Abstract
Tailed bacteriophages and herpes viruses use powerful ATP-driven molecular motors to translocate their viral genomes into a preformed capsid shell. The bacteriophage T4 motor, a pentamer of the large terminase protein (gp17) assembled at the portal vertex of the prohead, is the fastest and most powerful known, consistent with the need to package a ∼170-kb viral genome in approximately 5 min. Although much is known about the mechanism of DNA translocation, very little is known about how ATP modulates motor–DNA interactions. Here, we report single-molecule measurements of the phage T4 gp17 motor by using dual-trap optical tweezers under different conditions of perturbation. Unexpectedly, the motor pauses randomly when ATP is limiting, for an average of 1 s, and then resumes translocation. During pausing, DNA is unpackaged, a phenomenon so far observed only in T4, where some of the packaged DNA is slowly released. We propose that the motor pauses whenever it encounters a subunit in the apo state with the DNA bound weakly and incorrectly. Pausing allows the subunit to capture ATP, whereas unpackaging allows scanning of DNA until a correct registry is established. Thus, the “pause-unpackaging” state is an off-translocation recovery state wherein the motor, sometimes by taking a few steps backward, can bypass the impediments encountered along the translocation path. These results lead to a four-state mechanochemical model that provides insights into the mechanisms of translocation of an intricately branched concatemeric viral genome.
Keywords: virus assembly, DNA translocase, optical trap
Tailed bacteriophages are considered to be the most abundant forms of life on earth (1). In late stages of their life cycle, the newly replicated viral genome is translocated into a preformed “head” through a special portal vertex and compacted to near crystalline density (∼500 mg/mL) (2). Powerful molecular motors are used to drive this process, which can generate forces >80 pN to overcome bending and repulsive forces that resist DNA confinement and compaction (3). The phage T4 motor, which packages at a rate of up to ∼2,000 bp/s and has a power density of ∼5,000 kW/m3, is one of the fastest and most powerful molecular motors known (4). The T4 motor is also one of the best studied viral packaging machines, with extensive biochemical characterizations coupled with genetic analyses, atomic structures, and structural modeling (5, 6). It is a prototype for the packaging motors of phages and herpes viruses and belongs to the diverse additional strand, conserved E (ASCE) superfamily of homooligomeric motors, which includes hexameric helicases, protein translocases, and type III restriction enzymes (7).
The T4 packaging motor is a pentamer of the large terminase protein, gp17 (70 kDa), assembled at the portal vertex of the empty prohead (Fig. 1A) (6, 8, 9). It is a key component of the packaging machine that consists of two additional components: the dodecameric portal (61 kDa gp20) that provides the ∼35 Å central channel through which DNA enters and exits (10), and the 11- or 12-meric small terminase (18- kDa gp16), which regulates the three activities of the motor (11): ATPase, nuclease, and translocase. gp17 consists of two domains: an N-terminal ATPase domain that provides energy and a C-terminal translocase domain that moves DNA (Fig. 1A) (12, 13). Based on structural models of gp17, it has been proposed that the motor alternates between two conformational states, a relaxed ATP-bound state in which the domains are separated by ∼7 Å and a tensed state in which ATP is hydrolyzed, bringing the two domains into close contact through complementary charged pair interactions. For each ATP hydrolyzed, approximately 2 bp of DNA are thought to be translocated into the capsid in a piston-like fashion (6).
Fig. 1.
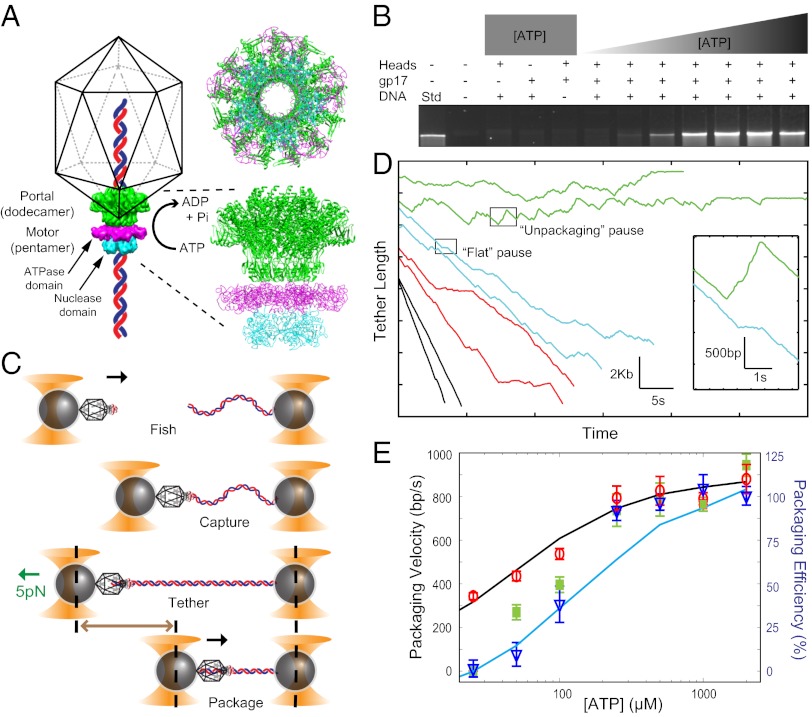
Reducing ATP concentration introduces pauses in packaging. (A) Schematic of the DNA packaging machine assembled at the portal vertex of the capsid (6). It consists of the pentameric gp17 (comprising the ATPase domain in magenta and the nuclease domain in cyan), the dodecameric portal protein [green; the structural model of the dodecameric phage SPP1 portal (32) has been used]. The top and side views of the packaging machine are shown. Schematic not to scale. (B) Results from bulk assays show that packaging efficiency decreases as ATP concentration is decreased. (C) In single-molecule experiments, tethers are formed by approaching the two beads, one containing the immobilized T4 packaging machine and another with tethered DNA, close to each other for ∼1 s in the presence of ATP and moving the beads apart. Tethers are typically stretched to a fixed external load of 5 pN. Schematic not to scale. (D) Representative traces across different ATP concentrations [green, cyan, red, and black correspond to 25 μM (n = 21), 50 μM (n = 17), 100 μM (n = 27), and 1,000 μM (n = 96) ATP, respectively]. Two distinct types of pauses are observed, flat pauses (cyan; Inset) and unpackaging pauses (green; Inset). (E) The packaging velocity after removing pauses (red circles) decreases only modestly with ATP. The velocities with pauses included (green squares) match the packaging efficiencies observed in bulk assays (blue triangles). Error bars represent SD. Kinetic simulations detailed in the text (black line, without pauses; cyan line, with pauses) reproduce the trends observed experimentally.
