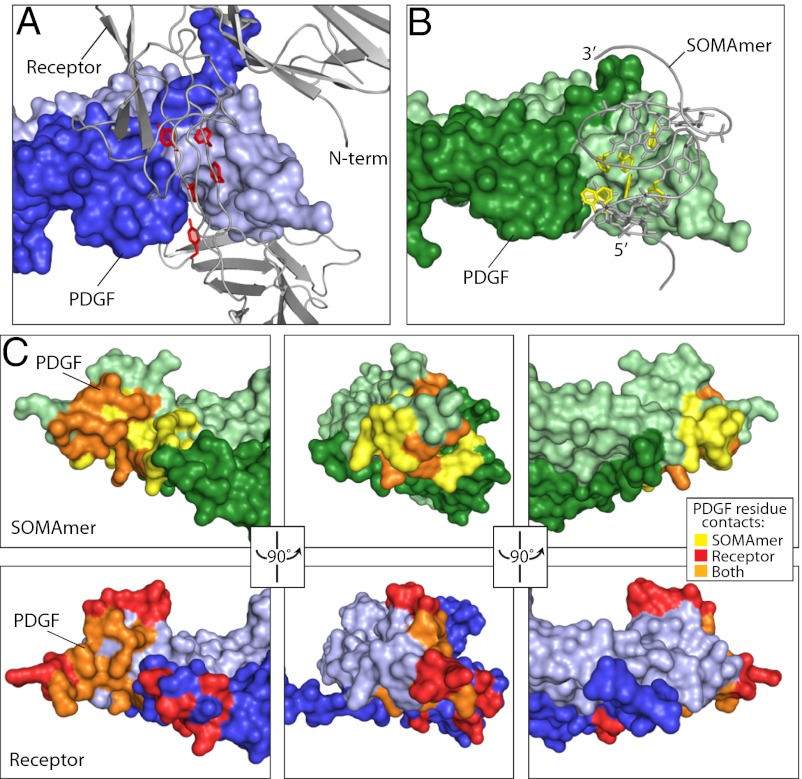
SOMAmer Binding to PDGF Mimics Receptor Binding.
A notable feature of the PDGF-BB:SL5 structure is the degree to which the SOMAmer mimics PDGFRβ. The receptor binds to PDGF primarily through hydrophobic interactions, including seven hydrophobic amino acids at the PDGF interface (35). SL5 binding site largely overlaps that of the receptor with the Bn-dU aromatic rings occupying the same hydrophobic groove on the protein (Fig. 5). PDGF contacts both the receptor and SL5 with 24 residues, of which 10 are shared. These shared, or “promiscuous,” residues likely represent a hot spot of binding energy on the surface of PDGF (32, 36). However, compared with PDGFRβ, SL5 exhibits a 10-fold higher affinity for PDGF-BB (37). Consistent with these observations, SL5 is a potent inhibitor of PDGF-BB (Fig. 1A and Fig. S1).
Fig. 5.
Comparison of SL5 and PDGFRβ binding to PDGF-BB. (A) Receptor cocrystal showing PDGF homodimer (chain 1, dark blue; chain 2, light blue) and receptor extracellular domain colored gray (35), with aromatic rings of hydrophobic amino acids highlighted red. (B) Complex of PDGF homodimer (chain 1, dark green; chain 2, light green) and SL5 (gray), with aromatic rings of modified nucleotides highlighted yellow. (C) Comparison of SL5 and PDGFRβ binding footprints. PDGF is shown in three orientations as a surface representation, with residues that make 4 Å contact only with SL5 or PDGFRβ or both (“overlap” residues) shown in yellow, red, or orange, respectively.
Discussion
Hydrophobic interactions have long been recognized as a fundamental driver of macromolecular stability and complex formation (6, 36, 38). We have shown here that the hydrophobic functional groups of SOMAmers augment the contours of the binding surface and its physicochemical complexity beyond that which can be achieved with natural nucleic acids. This situation is similar to antibodies where relatively limited shape diversity of the canonical main-chain conformations in CDRs (39) is enhanced with amino acids that can sweep out large volumes of space, interspaced with smaller amino acids that allow such motions to take place (15, 16). Indeed, CDR libraries composed of only two amino acids, tyrosine and serine, which are overrepresented in CDRs, have been used to derive specific antibodies with relatively high affinity (40). However, such a minimalist approach is not always sufficient for certain antigens (analogous to difficult targets for aptamers) and additional diversity is needed for high-affinity binding to a larger fraction of epitopes (40).