Although considerable information has accumulated on the functional motifs (14–16) and the basic mechanism of DNA translocation (6, 17, 18), very little is known about how DNA translocation and motor–DNA interactions are modulated by ATP. Here, we report that the phage T4 motor exhibits a surprising structural/functional state, the pause-unpackaging state. This state was observed in single-molecule measurements of T4 motor complexes including the prohead and gp17 (but lacking small terminase gp16) packaging at varying concentrations of ATP and analogs. Our results show that the T4 motor, in contrast to another well-characterized packaging motor from phage φ29 (18, 19), pauses stochastically when ATP fuel is limiting. Importantly, the T4 motor displays behavior when paused that has not been observed in any other motor: “unpackaging,” or controlled release of packaged DNA. Unpackaging is linked to the affinity of the motor subunits to DNA that, in turn, depends on nucleotide occupancy. We show that the ATP-bound state provides the highest affinity with little or no unpackaging, and the apo- or ADP-bound states provide the least affinity with the highest rate of unpackaging. These results implicate a tight coordination between the energy-supplying and translocating domains of the motor. We propose that the motor enters the pause-unpackaging state when an ATP site is unoccupied or incorrectly occupied, putting the bound DNA out of proper register: i.e., disengaged from the motor and unable to be translocated. We propose a minimal model that reproduces the trends observed in our measurements and speculate that the pause-unpackaging state may provide a mechanism by which the T4 motor can respond to various perturbations in its translocation path during packaging of a complex concatemeric genome in vivo.
Results
Limiting the ATP Fuel for the Phage T4 DNA Packaging Motor Induces Pauses.
Defined in vitro packaging assays show that the T4 packaging motor reaches a maximum packaging activity at 1–2 mM ATP (20). Decreasing the ATP concentration reduces the packaging activity, reaching negligible levels at 25 µM ATP (Fig. 1B). This decrease in packaging activity could be due to a reduced DNA translocation rate, a lower efficiency of packaging initiation, or motor stalling. To distinguish between these mechanisms, single-molecule packaging experiments were performed by using dual-trap optical tweezers (21).
In the dual-trap tweezers, one trap held a bead coated with a 10-kb λ DNA molecule while the other held a bead coated with the T4 packaging machine—consisting of the prohead and large terminase gp17 (but lacking the small terminase gp16)—stalled in the presence of ATP-γS (Fig. 1C) (see Materials and Methods for details). In a sample chamber, a single tether was formed between the beads in the presence of ATP by the capture of a free DNA end by the T4 motor assembled at the portal vertex of the empty prohead. Packaging activity was detected from the decrease in the tether length between the two beads as DNA was translocated into the capsid under a constant tension of 5 pN (maintained by using “force–feedback”; Materials and Methods) (Fig. 1C). Packaging traces at different ATP concentrations showed surprising characteristics that have not been observed in other viral motors (18). As ATP concentration decreased, the T4 motor paused frequently (Fig. 1D), a pause defined as any event greater than or equal to 0.1 s during which the translocation velocity was less than 50 bp/s (Materials and Methods). Many of these pauses exhibited unpackaging or slow release of packaged DNA (Fig. 1D, Inset; see below). Unpackaging is a previously unreported property of this motor; prior measurements of T4 at saturating ATP detected pauses but no unpackaging (4). This feature has also not been observed in the φ29 motor under varying ATP concentrations (17, 18).
Pausing and the associated unpackaging became increasingly severe with decreasing ATP concentrations until no net packaging was observed at 25 µM ATP (below this concentration, the frequency of tether formation, an indicator of packaging initiation, was reduced to near zero). Despite this dramatic effect of pauses on packaging, the translocation velocity—i.e., the packaging velocity with pauses removed—decreased only modestly, by a factor of approximately 2 to ∼400 bp/sec at the lowest concentration assayed (Fig. 1E, red circles). The dependence of translocation rate on ATP was well described by simple Michaelis–Menten kinetics, yielding a KM of ∼50 µM and a Vmax of ∼800 bp/s, in reasonable agreement with previous bulk and single-molecule packaging studies and bulk ATPase activity studies (4, 22). The measured KM is also comparable to values determined for other packaging systems (18, 23, 24). Although the net packaging rate including pauses exhibited the same Vmax at saturating ATP, it dropped precipitously to 0 bp/s at 25 µM ATP (Fig. 1E, green squares), indicating that the effect of pausing was prominent only at nonsaturating ATP concentrations. Indeed, the net packaging rates from the single-molecule assays duplicated the trend in packaging efficiencies observed in bulk at different ATP concentrations (Fig. 1E, blue triangles), suggesting that the reduced packaging activity in bulk assays may reflect similar changes in the packaging dynamics: increased pausing and unpackaging rather than an overall reduction in translocation rate, as observed in the φ29 packaging motor (18).