With traditional aptamers, and for a subset of targets, good success has been achieved to date with relatively limited functional group diversity inherent to natural nucleic acids (compared with proteins), but compensated for by a much larger shape repertoire (compared with antibodies) provided by random libraries of enormous size (∼1015 molecules). Conformational flexibility of nucleic acids is intrinsically far greater than that of proteins because the backbone of each monomer has seven rotatable bonds, compared with two in proteins (φ and ψ). The addition of novel functional groups to nucleic acid libraries results in a unique type of ligand that incorporates the best attributes of two types of biopolymers: the huge conformational repertoire of nucleic acids combined with diversity-enhancing protein-like side chains. This has demonstrably translated into considerable improvements in target binding affinity as well as expansion of protein targets accessible to SELEX (9). The incorporation of various functional groups into nucleic acids has also been used to enhance nucleic acid-mediated catalysis (41, 42). The cocrystal structures described in this report provide a glimpse into the astonishing array of unique structural themes that can be achieved with modifications to standard nucleotide chemistry.
Many protein targets that were previously resistant to aptamer selection have proven amenable to SOMAmer selection (9), and this has allowed us to identify SOMAmers to over 1,000 human proteins (9). We are currently investigating the potential of this nascent technology for applications ranging from biomarker discovery and diagnostics (43, 44) to histochemistry and imaging (45). With the well-established role of PDGF-BB in proliferative disorders including atherosclerosis, fibrosis, macular degeneration, and cancer (11, 46, 47), the SOMAmers described here also have clear potential utility as therapeutic agents.
Materials and Methods
SOMAmers were selected and synthesized by methods described previously (9). PDGF-BB was obtained from Creative BioMart. Crystals of SL4:PDGF-BB and SL5:PDGF-BB diffracted to 2.3 and 2.2Å resolution, respectively, and structures were solved by molecular replacement and refined to final Rwork/Rfree values of 23.9/27.9% and 22.5/26.2%, respectively. Binding assays were performed using radiolabeled SOMAmer and capture of protein-bound SOMAmer by Zorbax beads. Cellular phosphorylation assays were performed in human fibroblasts cells. Results were quantified by phospho-specific ELISA. Sequence analysis of SELEX pools was performed using 454 pyrosequencing data. Detailed materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues at SomaLogic, including Sheri Wilcox, Deborah Ayers, and Andrew Dalby, for performing SELEX; Allison Weiss and Alex Stewart for sequence analysis of the SELEX pool; Dom Zichi for Tm determinations; and Michael Mehan for assistance with data analysis and graphing. We thank the staff of Emerald BioStructures’ macromolecular crystallization core group, particularly Shelly Dieterich, Jeff Christensen, and Jan Abendroth, for their support. We thank Adam Steinberg (artforscience) for assistance in preparation of the figures.
Footnotes
Conflict of interest statement: D.R.D., A.B.B., T.E.E., and L.J.S. are employees and/or shareholders of Emerald BioStructures, Inc. A.D.G., C.Z., J.C.R., J.D.C., D.O., S.M.W., S.K.W., W.S.M., L.G., N.J., and T.C.J. are employees and/or shareholders of SomaLogic, Inc.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HQX and 4HQU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213933109/-/DCSupplemental.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4(8):588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 4.Patel DJ, Suri AK. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J Biotechnol. 2000;74(1):39–60. doi: 10.1016/s1389-0352(99)00003-3. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh G, Huang DB, Huxford T. Molecular mimicry of the NF-kappaB DNA target site by a selected RNA aptamer. Curr Opin Struct Biol. 2004;14(1):21–27. doi: 10.1016/j.sbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Kellis JT, Jr, Nyberg K, Sali D, Fersht AR. Contribution of hydrophobic interactions to protein stability. Nature. 1988;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- 7.Pace CN, et al. Contribution of hydrophobic interactions to protein stability. J Mol Biol. 2011;408(3):514–528. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 9.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaught JD, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132(12):4141–4151. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 11.Östman A, Heldin CH. Involvement of platelet-derived growth factor in disease: development of specific antagonists. Adv Cancer Res. 2001;80:1–38. doi: 10.1016/s0065-230x(01)80010-5. [DOI] [PubMed] [Google Scholar]
- 12.Claesson-Welsh L. Platelet-derived growth factor receptor signals. J Biol Chem. 1994;269(51):32023–32026. [PubMed] [Google Scholar]