Pauses Are Off-Pathway and an Intrinsic Feature of the Phage T4 Packaging Motor.
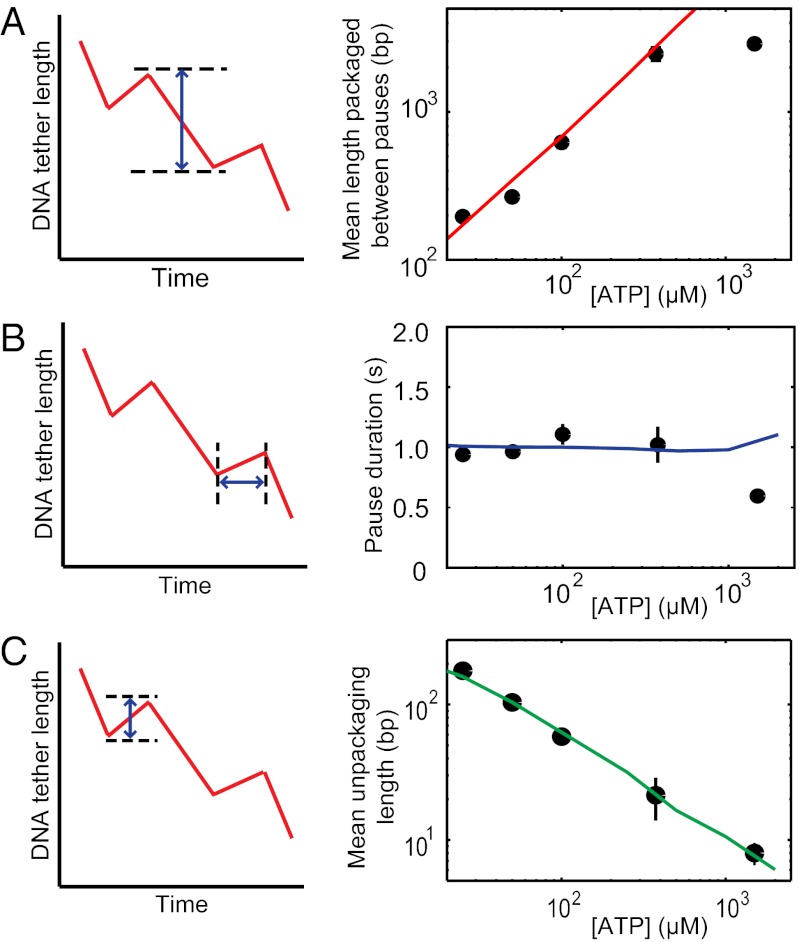
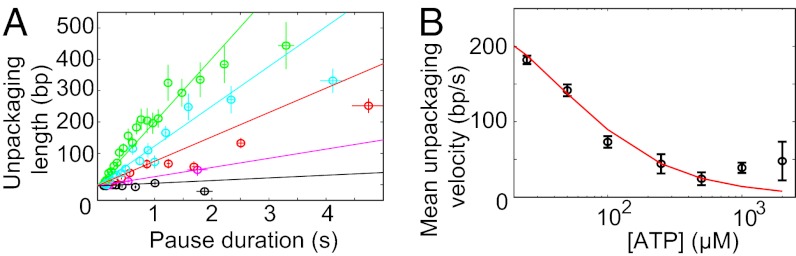
The mean length of DNA translocated between two successive pauses varied in a linear fashion with ATP concentration, ranging from 200 bp at 25 µM ATP to ∼2.7 kb at 250 µM ATP (Fig. 2A). Above 250 µM ATP, the packaged length appeared to plateau to 2.7 kb, consistent with previous measurements of T4 pausing at saturating ATP (2.3 kb; ref. 4). This plateau might represent a basal frequency of pausing by the T4 motor that is independent of ATP. Pausing thus appears to be an intrinsic feature of the T4 packaging motor. First, basal pausing is more prevalent in T4, occurring once in 2.7 kb vs. once in 12 kb in φ29 (3). Second, the ATP-dependent pausing has so far only been observed in T4; for instance, the pause frequency of φ29 motor does not change significantly even at very low ATP concentration (5 µM) (17). Relative to the translocation cycle, which is thought to occur in steps of d = 2 bp per ATP hydrolyzed, pausing is an infrequent event. At the lowest ATP concentration assayed, 25 µM, where there was no net packaging, pauses occurred only once in every ∼100 translocation steps. The rarity of pausing events compared with translocation is indicative of an off-pathway process.
Fig. 2.
Pause characteristics over different ATP concentrations. (A) The mean length packaged between pauses (Left) decreases as ATP concentration is reduced (black circles; Right). (B) The average pause duration (Left) across ATP concentrations remains approximately the same (black circles; Right). (C) The unpackaged DNA length (Left) increases with decreasing ATP concentration (black circles; Right). Error bars represent SD. Kinetic simulations (red, blue, and green lines in A, B, and C, respectively) reproduce the observed experimental data.
Pause Duration Is Independent of ATP Concentration.
The average duration of the individual pauses was on the order of ∼1 s and was largely unaffected by ATP concentration (Fig. 2B). This average duration is far greater than the expected binding time of ATP. The Vmax/KM ratio provides an estimate of the second-order ATP binding rate. Based on our analysis of the translocation rate, Vmax/KM is ∼8 µM-1⋅s−1. Even at the lowest concentration of ATP used (25 µM), an ATP molecule binds to the motor on average every 5 ms. Thus, the duration of a pause event is far longer than can be accounted for by each motor subunit simply waiting to capture ATP, further supporting the hypothesis that pauses are off-pathway kinetic events separate from the translocation cycle of the motor. In addition, the pause durations are exponentially distributed (Fig. S1), indicating that a single rate-limiting step determines the time scale for rescue out of the paused state into the translocation pathway.
Unpackaging During Pausing Is Unique to the Phage T4 Motor.
When paused at low ATP concentrations, the phage T4 motor exhibits unpackaging, which has not been observed in other packaging motors. It is distinct from the relatively rare “slipping” events observed in all motors where hundreds of base pairs of DNA are rapidly expelled from the capsid (4, 19). The average unpackaged length was inversely proportional to the ATP concentration ranging from almost no unpackaging at saturating ATP concentrations to ∼200 bp at 25 µM ATP (Fig. 2C). Although many individual pause events did not show any unpackaging, an increasing number of pauses displayed unpackaging as ATP concentration was decreased, leading to the observed trend in the average. Analyzing individual pause events, a linear correlation was observed between the length of DNA unpackaged and the corresponding pause duration. Longer pauses were observed to unpackage more, indicating a constant rate of unpackaging (Fig. 3A). Furthermore, the unpackaging rate increased with decreasing ATP concentration. Plotting the average unpackaging velocity measured from individual pauses at each ATP concentration also corroborated this dependence (Fig. 3B).
Fig. 3.
Unpackaging depends on ATP concentration. (A) The unpackaged DNA length correlates with the pause duration; long (short) pauses unpackage more (less). Each data point represents the average length of unpackaged DNA from a fixed number (n = 20) of pauses with similar durations. Colors correspond to different ATP concentrations: 25 µM (green), 50 µM (cyan), 100 µM (red), 250 µM and 500 µM (averaged together, magenta), and 1,000 µM and 2,000 µM (averaged together, black). Unpackaging appears to occur at a constant rate (solid lines) dependent on ATP concentration. (B) The mean unpackaging velocity increases as ATP concentration is decreased (black circles). Error bars represent SD. Kinetic simulations (red line in B) reproduce the experimentally observed trend.
The long duration and finite speed of unpackaging pauses distinguish these events from slipping, which results from complete, however transient, disengagement of DNA from the motor (4). Slipping events terminate when the motor quickly reengages and resumes packaging. In contrast, unpackaging appears to involve numerous small DNA release events. In many instances, any unpackaging “steps” are smaller than the spatial resolution of our assay. It is highly unlikely that unpackaging is a reversal of translocation, or active translocation in the reverse direction from packaging, as seen from the fact that the unpackaging speed increases with decreasing ATP, opposite of that expected for an active mechanism. Thus, unpackaging likely involves serial release of DNA, and its mechanism must be strongly linked to the motor’s affinity to DNA. The unpackaging velocity may thus reflect how strongly the motor subunit(s) “holds” the DNA when paused. Furthermore, the ATP dependence of unpackaging highlights an important link between the nucleotide-binding state of the motor and its DNA affinity. Specifically, the data suggest that motor subunits in the apo state are more likely to release DNA, leading to unpackaging.
Unpackaging Velocity Strongly Depends on Applied Force.
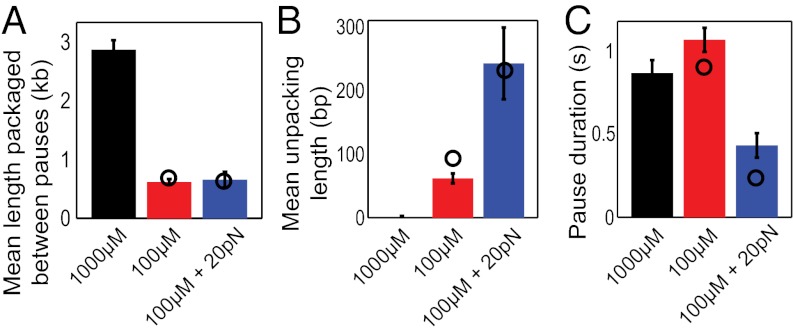
Increasing the applied force from 5 to 20 pN had no discernible effect on the mean length packaged between pauses (Fig. 4A), indicating that entry into the paused state was unrelated to the tension opposing packaging. In contrast, the unpackaging velocity increased almost fivefold (Fig. S2, Inset) and the amount of unpackaged DNA increased greatly at the higher tension (Fig. 4B). This behavior is consistent with a mechanism in which higher tensions disrupt the already weakened motor–DNA interactions, causing the motor subunits’ grip on DNA to be diminished further when ATP is limiting and DNA to be released at a higher rate. Surprisingly, however, the pause durations were significantly shorter at higher tensions (Fig. 4C). Pauses at 20 pN were less than half as long as those observed at 5 pN, indicating that the higher force enabled faster rescue of motor out of the pause state.
Fig. 4.
Higher forces increase unpackaging and enable faster rescue. (A) Packaging against a higher tension of 20 pN (blue bar) does not increase the pausing frequency, because the mean packaged length at 5 pN for the same 100 µM ATP concentration (red bar) remains unchanged. Data at 1,000 µM ATP (black bar) are shown for reference. (B) The mean length of unpackaged DNA increases as force is increased. (C) The mean pause duration decreases as force is increased. Error bars represent SD. Black circles represent results from kinetic simulations.