- 13.Green LS, et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35(45):14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad KM, et al. Probing the limits of aptamer affinity with a microfluidic SELEX platform. PLoS ONE. 2011;6(11):e27051. doi: 10.1371/journal.pone.0027051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217(1):133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 16.Ramaraj T, Angel T, Dratz EA, Jesaitis AJ, Mumey B. Antigen-antibody interface properties: Composition, residue interactions, and features of 53 non-redundant structures. Biochim Biophys Acta. 2012;1824(3):520–532. doi: 10.1016/j.bbapap.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsch ME, Snyder SA, Stockwell BR. Privileged scaffolds for library design and drug discovery. Curr Opin Chem Biol. 2010;14(3):347–361. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham BC, Jhurani P, Ng P, Wells JA. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243(4896):1330–1336. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- 19.Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11(11):3921–3926. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aalberts DP, Hodas NO. Asymmetry in RNA pseudoknots: Observation and theory. Nucleic Acids Res. 2005;33(7):2210–2214. doi: 10.1093/nar/gki508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3(6):e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonin-Lecomte S, Felden B, Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: New insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34(6):1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I., Jr Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc Natl Acad Sci USA. 2009;106(31):12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michiels PJ, et al. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J Mol Biol. 2001;310(5):1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein DJ, Edwards TE, Ferré-D’Amaré AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16(3):343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadonosono T, Chatani E, Hayashi R, Moriyama H, Ueki T. Minimization of cavity size ensures protein stability and folding: Structures of Phe46-replaced bovine pancreatic RNase A. Biochemistry. 2003;42(36):10651–10658. doi: 10.1021/bi034499w. [DOI] [PubMed] [Google Scholar]
- 27.Lin HJ, et al. Naturally occurring Phe151Leu substitution near a conserved folding module lowers stability of glutathione transferase P1-1. Biochim Biophys Acta. 2003;1649(1):16–23. doi: 10.1016/s1570-9639(03)00149-3. [DOI] [PubMed] [Google Scholar]
- 28.Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19(4):631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyle AM, Shakked Z. The ever-growing complexity of nucleic acids: From small DNA and RNA motifs to large molecular assemblies and machines. Curr Opin Struct Biol. 2011;21(3):293–295. doi: 10.1016/j.sbi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Tzeng S-R, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488(7410):236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 31.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc Natl Acad Sci USA. 1999;96(18):9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 33.Sali D, Bycroft M, Fersht AR. Surface electrostatic interactions contribute little of stability of barnase. J Mol Biol. 1991;220(3):779–788. doi: 10.1016/0022-2836(91)90117-o. [DOI] [PubMed] [Google Scholar]
- 34.Tang Q, Su X, Loh KP. Surface plasmon resonance spectroscopy study of interfacial binding of thrombin to antithrombin DNA aptamers. J Colloid Interface Sci. 2007;315(1):99–106. doi: 10.1016/j.jcis.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 35.Shim AH, et al. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci USA. 2010;107(25):11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 37.Lokker NA, et al. Functional importance of platelet-derived growth factor (PDGF) receptor extracellular immunoglobulin-like domains. Identification of PDGF binding site and neutralizing monoclonal antibodies. J Biol Chem. 1997;272(52):33037–33044. doi: 10.1074/jbc.272.52.33037. [DOI] [PubMed] [Google Scholar]
- 38.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 39.Chothia C, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342(6252):877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 40.Fellouse FA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373(4):924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389(6646):54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 42.Perrin DM, Garestier T, Hélène C. Bridging the gap between proteins and nucleic acids: A metal-independent RNAseA mimic with two protein-like functionalities. J Am Chem Soc. 2001;123(8):1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]
- 43.Ostroff RM, et al. Unlocking biomarker discovery: Large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS ONE. 2010;5(12):e15003. doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehan MR, et al. Protein signature of lung cancer tissues. PLoS ONE. 2012;7(4):e35157. doi: 10.1371/journal.pone.0035157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, et al. Rapid histochemistry using slow off-rate modified aptamers with anionic competition. Appl Immunohistochem Mol Morphol. 2011;19(3):273–278. doi: 10.1097/PAI.0b013e3182008c29. [DOI] [PubMed] [Google Scholar]
- 46.Appelmann I, Liersch R, Kessler T, Mesters RM, Berdel WE. Angiogenesis inhibition in cancer therapy: Platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) and their receptors: biological functions and role in malignancy. Recent Results Cancer Res. 2010;180:51–81. doi: 10.1007/978-3-540-78281-0_5. [DOI] [PubMed] [Google Scholar]
- 47.Trojanowska M. Role of PDGF in fibrotic diseases and systemic sclerosis. Rheumatology (Oxford) 2008;47(Suppl 5):v2–v4. doi: 10.1093/rheumatology/ken265. [DOI] [PubMed] [Google Scholar]
- 48.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7(4):499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.