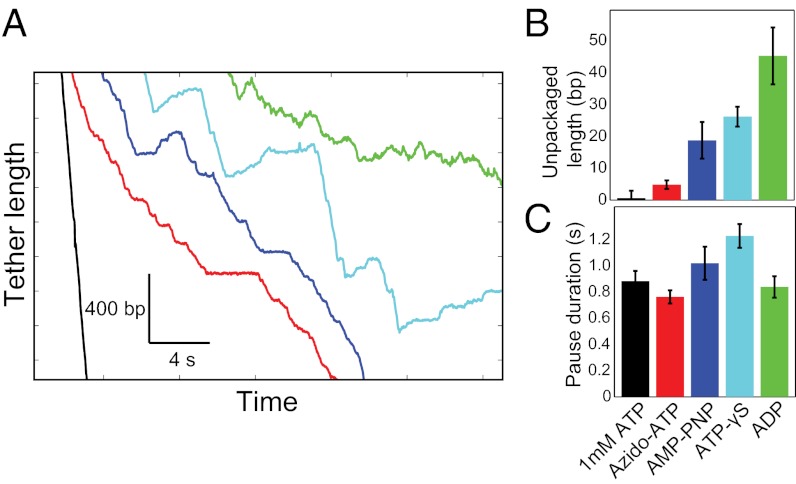
Nonhydrolyzable ATP Analogs Produce Stalled, but Not Unpackaging, Pauses.
Because unpackaging is linked to the lower affinity of the motor subunit(s) to DNA in the apo state, we tested the effect of nonhydrolyzable analogs. ATP-γS or AMP-PNP (100 µM) was added to the packaging buffer containing a saturating concentration of ATP (1 mM), and packaging parameters were quantified. The ATP analogs, as would be expected, also caused the motor to pause (Fig. 5A). However, in contrast to the unpackaging pauses observed at low ATP concentrations, the analog-dependent pauses showed little to no unpackaging (Fig. 5B). The same was observed with the addition of 100 µM of a slowly hydrolyzable analog, 8-azido-ATP, which has been shown to be hydrolyzed 260-fold slower than ATP by the T4 packaging ATPase gp17 (22). However, 100 µM of the product ADP mixed with 1 mM ATP caused unpackaging with an average unpackaged length of ∼50 bp, equivalent to that observed with 100 µM ATP by itself. The pause durations were similar to those observed at low ATP concentrations (Fig. 5C). Taken together, these results demonstrate that the motor enters into two types of pauses during which the motor either unpackages or stalls depending on its nucleotide-bound state.
Fig. 5.
Nonhydrolyzable ATP analogs induce flat pauses. (A) Representative packaging traces with 1 mM ATP (black), or 1 mM ATP mixed with 100 μM of azido-ATP (red), AMP-PNP (blue), ATP-γS (cyan), or ADP (green). (B) The amount of DNA unpackaged depends on the ATP analog. Little unpackaging was observed with most analogs, but the amount of unpackaging observed with ADP was comparable to that observed with 100 µM ATP. (C) The pause durations are similar to those observed across ATP concentrations, on the order of ∼1 s.
Analysis of pausing events under these varying conditions show that the unpackaging velocities fall in the following descending order: apo state (low ATP concentrations) > ADP state > ATP analog state (nonhydrolyzable or slowly hydrolyzable ATP analogs) > ATP state. This ordering indicates that the ATP bound state has the highest DNA binding affinity and the apo state the least affinity. Interestingly, the nucleotide dependence of the motor-DNA affinity matches that deduced in φ29 (18), although unpackaging pauses were not observed in that system.
Discussion
The phage T4 DNA packaging motor has been extensively characterized in recent years, both biochemically and structurally, leading to the proposal of an electrostatic-force driven mechanism for viral DNA translocation (6). A key feature of this mechanism is that the motor exists in two conformational states, relaxed and tensed. In this proposed mechanism, ATP binds to the motor (N-terminal ATPase domain of gp17) in the relaxed state, increasing its affinity to DNA at the C-terminal DNA binding groove. ATP hydrolysis triggers a conformational change pulling the C-domain–DNA complex upwards by 7 Å (tensed state). Two base pairs of DNA are translocated into the capsid, bringing the DNA into register with another subunit of the motor. Although several features of this mechanism have been supported by biochemical evidence, the detailed mechanism by which ATP modulates DNA translocation and motor–DNA interactions remain poorly understood. The unexpected behaviors observed in this single-molecule study identifies a unique state of the motor, the pause-unpackaging state, that provides insights on the motor’s interactions with ATP and DNA in both on- and off-translocation pathways and how these might regulate packaging of a complex viral genome in vivo.
The key distinguishing features of the T4 motor are random pausing and unpackaging during pausing. That the pauses are persistent (lasting ∼1 s) and relatively infrequent (at best once in every ∼100 translocation cycles) indicates that pausing occurs off the primary translocation pathway. Our data lead to a model in which the motor can switch between two stable structural states, one that is translocation competent and a second that is paused, through which unpackaging can occur. However, a simple two-state model would be inadequate to fully describe the ATP dependence of the data. First, the observed Michaelis–Menten kinetics of the translocation cycle (Fig. 1E) suggests that there must be a minimum of two rate-limiting steps determining the speed: an ATP binding step with second-order rate constant kb = Vmax/KM/d and a translocation step with rate constant ktranslocation = Vmax/d. Thus, a minimal description of the translocation cycle (Fig. 6) must include (i) an apo state (ApoT) in which the motor waits to bind an ATP molecule, and (ii) an ATP-bound state (ATPT) in which catalysis (ATP hydrolysis) and translocation occur. Second, in the paused state, the motor can either simply stall or unpackage. Based on our observations that suggest a correlation between unpackaging and the nucleotide-binding state of the motor, we propose that there are two paused states: (i) an apo state (ApoP) that leads to unpackaging pauses, and (ii) nucleotide-bound state (ATPP) that leads to stalled pauses. Based on this reasoning, we devised a minimal four-state kinetic model to describe the trends in the data (Fig. 6) and performed stochastic simulations (Materials and Methods) to reproduce the dependences in the data (Figs. 1E, black and blue green circles and lines; 2, red, blue, and green circles and lines; 3B, red circles and line; and 4, black circles; and Fig. S3). Indeed, the simulated traces of packaging generated at different concentrations of ATP (Fig. S3) matched with the experimental traces obtained at the same ATP concentrations (Fig. 1D).
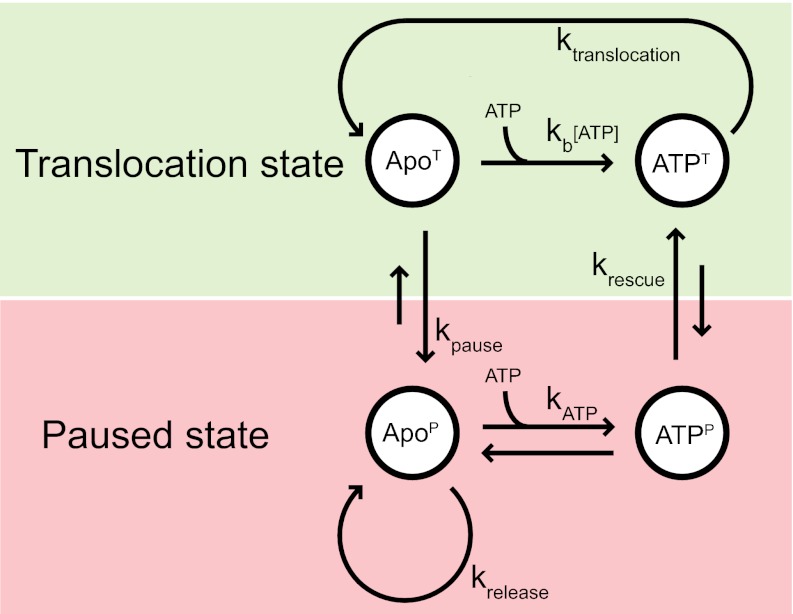
Fig. 6.
A minimal four-state mechanochemical model for the T4 DNA packaging motor. T4 DNA packaging involves two distinct cycles, translocation (green shaded area), and pause-unpackaging (red shaded area). The apo and ATP states within each cycle correspond to the relaxed and tensed states of gp17 (6), resulting in a total of four distinct states. Translocation follows Michaelis–Menten kinetics and is governed by the rate of ATP binding (kb) and packaging (ktranslocation) by the motor. The paused state is reached by an improper association of the motor, ATP and DNA. The frequency of pausing is governed by a kinetic competition between ATP binding (kb) and entry into the paused state (kpause). Within the paused state, interaction of DNA with an apo subunit results in unpackaging, at a rate given by krelease. Rescue from the pause-unpackaging state involves ATP binding to the apo subunit (kATP) and realignment of the motor and DNA (krescue). Short and long arrows represent transitions with small and high rate constants, respectively.
According to this model, entry into the paused state occurs through the ApoT state (Fig. 6). The probability of pausing per translocation cycle Ppause is determined by the kinetic competition between binding ATP in the ApoT state (with rate constant kb[ATP]) and transitioning into the ApoP state (with rate constant kpause). If kpause is small (as expected because pauses occur less than once per 100 translocation cycles), 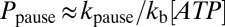 . The average length packaged between consecutive pauses is simply the step size d multiplied by the number of translocation cycles that occur between pauses, which is the inverse of the pausing probability, i.e.,
. The average length packaged between consecutive pauses is simply the step size d multiplied by the number of translocation cycles that occur between pauses, which is the inverse of the pausing probability, i.e., 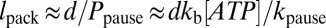 . Thus, the model reproduces the linear ATP dependence of the average packaged length (Fig. 2A, red line).
. Thus, the model reproduces the linear ATP dependence of the average packaged length (Fig. 2A, red line).
Unpackaging is correlated to motor-DNA affinity. When the motor is occupied with ATP or an ATP analog in the paused state (ATPP), the majority of pauses (∼80% at saturating ATP) showed little to no unpackaging. However, in the ApoP state, the motor subunit has a lower affinity for DNA and, hence, unpackages (∼80% pauses unpackage at 25 μM ATP). These features are captured in our kinetic model by including an unpackaging pathway while in the ApoP state (Fig. 6) (in contrast, the ATPP state is not connected directly to such a pathway). The rate constant krelease represents the rate at which the motor disengages from the DNA, allowing its serial release. Despite the two distinct paused states, ApoP and ATPP, we propose that transitions between the two occur frequently through binding and release of ATP. A result of this feature of the model is that the rate of unpackaging is determined not only by krelease, but also by the equilibrium between the low DNA-affinity ApoP state and high DNA-affinity ATPP state. If the transitions between the two states are faster than the DNA release rate, then the unpackaging velocity is given by 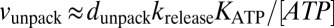 , where dunpack is the average length of DNA released during each unpackaging cycle and KATP is the equilibrium constant for the transition between the ApoP and ATPP states. As ATP is decreased, the equilibrium is shifted away from the ATPP state toward the ApoP state, leading to an increased unpackaging velocity, as observed in our measurements (Fig. 3B). The increase in the unpackaging velocity with force (Fig. S2, Inset) is included in our model by making krelease force dependent; increased force destabilizes motor–DNA interactions, leading to a higher release rate.
, where dunpack is the average length of DNA released during each unpackaging cycle and KATP is the equilibrium constant for the transition between the ApoP and ATPP states. As ATP is decreased, the equilibrium is shifted away from the ATPP state toward the ApoP state, leading to an increased unpackaging velocity, as observed in our measurements (Fig. 3B). The increase in the unpackaging velocity with force (Fig. S2, Inset) is included in our model by making krelease force dependent; increased force destabilizes motor–DNA interactions, leading to a higher release rate.
In principle, rescue out of a pause may occur through two pathways: from the ApoP state into the ApoT state or from the ATPP state into the ATPT state (Fig. 6). We favor the latter mechanism because it predicts that the number of unpackaging cycles completed during a pause is determined by the kinetic competition between releasing DNA vs. binding ATP while in the ApoP state. Thus, the mean length unpackaged—the number of cycles multiplied by the length of DNA released per cycle, dunpack—varies inversely with ATP, in agreement with observation (Fig. 2C). Under this preferred pathway, the pause duration is determined by the transition rate between the ATPP and ATPT states, krescue. Provided that this is a slow, rate-limiting step, the mean pause duration is simply given by 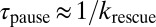 . The dual observations of an ATP-independent mean pause duration and exponential distribution of pause durations in the data, which indicate a single rate-limiting process, are again reproduced by the model. Although this minimal model reproduces all of the trends observed in our data, it cannot define the structural states of the motor. Structural evidence to date shows two conformational states, relaxed and tensed, both of which, as described above, relate to the translocation state of the motor (Fig. 6) (6). The relaxed state in which the ATPase and DNA binding domains are separated by ∼7 Å likely represents the ApoT state, and the tensed state in which the domains are in close contact represents the ATPT state. We speculate that the paused states, ApoP (unpackaging) and ATPP (stalling), might be similar to these states, except that the DNA is out of register, leading to disruption of the tight coordination required for DNA translocation. This situation might arise from incorrect binding of DNA to a motor subunit in apo state, which exists at higher probability when the ATP concentration is limiting. In contrast, under saturating ATP concentrations, the translocating DNA rarely encounters such a state because the rate of ATP binding is faster than switching into pause state (Fig. 6).
. The dual observations of an ATP-independent mean pause duration and exponential distribution of pause durations in the data, which indicate a single rate-limiting process, are again reproduced by the model. Although this minimal model reproduces all of the trends observed in our data, it cannot define the structural states of the motor. Structural evidence to date shows two conformational states, relaxed and tensed, both of which, as described above, relate to the translocation state of the motor (Fig. 6) (6). The relaxed state in which the ATPase and DNA binding domains are separated by ∼7 Å likely represents the ApoT state, and the tensed state in which the domains are in close contact represents the ATPT state. We speculate that the paused states, ApoP (unpackaging) and ATPP (stalling), might be similar to these states, except that the DNA is out of register, leading to disruption of the tight coordination required for DNA translocation. This situation might arise from incorrect binding of DNA to a motor subunit in apo state, which exists at higher probability when the ATP concentration is limiting. In contrast, under saturating ATP concentrations, the translocating DNA rarely encounters such a state because the rate of ATP binding is faster than switching into pause state (Fig. 6).
Once switched to the pause state, the motor needs not only to realign the DNA in the right register but also, in some scenarios, fill the nucleotide site with ATP to return to the translocating ATPT state. The latter requirement for the correct subunit to have a nucleotide bound to it to reengage with DNA likely explains why pauses persist for distances longer than the ∼10-bp helical pitch of DNA. Our data and the model described above suggest that pauses coupled with unpackaging allow the motor to “scan” across the DNA, albeit relatively slowly (10–200 bp/sec), until it finds the proper register, hence the persistence of the pauses. The pause duration can however be minimized, as has been observed, if the rate of scanning could be accelerated by applied force.
Our findings suggest a different coordination mechanism in T4 compared with other phage packaging systems. Although the φ29 motor’s affinity for DNA displays the same dependence on ATP as for T4, decreasing the ATP concentration does not induce pausing or unpackaging (17, 18). Instead, decreased ATP causes φ29 to slip, releasing large lengths of DNA at once. Moreover, φ29 has been shown to exhibit a biphasic “dwell-burst” translocation mechanism (17), during which the motor loads four ATPs (a “dwell”) and rapidly hydrolyzes them in succession, translocating DNA in 10-bp increments (a “burst”). The φ29 motor is believed to make strong, electrostatic contacts to the DNA during the dwell phase that are critical for coupling the mechanical and chemical cycles of the motor (25). Although our data do not strictly preclude a biphasic mechanism in T4 because of our inability to resolve motor stepping dynamics, we find it unlikely that T4 makes similarly strong contacts to DNA as it waits to bind ATP in the apo state. Thus, a more plausible scenario is that the T4 motor subunits interact differently with DNA, although more studies will be necessary to fully answer this question. A different coordination mechanism may have been evolved for the T4 packaging motor, which operates 7–10 times faster than the φ29 motor.
Our data identify a unique state of the T4 DNA packaging motor, the dynamic pause-unpackaging state induced by low ATP and misregistration between DNA and motor. In the cell, it is not known whether ATP levels ever decrease below 100 μM during phage infection. Also unknown is whether the small terminase, gp16, that is lacking in the motor complexes in our experiments regulates the pause-unpackaging state. However, we suspect that in vivo the pause-unpackaging state could arise during “normal” translocation when the motor encounters perturbations in the DNA that may affect registry with the motor such as accumulated torsion, variations in helical pitch, or “roadblocks” in the newly replicated viral genome (e.g., nicks, branches, tightly bound proteins). In φ29, perturbations in the DNA structure (including ssDNA gaps and bulges) that presumably disrupt motor-DNA registry are known to induce pausing (but not unpackaging) (25). Unpackaging may allow the motor, by taking a few steps backward, to bypass the impediments encountered along the translocation path and reset. Capsid internal pressure, which is expected to be important (>10 pN), especially in the late stages of packaging when DNA is highly compacted (26), may help drive DNA release during pauses. In addition, because the viral genome in vivo is engaged with various force-generating replicases, transcriptases, and recombinases, these enzymes might also assist the motor to release DNA out of capsid when it pauses at a roadblock (27). This mechanism would not only help restore the DNA register more rapidly but also provide an opportunity for repair enzymes to correct the abnormality. A key T4 recombinase, gp49, which resolves recombinational branched intermediates, is reported to be physically associated with the packaging machinery (28). Mutants in this gene (29), or introduction of a branch into the packaging substrate in vitro (30), dynamically stall the T4 motor at the branch point, as the motor makes repeated, but unsuccessful, attempts to translocate the abnormal DNA (30). Thus, the pause-unpackaging state might be critical for motor function and evolved under strong selection pressure because otherwise the motor might unproductively stall and generate incompletely packaged noninfectious viral particles. This mechanism is particularly critical for a highly recombinogenic virus such as phage T4 that depends almost entirely on recombination to initiate DNA replication and generates an intricately branched concatemeric genome substrate for packaging (31).
Materials and Methods
Single-molecule optical tweezers experiments were carried out with stalled T4 complexes consisting of empty purified heads, purified gp17 monomers, and 120 bp dsDNA. Measurements were conducted in a buffer containing 30 mM Tris HCl pH 7.6, 5 mM MgCl2, and 80 mM NaCl, and varying concentrations of ATP, ATP analog, or ADP as detailed in the text. Bulk packaging assays, single molecule optical tweezers assays, Matlab analyses, and kinetic simulation models are described in more detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the laboratories of Y.R.C. (Rustem Khafizov and Matthew Comstock) and V.B.R. (Zhihong Zhang, Marthandan Mahalingam, and Victor Padilla-Sanchez) for providing help with experiments, figures, and data analysis. Work in the Y.R.C. laboratory is supported by National Science Foundation Grant MCB 09-52442 (CAREER), the Burroughs Wellcome Fund—Career Awards at the Scientific Interface, and the Alfred P. Sloan Research Fellowship. Work in the V.B.R. laboratory is supported by National Science Foundation Grant MCB-0923873 and National Institute of Allergy and Infectious Diseases Grant R01AI081726.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209214109/-/DCSupplemental.
References
- 1.Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75(10):4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 3.Smith DE, et al. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413(6857):748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 4.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci USA. 2007;104(43):16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J Mol Biol. 2001;314(3):401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135(7):1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 8.Hegde S, Padilla-Sanchez V, Draper B, Rao VB. 2012. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicates a molecular lever mechanism for coupling ATPase to DNA translocation. J Virol 86(8):4046–4057. [DOI] [PMC free article] [PubMed]
- 9.Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: Endonuclease VII resolvase release of arrested Y-DNA substrates. J Biol Chem. 2011;286(21):18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S, et al. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci USA. 2012;109(3):817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279(39):40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 13.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25(6):943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30(18):4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetzinger KR, Rao VB. Defining the ATPase center of bacteriophage T4 DNA packaging machine: Requirement for a catalytic glutamate residue in the large terminase protein gp17. J Mol Biol. 2003;331(1):139–154. doi: 10.1016/s0022-2836(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 16.Alam TI, Rao VB. The ATPase domain of the large terminase protein, gp17, from bacteriophage T4 binds DNA: Implications to the DNA packaging mechanism. J Mol Biol. 2008;376(5):1272–1281. doi: 10.1016/j.jmb.2007.12.041. [DOI] [PubMed] [Google Scholar]
- 17.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457(7228):446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chemla YR, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122(5):683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Fuller DN, et al. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373(5):1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363(4):786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante C, Chemla YR, Moffitt JR. High resolution dual trap optical tweezers with differential detection. In: Selvin P, Ha TJ, editors. Single-Molecule Techniques: A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Lab Press; 2007. pp. 297–324. [Google Scholar]
- 22.Al-Zahrani AS, et al. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J Biol Chem. 2009;284(36):24490–24500. doi: 10.1074/jbc.M109.025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J Mol Biol. 2005;347(3):523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J Biol Chem. 2006;281(31):21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 25.Aathavan K, et al. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461(7264):669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purohit PK, et al. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88(2):851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosig G. Homologous recombination. In: Karam JM, editor. Molecular Biology of Bacteriophage T4. Washington: Am Soc Microbiol; 1994. pp. 54–82. [Google Scholar]
- 28.Golz S, Kemper B. Association of holliday-structure resolving endonuclease VII with gp20 from the packaging machine of phage T4. J Mol Biol. 1999;285(3):1131–1144. doi: 10.1006/jmbi.1998.2399. [DOI] [PubMed] [Google Scholar]
- 29.Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28(2):643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381(1):61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreuzer KN, Brister JR. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: A review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J. 2010;7:358. doi: 10.1186/1743-422X-7-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26(7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.